Abstract
Bud-break is an environmentally and economically important trait in trees, shrubs and vines from temperate latitudes. Poor synchronization of bud-break timing with local climates can lead to frost injuries, susceptibility to pests and pathogens and poor crop yields in fruit trees and vines. The rapid climate changes outpace the adaptive capacities of plants to respond through natural selection. This is particularly true for trees which have long generation cycle and thus the adaptive changes are significantly delayed. Therefore, to devise appropriate breeding and conservation strategies, it is imperative to understand the molecular underpinnings that govern dormancy mechanisms. We have recently identified and characterized the poplar EARLY BUD-BREAK 1 (EBB1) gene. EBB1 is a positive regulator of bud-break and encodes a transcription factor from the AP2/ERF family. Here, using comparative and functional genomics approaches we show that EBB1 function in regulation of bud-break is likely conserved across wide range of woody perennial species with importance to forestry and agriculture.
Keywords: climate change, dormancy, phenology
Dormancy is an adaptive mechanism in trees from temperate latitudes that allows survival the dehydration and freezing stress during winter months through a temporary suspension of growth. The process encompasses several distinct developmental, growth and physiological stages. Specifically, in the fall shoot growth is terminated, followed by transformation of the growing apex into a dormant bud. Buds undergo a number of physiological changes culminating into what is known as endodormancy. Endodormant buds can no longer respond to growth promoting signals and require exposure to several weeks of near freezing temperatures (known as meeting a chilling requirement) before they become again competent to regrow in the presence of inductive signals.1-4 Once the chilling requirement is met, regrowth is almost exclusively dependent on high temperatures. The timing of each of these processes is synchronized with the local climates. Poor synchronization can lead to frost damages by either late spring frosts around the time of bud-break, early fall frosts around the time of growth cessation and/or poor, prolonged and uneven bud-break when chilling requirement is not met due to warm winter temperatures.5,6
Dormancy traits are polygenic, and a large number of independent genes can control the onset and release from dormancy.7,8 These genes and mechanisms are still poorly understood and generally dissected in one or a few species. Thus, it is unclear if the genes and mechanisms that control dormancy characteristics are conserved and to what extent among different taxonomic lineages. Development of genomics and functional genomics resources in many species, including woody perennials trees from temperate latitudes with a dormancy cycle9-13 enables translational approaches for validation the conservation of these mechanisms.
We have previously identified and shown that the EARLY BUD-BREAK 1 (EBB1) gene is a positive regulator of bud-break in poplar.14,15 The gene encodes a transcription factor from the AP2/ERF family. EBB1 is involved in regulation of bud-break in Populus but the importance of the gene in regulation of bud phenology in other woody perennial species is still unclear. Using homology searches and taking advantage of a number of fully sequenced and annotated genomes, close homologs of EBB1 were found in several woody perennial species. We specifically selected species from the temperate latitudes that display cycling between dormancy and active growth (Fig. 1A). These included Prunus persica, Malus domestica, Vitis vinifera and Picea abies. The four species span a wide range of taxonomies, life histories (trees vs. vine) and geographic ranges. Three species (Prunus persica, Malus domestica, Vitis vinifera) are of significance to the grape and fruit trees industry while spruce (Picea abies) is important for the forestry industry and is also a central component of many natural ecosystems in the Eurasian region. We found one ortholog of EBB1 in Prunus persica and Vitis vinifera. Three orthologs were discovered in spruce. All other species, including Arabidopsis had 2 close paralogs showing homology to EBB1. Comparative phylogenetic analyses showed that the Vitis ortholog was most similar to the poplar EBB1. Furthermore, all proteins from Rosacea species grouped in a separate lineage. Consistent with their evolutionary history, the gymnosperm spruce orthologs were different from all the angiosperm proteins.
Figure 1.
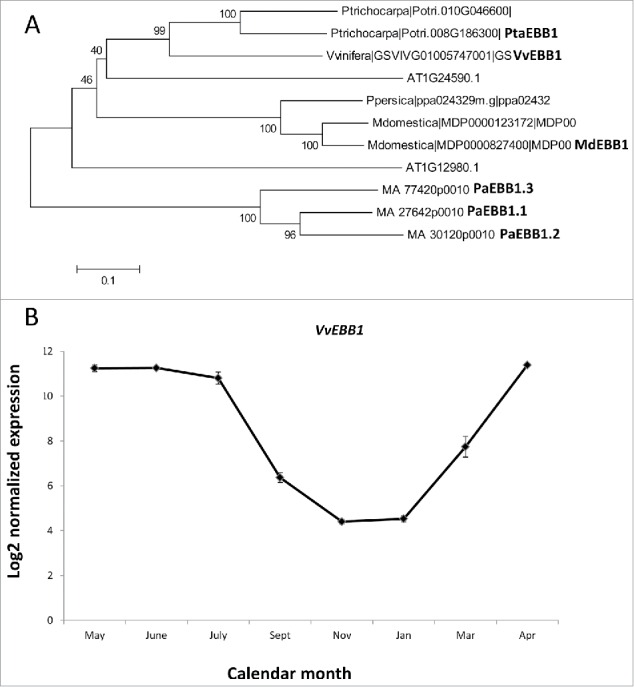
Phylogeny and expression of EBB1 orthologs. (A) Orthologs of poplar EBB1 were identified through homology searches. Arabidopsis orthologs were also included. Phylogenetic tree was constructed using Mega 4.1. Numbers in the branches show percent bootstrap support out of 1,000 iterations. (B) Expression of Vitis ortholog. Expression values are based on microarray data.17 Abbreviations used for the names of the genes in different species are as follows: Md-Malus domestica; Pa=Picea abies; Pta=Populus tremula X alba; Vv=Vitis vinifera.
We next looked for additional evidence to corroborate EBB1 orthologs' involvement in regulation of bud-break. Recent work in apple provides a strong evidence for conservation of EBB1 function.16 The C-repeat binding factor (CBF/DREB) transcriptional activator genes have been shown to have a critical function in acclimation to cold stress.16 Overexpression of a peach CBF gene in apple has indeed resulted in increased cold tolerance but also caused accelerated bud set, leaf senescence and delayed bud-break. This suggested that CBF may play a coordinating role between the physiological acclimation (e.g., cold tolerance) and the growth and developmental changes that occur during dormancy cycle (e.g., growth cessation, bud set, dormancy and bud-break). The expression of several critical regulators of these processes were compared between the CBF transgenics and WT plants, including one of the 2 apple EBB1 orthologs (MdEBB1). In WT apple plants, MdEBB1 showed nearly identical expression pattern in the transition between dormancy and re-initiation of growth (bud-break) as its poplar ortholog. MdEBB1 was barely detectable in the dormant buds but its transcript abundance highly increased prior to bud-break. The expression increase prior to bud-break was significantly delayed in the CBF overexpressing transgenics. Thus the delayed bud-break in CBF transgenics was almost perfectly correlated with a delayed increased in the expression of the MdEBB1 before bud-break.
In Vitis, recent study provided comprehensive view of the genome wide transcriptomic changes associated with different dormancy phases.17 Using this data, we studied the expression of the Vitis EBB1 ortholog (VvEBB1) during dormancy cycle. Consistent with the EBB1 expression in Populus, VvEBB1 was sharply downregulated during dormancy period and increased in the months prior to bud-break. The gene was expressed at high levels during the active growing season (Fig. 1B).
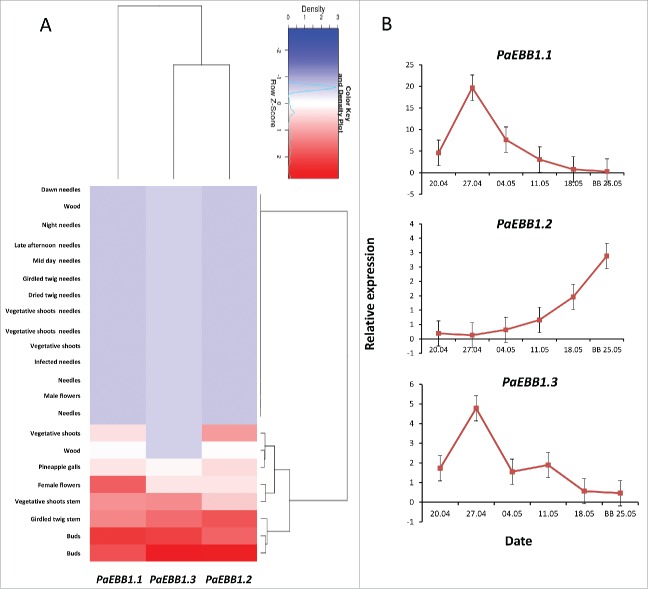
Finally, we used the congenie.org functional genome resource to study the expression of the 3 spruce EBB1 orthologs in various tissues.11 Among 22 different tissue types, the 3 orthologs were most highly expressed in vegetative buds (Fig. 2A), further supporting the conservation of EBB1 function. However, analyzes of the dynamics of the 3 genes expression in buds prior and around bud-break showed a much more complex dynamics (Fig. 2B), indicating putative differences between the role of these genes in angiosperms and gymnosperms. In angiosperm lineages, EBB1 orthologs appear to be down regulated in dormant and up regulated in actively growing apices. The expression of the gymnosperm orthologs appear to be more complex (Fig. 2B). One of the orthologs (PaEBB1.2) showed an expression similar to the ones observed in the angiosperm trees. In the other 2 genes (PaEBB1.1 and PaEBB1.3) we did detect a spike in the expression approximately 5 weeks before bud-break, but the increased in expression was transient (weeks) and reverted to low levels around the actual bud-break. Differences in expression patterns between deciduous angiosperms and evergreen gymnosperms could be related to fundamental difference in the biology of dormancy between these 2 groups. In spruces, epigenetic memory is set during embryogenesis and growth conditions during the first vegetation period considerably affect chilling requirement for bud-break.18 In addition, spruce, as an evergreen conifer, has photosynthetic area prior to budburst, which means that the involved signaling mechanisms could be quite different.
Figure 2.
Expression of EBB1 homologs in spruce (Picea abies). (A) Clustering was based on RNA-seq expression data11 and using the congenie.org's exHeatmap tool. The hierarchical clustering was performed using the Ward's method and Euclidean distances with default parameters. (B) Expression of the 3 EBB1 spruce orthologs in buds during bud-break. BB=bud -break (see Supplemental materials for details).
The sum of phylogenetic and expression analyses of EBB1 orthologs indicate that EBB1 is a part of a conserved mechanism for control of bud-break in trees and show the translational power of accumulated genomics and transcriptomics resources in a number of plant species. Our data also suggests that there could be significant differences or diversification of EBB1 function in relation to evolutionary history and specific biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
Funding
This work was supported in part by grants from the USDA-NRI Plant Genome program (2003–04345), the US Department of Energy, Poplar Genome Based Research for Carbon Sequestration in Terrestrial Ecosystems (DE-FG02–06ER64185, DE-FG02–05ER64113), the Consortium for Plant Biotechnology Research, Inc.. (GO12026–203A), the USDA Biotechnology Risk Assessment Research Grants Program (2004–35300–14687) and the USDA McIntire Stennis Fund. Elena Carneros was supported by a grant from Iceland, Liechtenstein and Norway through the EEA Financial mechanism. Operated by Universidad Complutense de Madrid
References
- 1.Rohde A, Bhalerao RP. Plant dormancy in the perennial context. Trends Plant Sci 2007; 12:217-23; PMID:17416545; http://dx.doi.org/ 10.1016/j.tplants.2007.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Horvath DP, Anderson JV, Chao WS, Foley ME. Knowing when to grow: Signals regulating bud dormancy. Trends Plant Sci 2003; 8:534-40; PMID:14607098; http://dx.doi.org/ 10.1016/j.tplants.2003.09.013 [DOI] [PubMed] [Google Scholar]
- 3.Cooke JEK, Eriksson ME, Junttila O. The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environm 2012; 35:1707-28; PMID:22670814; http://dx.doi.org/ 10.1111/j.1365-3040.2012.02552.x [DOI] [PubMed] [Google Scholar]
- 4.Brunner AM, Evans LM, Hsu CY, Sheng X. Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation?. Front Plant Sci 2014; 5:732; PMID:25566302; http://dx.doi.org/ 10.3389/fpls.2014.00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aitken SN, Yeaman S, Holliday JA, Wang T, Curtis-McLane S. Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol Appl 2008; 1:95-111; PMID:25567494; http://dx.doi.org/ 10.1111/j.1752-4571.2007.00013.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howe GT, Aitken S, Neale DB, Jermstad KD, Wheeler N, Chen TH. From genotype to phenotype: Unraveling the complexities of cold adaptation in forest trees. Can J Bot 2003; 81:1247-66; http://dx.doi.org/ 10.1139/b03-141 [DOI] [Google Scholar]
- 7.Rohde A, Storme V, Jorge V, Gaudet M, Vitacolonna N, Fabbrini F, Ruttink T, Zaina G, Marron N, Dillen S et al.. Bud set in poplar—genetic dissection of a complex trait in natural and hybrid populations. New Phytol 2011; 189:106-21; PMID:21039557; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03469.x [DOI] [PubMed] [Google Scholar]
- 8.Frewen BE, Chem THH, Howe GT, Davis J, Rohde A, Boerjan W, Bradshaw HD Jr. et al.. Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 2000; 154 837-45; PMID:10655234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A et al.. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006; 313:1596-604; PMID:16973872; http://dx.doi.org/ 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- 10.Yakovlev IA, Fossdal CG, Johnsen O, Junttila O, Skroppa T. Analysis of gene expression during bud burst initiation in Norway spruce via ESTs from subtracted cDNA libraries. Tree Genet Genom 2006; 2:39-52; http://dx.doi.org/ 10.1007/s11295-005-0031-z [DOI] [Google Scholar]
- 11.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al.. The Norway spruce genome sequence and conifer genome evolution. Nature 2013; 497:579-84; PMID:23698360; http://dx.doi.org/ 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- 12.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C et al.. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007; 449:463-7; PMID:17721507; http://dx.doi.org/ 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- 13.Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D et al.. The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 2010; 42:833-9; PMID:20802477; http://dx.doi.org/ 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- 14.Yordanov YS, Ma C, Strauss SH, Busov VB. EARLY BUD-BREAK 1 (EBB1) is a regulator of release from seasonal dormancy in poplar trees. Proc Natl Acad Sci USA 2014; 111:10001-6; PMID:24951507; http://dx.doi.org/ 10.1073/pnas.1405621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busov V, Yordanov Y, Gou J, Meilan R, Ma C, Regan S, et al.. Activation tagging is an effective gene tagging system in Populus. Tree Gen Genom 2010; http://dx.doi.org/ 10.1007/s11295-010-0317-7 [DOI] [Google Scholar]
- 16.Wisniewski M, Norelli J, Artlip T. Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci 2015; 6:85; PMID:25774159; http://dx.doi.org/ 10.3389/fpls.2015.00085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Riquelme J, Grimplet J, Martinez-Zapater JM, Carmona MJ. Transcriptome variation along bud development in grapevine (Vitis vinifera L.). BMC Plant Biol 2012; 12:181; PMID:23035802; http://dx.doi.org/ 10.1186/1471-2229-12-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yakovlev I, Fossdal CG, Skrøppa T, Olsen JE, Jahren AH, Johnsen Ø. An adaptive epigenetic memory in conifers with important implications for seed production. Seed Sci Res 2012; 22:63-76; http://dx.doi.org/ 10.1017/S0960258511000535 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.