Abstract
Cryptochromes are blue-light absorbing flavoproteins with multiple signaling roles. In plants, cryptochrome (cry1, cry2) biological activity has been linked to flavin photoreduction via an electron transport chain to the protein surface comprising 3 evolutionarily conserved tryptophan residues known as the ‘Trp triad.’ Mutation of any of the Trp triad residues abolishes photoreduction in isolated cryptochrome protein in vitro and therefore had been suggested as essential for electron transfer to the flavin. However, photoreduction of the flavin in Arabidopsis cry2 proteins occurs in vivo even with mutations in the Trp triad, indicating the existence of alternative electron transfer pathways to the flavin. These pathways are potentiated by metabolites in the intracellular environment including ATP, ADP, AMP, and NADH. In the present work we extend these observations to Arabidopsis cryptochrome 1 and demonstrate that Trp triad substitution mutants at W400F and W324F positions which are not photoreduced in vitro can be photoreduced in whole cell extracts, albeit with reduced efficiency. We further show that the flavin signaling state (FADH°) is stabilized in an in vivo context. These data illustrate that in vivo modulation by metabolites in the cellular environment may play an important role in cryptochrome signaling, and are discussed with respect to possible effects on the conformation of the C-terminal domain to generate the biologically active conformational state.
Keywords: cryptochrome, electron transfer, light signaling, photoreceptor, photoreduction, Trp triad
Introduction
Cryptochromes are blue-light absorbing flavoproteins occurring throughout the biological Kingdom with many important signaling roles. In plants, there are 2 homologous cryptochromes (cry1, cry2) which regulate de-etiolation, photomorphogenesis, the initiation of flowering, gene expression, and the entrainment of the circadian clock.1 Cryptochromes are highly conserved evolutionarily to photolyases, which are flavoproteins that catalyze the light dependent repair of lesions in UV-damaged DNA. Plant cryptochromes have lost DNA repair function but retain many of the photoreactions characterized in photolyases. These include the ability to undergo a process known as ‘photoactivation’, whereby oxidized FAD bound in a hydrophobic internal pocket in the resting (dark) state of the protein becomes reduced after absorption of a photon of light energy. This reaction occurs by electron transfer to the excited state flavin from a nearby Trp residue in the protein, which in turn is reduced by a chain of electron transfer through 3 suitably positioned Trp residues leading to the protein surface. This electron transfer chain is known as the Trp triad, and is highly conserved between cryptochromes from many sources and with photolyases.1
The signaling mechanism of plant cryptochromes has been shown to involve interaction with signaling partner molecules following light absorption. These include the transcription factors CIB1 and SPA1 in the case of Arabidopsis cry1 and cry2 cryptochromes.2,3 The partner proteins do not bind to the full-length dark-adapted (resting) state of these cryptochromes, but only to their illuminated (activated) form. The substrates however do also bind to the dark (unilluminated) truncated cryptochromes from which the C-terminal domains have been removed.2,3 These data imply that the C-terminal domain is somehow blocking correct docking of the partner proteins to plant or drosophila cry N-terminal domain in the inactive (dark) state, and that a conformational change at the C-terminal must occur in the course of light activation in order to provide substrate access to the N-terminal domain. Experiments showing altered proteolytic patterns in response to light and transient grating experiments are consistent with large structural changes which involve the C-terminal domain.4,5
To explain how cryptochromes are activated by light, many lines of evidence have shown that light-induced redox transitions in the bound flavin (from FAD to FADH°) are correlated with biological function,6-8 and to conformational change of the C-terminal domain.5 An abbreviated version of the resulting photocycle proposed for plant cryptochromes is summarized in Figure 1. Firstly, in the dark, the receptor is found with flavin in the oxidized redox state and the C-terminal domain folded into an inactive conformation adjacent to the flavin binding pocket. Upon illumination with blue light, flavin is reduced to the neutral radical state via multiple electron and proton transfer steps. In the course of these reactions conformational changes are initiated which release the C-terminal domain of cryptochrome from the protein surface and provide access to biological substrates.3,5 This relatively long-lived (several minutes)9 intermediate is the biologically active signaling state.
Figure 1.
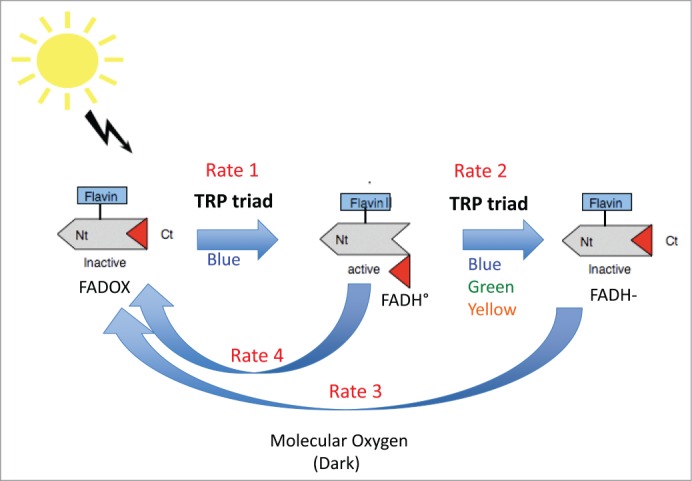
Presumed photocycle of plant cryptochromes. In the dark, cryptochromes are in the inactive state (C-terminal domain folded against the protein, flavin in the oxidized redox state). Upon illumination, flavin undergoes photoreduction to the FADH° redox form, which triggers conformational unfolding of the C-terminal domain to give the activated form of the receptor. Subsequent illumination of FADH° reduces the flavin further to the inactive (FADH-) redox form. Both radical (FADH°) and fully reduced (FADH-) flavin are reoxidized to the inactive (FAD) form by molecular oxygen.10 The receptor undergoes continuous cycling between redox forms under illumination and the degree of activation depends on the equilibrium reached (rates 1, 2, 3, 4) between reducing and oxidizing reactions. In this mechanism, the Trp triad mediates forward electron transfer driving forward redox state interconversion.
Further illumination of the flavin cofactor in cryptochrome results in full reduction of the flavin to a biologically inactive redox form (FADH-) which refolds the C-terminal domain and thereby returns the receptor to the biologically inactive conformational state. Both flavin radical (FADH°) and fully reduced (FADH-) flavin redox states undergo spontaneous reoxidation to the FAD (oxidized) form in a process that is independent of light and relies on the concentration of molecular oxygen.10 Thus the cryptochrome, once placed in continuous light, undergoes multiple rounds of light-dependent flavin photoreduction (from FAD to FADH° and FADH-), balanced by continuous reoxidation to FAD. The equilibrium reached between the different redox states (see Figure 1, rates 1–4) thereby determines the ultimate biological activity of cryptochrome by determining the concentration of the FADH° signaling state occurring at a given light intensity. In this mechanism, the role of the Trp triad is important in order to generate the FADH° (signaling state) (Fig. 1).
Recently, the role of the Trp triad in the formation of the FADH° signaling state has come under discussion, following the report that amino acid substitutions of cry2 at residues W321, W374, and W397 in the Trp triad still retained in vivo biological activity.11 These observations were for the most part based on an experimental artifact, as the reported in vivo cry2 Trp triad mutants phenotypes (hypocotyl inhibition, yeast 2 hybrid interaction with substrate) had also entirely lost light responsivity. The mutants were constitutively active even in red light and darkness, most likely due to altered folding of the protein (into the constitutively active conformation). From such biological readouts that have lost light responsiveness, no conclusions on the underlying photochemical properties of the receptor in response to blue light can be drawn. Nonetheless, one of the reported cry2 phenotypes (of cry2 protein degradation in response to blue light) did indeed showed blue light responsivity even in cry2 Trp triad mutants in vivo.11 Therefore an argument for biological activity in the absence of flavin photoreduction could be made. Similar arguments (of biological activity in Trp triad mutants in Drosophila, for instance) have been made in the case of insect cryptochromes.12
In order to resolve this issue, it was necessary to determine the redox state of cryptochrome flavin in the Trp triad mutants directly within the living cell. The possibility that in vivo photoreduction may occur more efficiently and/or by alternate pathways to in vitro photoreduction in these mutants was already suggested from indirect fluorescence monitoring of living SF21 insect cells expressing cryptochrome proteins.13 These studies showed that proteins with mutations in the Trp triad that rendered them insensitive to photoreduction in vitro, could nonetheless undergo photoreduction in living insect cells, albeit at reduced efficiency.13
This possibility was subsequently directly verified in the case of cry2 Trp triad mutants W321A, W374A and W397A.14 It was discovered that the presence of small metabolites in the cellular environment (ATP, ADP, NADP etc.) could potentiate alternate electron transfer pathways in the cryptochrome involving distinct (to the Trp triad) W and Y residues in vivo. Therefore, even cry2 Trp triad mutants lacking detectable flavin photoreduction activity in vitro could be photoreduced in a whole cell in vivo environment. The relative efficiency of in vivo photoreduction (highest for the cry2 W374A mutant, reduced in efficiency for W397A and W321A mutants) corresponds with the relative light-dependent biological activity (in vivo protein cry2 degradation assay) reported for these mutants11 and is thereby fully consistent with photoreduction of flavin as a requirement for cryptochrome signaling. These studies also highlight the pitfalls of applying conclusions from data obtained with isolated proteins toward biological function in vivo, and of interpretations based solely on mutagenic analyses, where secondary effects on protein structure may introduce artifactual findings.
In this Short Communication, we extend the observations of in vivo photoreduction in the Trp triad mutants of cry214 to Trp triad mutants of the cryptochrome-1 photoreceptor. It has previously been demonstrated15 that the Trp triad mutants W400F and W324F of Arabidopsis cry1 (homologues of the cry2 W397 and W321 residues) cannot be photoreduced in vitro. These cry1 Trp triad mutants also showed significantly reduced biological activity in vivo, consistent with a role for flavin reduction in biological activation. Nonetheless, biological activity was not completely null, leaving the possibility of residual flavin reduction occurring in vivo in these mutants in spite of disruption of the Trp triad.
Here we investigate directly the efficiency of photoreduction of the cry1 Trp triad mutations within whole cell extracts and show that partial photoreduction does indeed occur, in contrast to the isolated protein in vitro.15 In this respect, the W400F and W324F Trp triad mutants behave in an analogous fashion to the W397A and W321A mutants of cry2 which show measureable photoreduction activity in vivo.14 and also light dependent biological activity (albeit reduced as compared to wild type).11 We extend these observations further by showing that the the cellular (in vivo) environment also has a considerable effect on the rate of the back (reoxidation) reaction of cry1 and that these effects are also consistent with binding of small cellular metabolites in an in vivo context. Finally, we discuss these findings in the context of how the signaling state (in vivo and in vitro) may trigger the conformational change in the C-terminal domain, which is at the basis of biological activation.
Mutant constructs, methods for protein expression, production of whole cell extracts, and their analysis by UV/Vis spectroscopy are as previously described.14,15 The C-terminal fragment of cry1 (comprising amino acids 443–681) and including a 6-Histidine tag at the N-terminus was expressed in E. coli and purified by affinity column chromatography over nickel column by standard protocols.
CD spectroscopy: Protein samples were diluted to a final concentration of 1 µM (Full – Length cry1) and 6 µM (truncated C-terminal fragment of cry1) respectively in 25 mM sodium phosphate buffer at pH = 7.6. CD data was obtained using a Jasco J-810 spectropolarimeter with experimental conditions of 3 accumulations, D.I.T. of 1 second, 1.5 nm bandwidth, 0.1 nm data pitch, 100 nm/sec scan speed, and appropriate background subtraction. All samples were in 1 mm Helma Quartz cuvettes at held at 20 degree Celsius for the duration of the experiment. Analysis was performed with CDPro16 using the CONTIN algorithm17 and the reference data set SMP56.18
Cry1 Trp triad mutants W400F and W324F are photoreduced in vivo
Studies on isolated cry1 proteins have been previously reported with mutations in the Trp triad at positions W400F and W324F, which are found in the cry1 sequence and crystal structure16 at the homologous positions to W382 and W306 of E. coli photolyase, and of W397 and W321 of Arabidopsis cry2, respectively. The mutant proteins (W400F and W324F) thereby lack the predicted electron donor proximal to the flavin (W400) or exposed to the protein surface (W324) and should hence be impaired in electron transfer. Consistent with this expectation, the isolated proteins showed reduced electron transfer and could not be photoreduced in vitro.15 Biological activity in vivo was also significantly reduced in these mutant proteins, in keeping with the expectation of a requirement for photoreduction to produce the signaling state (FADH°). Nonetheless, there was evidence of some residual in vivo biological activity (10–20% under the tested conditions) even though the mutants showed no photoreduction activity in vitro. It therefore seemed possible that these cry1 Trp triad mutants could also undergo somewhat enhanced photoreduction in vivo. We therefore tested the cry1 Trp triad mutants for photoreduction activity in whole cell extracts.
As in our prior study14 we expressed the cry1 Trp triad mutants W400F and W324F, as well as wild type cry1, in insect cells at high concentration by baculovirus infection. Cell culture pellets were lysed and the crude extracts examined directly for flavin reduction using UV/Vis spectroscopy. Spectra were taken before (Dark) and after (Light) illumination with white light, and difference spectra (Light minus Dark) determined (Fig. 2). As a control, spectra were taken of cell extracts from insect cells that did not express recombinant cryptochrome.
Figure 2.
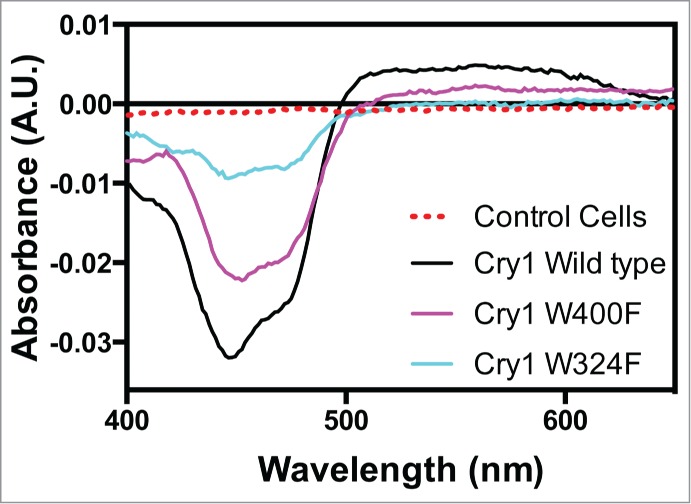
Flavin photoreduction of cry1 Trp triad mutants in whole cell extracts. Whole cell lysates from cultures expressing Arabidopsis cry1 wild type, W400F or W324F mutants were illuminated for 1 min with white light (600 μmolm−2sec−1) in PBS pH7.5, 5 mM DTT. Spectra taken after illumination were substracted from dark spectra to provide the (Dark minus Light) difference spectra shown in the figure, indicating the reduction of flavin from oxidized to radical (FADH°) state. No difference in absorption was seen under these conditions in whole cell extracts that were not expressing cryptochrome proteins (control cells).
The results show that, upon illumination, a clear decrease in signal at 450 nm indicative of flavin reduction can be seen in wild type cry1 samples (Fig. 2). This signal was specific to cryptochrome – bound flavin as no detectable difference was seen in (Light – Dark) difference spectra from extracts of non-expressing control cells (Fig. 2). However, in the case of both W400F and W324F mutant proteins, clear evidence of photoreduction is observed, albeit reduced as compared to that of the wild type protein. These data are consistent with alternate electron transfer pathways to the Trp triad being potentiated by components of the cellular environment, as previously shown for cry214 and for cry1 in vitro.20,21 Photoreduction was nonetheless decreased in comparison to the wild type in the W400F and especially W324F (Fig. 2), consistent with their reduced biological activity in vivo.15
Dark reoxidation of AtCry1 is altered in a whole cell in vivo context
In addition to their effects on potentiating forward electron transfer pathways, a role for ATP has been previously reported on the stability of the radical (FADH°) flavin redox state in plant cryptochrome.21 In particular, ATP greatly slows the rate of flavin reoxidation in vitro. This suggests that kinetics of reoxidation of cryptochrome may be likewise considerably altered in an in vivo context. To verify this possibility, we have determined the rate of reoxidation of wild type cry1 protein in whole cell extracts, and compared this rate to that of the isolated cry1 protein in vitro photoreduced under the same conditions (Fig. 3). Whole cell extracts expressing cry1 or purified samples of isolated cry1 protein were illuminated in white light for 1 minute. The samples were then transferred to darkness, and spectra were taken at increasing times. Reoxidation (from FADH° - FAD) was determined by absorbance change (increase at 450 nm) relative to the illuminated sample after increasing times in darkness and is expressed as the percentage of reoxidation over time to the maximal (fully oxidized) state (Fig. 3). In the case of the purified cry1, reoxidation occurred rapidly and was virtually complete within 5 minutes after the end of illumination. We next evaluated the rate of flavin reoxidation of Cry1 in whole cell lysates. Cell extracts expressing cry1 were first illuminated with white light for 1 min. Subsequently, samples were returned to darkness and spectra were taken at time intervals in darkness. As can be seen (Fig. 3), even after 30 minutes in darkness, there was no detectable increase in absorbance at 450 nm and therefore no detectable reoxidation of flavin in wild type cry1. Full reoxidation of cryptochrome in whole cell extracts could be accelerated by bubbling oxygen through the sample, confirming that the cryptochrome remained intact throughout this treatment and did not simply lose the flavin or undergo some other irreversible bleaching reaction (not shown). Therefore, components within the whole cell extracts must stabilize the flavin FADH° radical state and significantly slow the rate of reoxidation.
Figure 3.

Flavin reoxidation of cry1 in whole cell extracts. Isolated purified cry1 protein (10 μM) was photoreduced in PBS buffer at pH7.5, 5 mM DTT for 1 minute in white light (600 μmolm−2sec−1) in the absence or presence (+ ATP) of 1mM ATP. Upon return to darkness, scans were taken at increasing time intervals and the absorption at 450 nm (peak absorption of oxidized flavin) monitored. % reoxidation represents the increase in A450 expressed as a percentage of the dark (pre-illumination) A450 peak value. Reoxidation of flavin in cry1 expressed in whole cell lysates was analyzed in the same way.
To determine whether the decreased rate of flavin reoxidation can be explained by binding to metabolites in the cellular environment, we added ATP at a concentration of 1mM to isolated cryptochrome proteins and determined the rate of the dark reoxidation reaction subsequent to illumination. Indeed, addition of metabolite slowed the rate of flavin reoxidation to levels comparable to proteins analyzed in the context of whole cellular extracts. Taken together, these data indicate that both forward (light induced) and reverse (reoxidation) redox reactions of cry1 are modified by the cellular environment and can be explained by the effect of small metabolites.
The C-terminal domain of cryptochrome is implicated in structural changes related to signaling
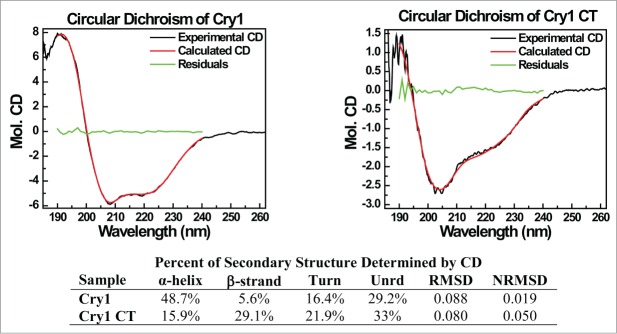
Ultimately, the light signaling reaction must induce some form of structural change in the C-terminal domain to initiate interaction with partner proteins. To probe the cryptochrome domain structure as a preliminary to identifying the structural changes that may be induced by light, we have performed CD spectral analysis on both full-length cry1 and of an isolated C-terminal fragment of cry1 which comprises the amino acids 443–681 beyond the photolyase-like region of homology (Fig. 4). The obtained spectra show a large increase in random structure as compared to that of the full-length protein for which a crystal structures is available.19 Unexpectedly, our experiments which analyzed amino acids 443–681 of the C-terminus also showed that the degree of unordered structure is significantly lower than what was reported previously19 using a smaller C-terminal fragment (amino acids 506–681), and that the percentage of α helical structure is increased. This suggests that a perturbation in the protein structure immediately adjoining the C-terminal domain (for instance amino acids 443–506) may contribute disproportionately to the overall structure, and may be affected by redox changes in the flavin state induced by light (Fig. 4).
Figure 4.
CD Spectra of Cry1. Upper Panel. CD Spectra of Full-Length AtCry1 (left) and truncated AtCry1 (amino acids 445–681 (right). Percentage α-helix include regular and distorted helices, percentage β-strands include regular and distorted β-strand while (N)RMSD = (normalized) root mean square deviation). Lower Panel: identification of the proportion of secondary structure within truncated and full-length intact protein.
In this study we have extended results from our prior work14 on the existence of alternate electron transfer pathways to the Trp triad pathway of cry2 to include analysis of Trp triad mutations of cry1. We show that photoreduction reaction rates in whole cell extracts are enhanced as compared to those of isolated W400F and W324F cry1 proteins, consistent with altered electron transfer routes potentiated by small metabolites found in these extracts. We note that W400F and W324F mutants both show considerably reduced biological activity in vivo.15 Therefore these alternate electron transfer routes are not as efficient as the Trp triad pathway under the relatively moderate light intensities used in the in vivo studies,15 and, at least in the case of cry1, probably do not contribute greatly to light sensing except under high light conditions. These results are furthermore consistent with those of the homologous Trp triad mutants W397F and W321F of cry2, which also show reduced light –dependent biological activity in vivo (the cry2 protein degradation assay) as compared to wild type.11 Taken together, the results of both cry1 and cry2 Trp triad mutant studies indicate a role for light-driven electron transfer in the biological activation of cryptochromes.
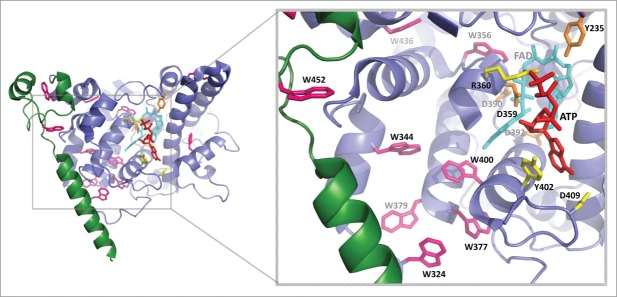
The data from both cry1 and cry2 Trp triad mutants is in agreement with our prior observations that electron transfer to the cry2 flavin in vivo is enhanced through the binding of small metabolites within the flavin pocket.14 In this former study we had identified a number of alternate electron transfer pathways as a result of analysis of additional cry2 mutants. These include a possible electron tunneling pathway via ATP through the Y399 residue (Y402 of cry1) that is directly potentiated by metabolite binding. In addition, electron transfer via aromatic amino acids W331 and W376 adjacent to the Trp triad (W334, W379 of cry1) is facilitated. ATP binding induces altered accessibility of the protein surface of both cry1 and cry2 to added protease,14,22 indicative of increased flexibility in the cry apoprotein.22,24 A consequence of greater protein flexibility would be improved alignment of alternate electron transfer residues with either the flavin – proximal W400 or flavin – distal W324, and possibly also of the Y402, resulting in more efficient electron transfer subsequent to illumination. A suggestion that ATP may potentiate electron transfer by increasing the pKa of an amino acid residue (D396 of cry1) at the flavin binding pocket20 is not supported by the data, as ATP effects on photoreduction are seen even at low pH14 and also if the D396 residue of cry1 is replaced by cysteine, which does not undergo efficient deprotonation.25 Furthermore, early conformational changes linked to cry1 activation occur in the absence of flavin protonation,25 further excluding that the protonation state of D396 is important for signaling. The positions of relevant amino acids in cry1 that may participate in these alternate electron transfer pathways in vivo are summarized in Fig. 5.
Figure 5.
Electron transfer pathways and possible C-terminal domain structure in A. thaliana cryptochrome 1. Structure was taken from PDB: 1U3D. Tryptophan residues are shown in pink, including positions of the classical Trp triad (FAD -W400 - W377 - W324) and residues implicated in alternate electron transfer pathways deduced from cry2 including aromatic amino acid residues near the Trp triad (cry1 W334, W379). The cry1 Y402 residue (homolog to cry2 Y399) is suitably positioned to form a potential electron tunneling pathway via ATP.14 The FAD is shown in cyan and the FAD binding amino acids, which may impact on structural changes in the protein are in orange. The ATP analog AMPPNP is in red. The fragment of C-T terminal analyzed in this study is in green and is lacking in the crystal structure. Created in the PyMOL Molecular Graphics System, Version 1.7.6.3 Schrödinger, LLC. Created in PyMOL.
We have extended these observations on the effects of the in vivo cellular environment on forward electron transfer to flavin to show that, cellular components also affect the rate of flavin reoxidation (from FADH° to FAD flavin). In fact, in purified isolated cry1 protein, the rate of flavin reoxidation is an order of magnitude more rapid than the rate of reoxidation of the protein under the identical reducing conditions in a whole-cell lysate. This effect of slowing of the reoxidation rate is fully consistent with the effect of metabolites on stabilizing the flavin neutral radical21 – see also Fig. 3.
We note that the flavin reoxidation rate determined for cry1 in lysed cellular extracts (Fig. 3) does not directly correspond to the in vivo lifetime of the activated (FADH°) flavin redox state determined directly in living intact cells in vivo.26 This in vivo lifetime was determined through whole-cell EPR spectroscopic methods in living intact insect cells, to give a half – life for cry reoxidation on the order of several minutes. The difference with conditions used in the current manuscript is that experiments in whole cell extracts were performed in the presence of a strong reductant (5 mM DTT), as AtCry1 is otherwise not efficiently reduced under aerobic (lysed cell) conditions. In the cellular environment, the biological in vivo reducing agents and cellular oxygen concentrations are as yet unknown and these may play a sigificant additional effect on the final rate of flavin reoxidation in vivo. What is clear from our study is that the flavin radical should be stabilized in vivo by the presence of metabolites and thereby contribute to the half-life of the activated signaling state also in living cells.
In addition to their well-established role as light receptors, cryptochromes have been suggested as a candidate for the elusive magnetic receptor in birds and other animals27,28 In birds, avian cryptochrome 1 has been found in the outer segments of UV cone receptor cells, an ideal location for a potential magnetic compass sensor based on photochemical reactions. Initial mutant studies show that magnetic field effects disappear in responses of plants29 and fruit flies lacking cryptochromes,30,31 but a puzzling observation showed that fruit flies with type 1 cryptochromes without a functional Trp triad still exhibited magnetic orientation responses.32 Our study here demonstrates that cry 1 Trp triad mutants can still be rendered active in in vivo environments by metabolites. It thus both provides an explanation for the fruit fly results and a caution for mutant magnetic orientation experiments without considering the influence of metabolites and the complete in vivo environment.
A continuing puzzle in the mechanism of cryptochrome activation concerns the nature of the conformational change undergone by the cryptochromes in the course of signaling, which seem to involve the C-terminal domain (see Fig. 1). To obtain insight on the possible structural changes, we have obtained a CD spectra of the full-length cry1 protein, and compared it to that of a truncated C-terminal fragment. The full-length protein shows a proportion of ordered and disordered regions that is not significantly different from the proportions derived from the known crystal structure of the N-terminal domain.22 This suggests that the C-terminal domain may not contribute disproportionately to the degree of unordered structure in the full-length protein in its resting (dark adapted) state.
By contrast, the isolated C-terminal domain (amino acids 443–681), had a higher proportion of unstructured regions and turns (55%). Nonetheless, the proportion of turns and unstructured regions were significantly lower than reported previously (80%) for a somewhat smaller fragment of the C-terminal domain (amino acids 506–681).22 Therefore the region from amino acids 443–681 may contribute disproportionately toward the establishment of conformational changes in response to light. The location and suggested structure of the C-terminal domain relative to the flavin binding domain is shown in Fig. 5.
How exactly the primary photoreactions triggers structural changes in the receptor is also currently a matter of debate. The available evidence is consistent with flavin reduction required for formation of the signaling state, and therefore a requirement for forward electron transfer to the flavin. The question is whether the primary photoreactions (electron and proton transfer) themselves trigger conformational change, or whether it is the flavin in its radical state that drives structural change. It has been previously shown that conformational change involving the cry1 C-terminal domain occurs on a millisecond time scale,33 which is too slow for triggering by direct electron and/or proton transfer events. More recent data suggests conformational change may be initiated already within the N-terminal domain at much more rapid time scales,34 consistent with triggers such as deprotonation of D396, an aspartic acid residue in proximity to the flavin.
However, this suggestion is excluded by results from the D396C mutation of cry1, for which infrared spectroscopic experiments demonstrate that early light-induced conformational changes of the N-terminal domain occur despite the absence of proton transfer.25 Instead, the lifetime of the conformational change in vitro25 and of the biologically active signaling state in vivo26 have both been correlated with the lifetime of the flavin radical, consistent with a requirement for the radical to initiate and/or maintain the signaling structure. Other suggestions as to alternate mechanisms of cryptochrome activation, for example that cry is in the radical redox state in the dark (resting) conformation35 and undergoes some (unknown) photoreaction are contradicted by experimental evidence. The action spectrum for plant cryptochromes (exclusively to blue/UV light) excludes neutral radical flavin in the resting state. Furthermore, the radical state in both plant and insect cryptochromes in vivo is not detected in the dark, but only subsequent to illumination.6,7,13 In sum, all of the available evidence suggests that the cryptochrome flavin radical, in some manner stabilizes the activated cryptochrome structure and that protonation is not required for this role in cry1.
Conclusions and Outlook
Trp triad mutants W400F and W324F of Arabidopsis cry1 are photochemically inactive in vitro yet undergo flavin photoreduction in whole cell extracts. Their efficiency of photoreduction is reduced as compared to wild type proteins, consistent with reduced in vivo biological activity.15 These results are consistent with those obtained for Trp triad mutants W397F and W321F of cry2,11,14 which likewise show in vivo but not in vitro photoreduction and reduced light dependent biological activity. Binding of small metabolites including ATP enhance the efficiency of alternate electron transfer pathways in cry2 Trp triad mutants and are conserved in cry1 (Fig. 5). ATP in addition extends the lifetime of the radical (signaling) state in vivo. These studies thereby resolve existing confusion concerning the role of the Trp triad and flavin reduction in biological activation of plant cryptochromes in vivo by showing that alternate electron transfer pathways can compensate in the absence of the Trp triad. Our results point to sequences immediately adjacent to the C-terminal domain as potentially implicated in the conformational change leading to signaling. Finally, the redox cycle of cryptochromes by itself generates reactive oxygen species (ROS) that may also play a part in signaling,36 independently of any conformational change.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the “Imagerie Paris Seine” imaging platform for confocal microscopy analyses. We thank Alain d'Harlingue, Pierre-Etienne Bouchet, Jacques Witzak and Claire Monné for excellent technical assistance.
Funding
We are grateful to AFOSR (FA9550-14-0-0409) for funding. M. Elesawi is the recipient of a French government postdoctoral fellowship.
References
- 1.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Ann Rev Plant Biol 2011; 62:335-64; PMID:21526969; http://dx.doi.org/ 10.1146/annurev-arplant-042110-103759 [DOI] [PubMed] [Google Scholar]
- 2.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue light induction of protein interactions in living cells. Nat Methods 2010; 7:973-5; PMID:21037589; http://dx.doi.org/ 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 2011; 25:1029-34; PMID:21511871; http://dx.doi.org/ 10.1101/gad.2025011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Partch CL, Clarkson MW, Ozgür S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 2005; 44:3795-805; PMID:15751956; http://dx.doi.org/ 10.1021/bi047545g [DOI] [PubMed] [Google Scholar]
- 5.Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol 2011; 413:128-37; PMID:21875594; http://dx.doi.org/ 10.1016/j.jmb.2011.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, Batschauer A. The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem 2007; 282:14916-22; PMID:17355959; http://dx.doi.org/ 10.1074/jbc.M700616200 [DOI] [PubMed] [Google Scholar]
- 7.Bouly JP, Giovani B, Djamei A, Mueller M, Zeugner A, Dudkin EA, Batschauer A, Ahmad M. Novel ATP-binding and autophosphorylation activity associated with Arabidopsis and human cryptochrome-1. Eur J Biochem 2003; 270:2921-8; PMID:12846824; http://dx.doi.org/ 10.1046/j.1432-1033.2003.03691.x [DOI] [PubMed] [Google Scholar]
- 8.Burney S, Wenzel R, Kottke T, Roussel T, Hoang N, Bouly JP, Bittl R, Heberle J, Ahmad M. Single amino acid substitution reveals latent photolyase activity in Arabidopsis cry1. Angew Chem Int Ed 2012; 51:9356-60; PMID:22890584; http://dx.doi.org/ 10.1002/anie.201203476 [DOI] [PubMed] [Google Scholar]
- 9.Herbel V, Orth C, Wenzel R, Ahmad M, Bittl R, Batschauer A. Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J 2013; 74:583-92; PMID:23398192; http://dx.doi.org/ 10.1111/tpj.12144 [DOI] [PubMed] [Google Scholar]
- 10.Müller P, Ahmad M. Light-activated cryptochrome reacts with molecular oxygen to form a flavin-superoxide radical pair consistent with magnetoreception. J Biol Chem 2011; 286:21033-40; PMID:21467031; http://dx.doi.org/ 10.1074/jbc.M111.228940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Wang Q, Yu X, Liu H, Yang H, Zhao C, Liu X, Tan C, Klejnot J, Zhong D, Lin C. Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc Natl Acad Sci USA 2011; 108:20844-49; PMID:22139370; http://dx.doi.org/ 10.1073/pnas.1114579108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oztürk N, Song SH, Selby CP, Sancar A. Animal type 1 cryptochromes. Analysis of the redox state of the flavin cofactor by site-directed mutagenesis. J Biol Chem 2008; 283:3256-63; PMID:18056988; http://dx.doi.org/ 10.1074/jbc.M708612200 [DOI] [PubMed] [Google Scholar]
- 13.Hoang N, Schleicher E, Kacprzak S, Bouly JP, Picot M, Wu W, Berndt A, Wolf E, Bittl R, Ahmad M. Human and Drosophila cryptochromes are light activated by flavin photoreduction in living cells. PLoS Biol 2008; 6(7): e160; PMID:18597555; http://dx.doi.org/ 10.1371/journal.pbio.0060160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhard C, Wang X, Robles D, Moldt J, Essen L-O, Batschauer A, Bittl R, Ahmad M. Cellular metabolites enhance light sensitivity through alternate electron transfer pathways in Arabidopsis cryptochrome. Plant Cell 2014; 26:4519-31; PMID:25428980; http://dx.doi.org/ 10.1105/tpc.114.129809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeugner A, Byrdin M, Bouly JP, Bakrim N, Giovani B, Brettel K, Ahmad M. Light-induced electron transfer in Arabidopsis cryptochrome-1 correlates with in vivo function. J Biol Chem 2005; 280:19437-40; PMID:15774475; http://dx.doi.org/ 10.1074/jbc.C500077200 [DOI] [PubMed] [Google Scholar]
- 16.Stevens JA, Link JJ, Zang C, Wang L, Zhong D. “Ultrafast Dynamics of Nonequilibrium Resonance Energy Transfer and Probing Globular Protein Flexibility of Myoglobin”. J Phys Chem A 2012; 116:2610-19; PMID:21863851; http://dx.doi.org/ 10.1021/jp206106j [DOI] [PubMed] [Google Scholar]
- 17.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 1981; 20:33-37; PMID:7470476; http://dx.doi.org/ 10.1021/bi00504a006 [DOI] [PubMed] [Google Scholar]
- 18.Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Prtoein Sci 2004; 13:100-12; PMID:14691226; http://dx.doi.org/ 10.1110/ps.03258404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA 2004; 101:12142-7; PMID:15299148; http://dx.doi.org/ 10.1073/pnas.0404851101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller P, Bouly JP, Hitomi K, Balland V, Getzoff ED, Ritz T, Brettel K. ATP binding turns plant cryptochrome into an efficient natural photoswitch. Sci Rep 2014; 4:5175; PMID:24898692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Immeln D, Schlesinger R, Heberle J, Kottke T. Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem 2007; 282:21720-8; PMID:17548357; http://dx.doi.org/ 10.1074/jbc.M700849200 [DOI] [PubMed] [Google Scholar]
- 22.Partch CL, Clarkson MW, Ozgür S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry 2005; 44:3795-805; PMID:15751956; http://dx.doi.org/ 10.1021/bi047545g [DOI] [PubMed] [Google Scholar]
- 23.Burney S, Hoang N, Caruso M, Dudkin EA, Ahmad M, Bouly JP. Conformational change induced by ATP binding correlates with enhanced biological function of Arabidopsis cryptochrome. FEBS Lett 2009; 583:1427-33; PMID:19327354; http://dx.doi.org/ 10.1016/j.febslet.2009.03.040 [DOI] [PubMed] [Google Scholar]
- 24.Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol 2004; 51:299-321; PMID:15218531 [PubMed] [Google Scholar]
- 25.Hense A, Herman E, Oldemeyer S, Kottke T. Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome. J Biol Chem 2015; 290:1743-51; Epub 2014 Dec 3; PMID:25471375; http://dx.doi.org/ 10.1074/jbc.M114.606327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbel V, Orth C, Wenzel R, Ahmad M, Bittl R, Batschauer A. Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J 2013; 74:583-92; PMID: 23398192; http://dx.doi.org/ 10.1111/tpj.12144 [DOI] [PubMed] [Google Scholar]
- 27.Ritz T, Adem S, Schulten K. A model for photoreceptor-based magnetoreception in birds. Biophys. J. 2000; 78:707-18; PMID:10653784; http://dx.doi.org/ 10.1016/S0006-3495(00)76629-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritz T, Yoshii T, Foerster C, Ahmad M. Cryptochrome: A photoreceptor with the properties of a magnetoreceptor? Comm Integr Biol 2009; 3:24-7; PMID: 20539777; http://dx.doi.org/ 10.4161/cib.3.1.9865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad M, Galland P, Ritz R, Wiltschko R, Wiltschko W. Magnetic intensity affects cryptochrome-dependent responses in Arabidopsis thaliana. Planta 2007; 225: 615-24; PMID:16955271; http://dx.doi.org/ 10.1007/s00425-006-0383-0 [DOI] [PubMed] [Google Scholar]
- 30.Gegear R. J, Casselman A, Waddell S, Reppert S M. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 2008; 454; 1014-1018; PMID:18641630; http://dx.doi.org/ 10.1038/nature07183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshii T, Ahmad M, Helfrich-Forster C. Cryptochrome mediates light-dependent magnetosensitivity of Drosophila's circadian clock. PLoS Biol. 2009: 7:813-9; http://dx.doi.org/ 10.1371/journal.pbio.1000086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gegear R J, Foley L E, Casselman A, Reppert S M. Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 2010; 463; 804-807; PMID:20098414; http://dx.doi.org/ 10.1038/nature08719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondoh M, Shiraishi C, Müller P, Ahmad M, Hitomi K, Getzoff ED, Terazima M. Light-induced conformational changes in full-length Arabidopsis thaliana cryptochrome. J Mol Biol 2011; 413:128-37; PMID: 21875594; http://dx.doi.org/ 10.1016/j.jmb.2011.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thöing C, Oldemeyer S, Kottke T. Microsecond deprotonation of aspartic acid and response of the subdomain precede C-terminal signaling in the blue light sensor plant cryptochrome. J Am Chem Soc 2015; 137:5990-9; PMID:25909499; http://dx.doi.org/ 10.1021/jacs.5b01404 [DOI] [PubMed] [Google Scholar]
- 35.Liu B, Liu H, Zhong D, Lin C. Searching for a photocycle of the cryptochrome photoreceptors. Curr Opin Plant Biol 2010; 13:578-86; PMID:20943427; http://dx.doi.org/ 10.1016/j.pbi.2010.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consentino L, Lambert S, Martino C, Jourdan N, Bouchet PE, Witczak J, Castello P, El-Esawi M, Corbineau F, d'Harlingue A, Ahmad M. Blue-light dependent reactive oxygen species formation by Arabidopsis cryptochrome may define a novel evolutionarily conserved signaling mechanism. New Phytol 2015; Feb 26. 206(4):1450-62; PMID:25728686; http://dx.doi.org/ 10.1111/nph.13341 [DOI] [PubMed] [Google Scholar]