Abstract
Steroidogenic acute regulatory related transfer (StART) proteins that are involved in transport of lipid molecules, play a myriad of functions in insects, mammals and plants. These proteins consist of a modular START domain of approximately 200 amino acids which binds and transfers the lipids. In the present study we have performed a genome-wide search for all START domain proteins in chickpea. The search identified 36 chickpea genes belonging to the START domain family. Through a phylogenetic tree reconstructed with Arabidopsis, rice, chickpea, and soybean START proteins, we were able to identify four transmembrane START (TM-START) proteins in chickpea. These four proteins are homologous to the highly conserved mammalian phosphatidylcholine transfer proteins. Multiple sequence alignment of all the transmembrane containing START proteins from Arabidopsis, rice, chickpea, and soybean revealed that the amino acid residues to which phosphatidylcholine binds in mammals, is also conserved in all these plant species, implying an important functional role and a very similar mode of action of all these proteins across dicots and monocots. This study characterizes a few of the not so well studied transmembrane START superfamily genes that may be involved in stress signaling. Expression analysis in various tissues showed that these genes are predominantly expressed in flowers and roots of chickpea. Three of the chickpea TM-START genes showed induced expression in response to drought, salt, wound and heat stress, suggesting their role in stress response.
Keywords: abiotic stress, chickpea, lipid binding, signaling, START domain
Introduction
Steroidogenic acute regulatory (StAR) proteins in mammals bind and transfer lipid molecules such as cholesterol between the intracellular membranes in humans.1 The START domain (StAR -related lipid transfer protein) is a lipid binding and transfer domain of around 200 amino acid in length.2 X-ray crystallography revealed a helix grip fold secondary structure for the START domain.3,4 Ligands like lipids are important for metabolism, act as signaling molecule, assist in membrane stability5 etc. A few StAR proteins have been reported to bind to cholesterol, phosphotidylcholine, ceramide6 and the carotenoid lutein.7
START domain containing genes are present in a few genera of bacteria, unicellular protists but absent in archaea and yeast. Drosophila melanogaster, Caenorhabditis elegans, and Bombyx mori also possess a few START proteins, while humans and mice have 15 each.2 In humans, most of the START proteins are multi-domain proteins. Mutations in the START domain results in congenital adrenal hyperplasia, and in one case,8 a protein homologous to StAR was found to be over-expressed in breast cancer. In Drosophila, START1 gene is a key regulator of ecdysteroid synthesis.9
In plants, START domain containing proteins are commonly found to have a homeodomain, a DNA-binding domain (HD-START). Two other domains, the pleckstrin homology (PH) domain and the DUF 1336 (domain of unknown function), are also present along with the START domain in some proteins.2 Apart from these two classes, two more viz., the START minimal protein class containing only the START domain, and transmembrane domain (TM-START) containing START proteins are also present in plants. The number of START proteins present in Arabidopsis and rice is 35 and 29, respectively.2 In plants, START domain containing proteins have been reported to perform varied functions. In Arabidopsis, ATML1 and PDF2 HD-START proteins are required for epidermal differentiation during embryogenesis.10 In rice, the ortholog of ATML1, rice outermost cell specific gene 1 (Roc1), has an analogous function, and is developmentally regulated during early embryogeneiss.11 Arabidopsis Anthocyaninless 2, a HD-START protein, affects anthocyanin accumulation and cellular organization in roots.12 Another HD-START protein, GL2, is involved in trichome development and is a negative regulator of phopholipid signaling in the roots.13 Mutations in two HD-ZIP START genes, PHABULOSA and PHAVOLUTA of Arabidopsis, which are implicated in the perception of radial positional information in the leaf primordium, cause a dramatic transformation of abaxial leaf fates into adaxial leaf fates.14
The function of START proteins with domains like HD and PH has been mostly related to plant development. Also, mutants of these genes and genes involved in sterol biosynthesis have similar phenotypes. One of the START proteins, EDR2, containing the DUF 1336 and PH domains was reported to act as a negative regulator of pathogen induced resistance.15 According to Tang et al. (2005) the EDR2 protein may become functional with the binding of a lipid ligand and that it may provide the missing links between lipid signaling, mitochondria and activation of programmed cell death in plants. Most of the studies hitherto mentioned have been on START proteins that are associated with the HD/PH/DUF 1336 domains. To the best of our knowledge, no study has thus far been conducted on the transmembrane containing START proteins. Generally, membrane proteins are involved in diverse processes such as cell signaling, transport of membrane-impermeable molecules, cell-cell communication, cell recognition and cell adhesion.16 In plants also, transmembrane domain containing genes have been implicated in a variety of processes. For example, ethylene signal transduction pathway consists of a receptor containing transmembrane domain at the N-terminal region for ethylene binding and is localized in the endoplasmic reticulum membrane.17 Brassinosteroid insensitive (BRI1), receptor for a steroid, consists of a transmembrane domain localized in the plasma membrane.18 In addition, inside a cell, membrane lipid and protein dynamics is thought to be important for stress perception and signaling. There are several proteins associated with the membrane by means of transmembrane domain. In Arabidopsis, several transcription factors such as the NAC family, which possess transmembrane domain have been identified that are reported to function in stress response as and when the membrane profiles change.19 All examples cited above describe protein activation upon the binding of a ligand followed by the transduction of stress signal. The protein could act as a transcription factor which triggers a pathway or as a component of a wider signal transduction pathway by taking part in protein-protein interactions.
The presence of transmembrane domain in a protein along with a ligand binding domain like START could be indicative of the important roles these proteins play in various processes in plant such as growth, development, and the ability to survive harsh conditions. Schrick et al.2 reported that one of the classes of START proteins named phosphatidylcholine transfer proteins were conserved from animals to plants. All the proteins of this class also contain transmembrane domain(s). Since, the START proteins with transmembrane domains are highly conserved, we have undertaken a study of this group of proteins in chickpea. The availability of the draft sequence of its genome20,21 facilitates in silico identification and characterization of this important group of genes. In our study, we have identified START domain containing proteins, particularly the ones that contain transmembrane domains, and carried out expression and sequence analyses.
Materials and methods
Identification of START domain genes in the whole genome sequence
The START domain of the Arabidopsis gene AT5G54170 (∼200 amino acids) was used as query sequence in TBLASTN search of whole genome shotgun contigs database of desi chickpea variety to identify putative chickpea START proteins on NCBI (taxid:3827). Gene prediction programmes Augustus,22 GeneMark23 and Genscan24 were utilized for the prediction of genes and protein sequences. A manual analysis of the gene predictions was performed and the longest protein sequence was taken for further analysis. Pfam analysis25 and SMART26 program were utilized to find out the START domain in the predicted sequences.
Domain analysis, motif and phylogenetic tree construction
Transmembrane helices in the predicted proteins were identified using TMHMM server,27 v.2.0 Kyte Doolittle Hydropathy Plot,28 and the PSIPRED Protein Sequence Analysis Workbench.29 Multiple alignments of selected proteins were carried out using MUSCLE program. After manual editing, MEGA (version 6.0) was used for construction of, neighbor joining tree with following parameters: Poisson correction, Pairwise-Deletion, and Bootstrap (1500 replicates).30 MEME analyses were performed to identify conserved motif in the sequences. The search parameters were: Motif width range: 6–50 with maximum 20 motifs for discovery.31 The cis-regulatory elements in the promoter were analyzed using PLANTCARE database32 in the 2Kb upstream sequences of the transmembrane START genes.
Putative START domain containing transcripts of chickpea were obtained from CTDB through providing Pfam domain id33 (PF). BlastN search of predicted CDS from the genome sequence was performed to identify its corresponding transcript in the CTDB. Transcript identifier were used to retrieve the normalized read counts of the transcriptome data in different tissues. These read values were used for the construction of heat map with the Heatplus package available on Bioconductor using the function regHeatmap in the R statistical environment.
Plant material and stress treatments
Samples from five different tissues were collected from desi Chickpea (variety ICC4958) grown in the field. Line sowing of seeds was done during last week of October, 2013 with each line consisting of up to 15 seedlings. During the pod development stage (around 2nd week of February), shoots, roots, flowers and pods were collected from the plants and immediately frozen in liquid nitrogen and stored in −80°C for RNA isolation.
For studies involving stress treatment, 15 day-old-chickpea seedlings grown in pots were used. The pots were kept in glass house maintained at 22 ± 2°C with 14 hours light condition. The leaflets and the shoot of the seedlings were subjected to wounding with tweezers; cold and heat stress were given by shifting seedlings to incubator at 4 ± 1°C and 37 ± 1°C respectively. Dehydration stress was given by uprooting the seedlings and washing the roots in tap water and placing the seedlings on the tissue paper. Salt stress was given by uprooting, washing and placing the seedlings in a solution of 200 mM NaCl. Seedling samples were collected at different time intervals after stress treatment viz., 15 min, 1 hr, 6 hr and 24 hr along with untreated plants that served as control. The samples were rapidly frozen in liquid nitrogen and stored at −80°C.
RNA isolation, cDNA synthesis and Real time PCR
Total RNA was isolated using Trizol method. The integrity and quality of RNA was analyzed through agarose gel (1% w/v) and nano drop ND-1000 UV-Vis spectrophotometer, respectively. 10 µg of RNA was used for the DNase I treatment. DNase I treated RNA was used for the first strand cDNA synthesis using Superscript III Reverse transcriptase (Invitrogen). Quantitative Real Time PCR was done in StepOnePlus instrument (ABI). Three biological replicates were used for the different tissues and stress samples. The gene, GADPH2, was used for normalization of cDNA samples among different stress conditions in all time points and in the tissue samples.34 Each reaction contains 5 µL 2×SYBR Master mix reagent (Genetix), 1 µL cDNA and 200 nM of gene specific primers in a final volume of 10 µL. Each pair of primers were designed from Primer3plus program with an amplicon size ranging from 150–200 bp. The sequence of the primers are provided in Table S2. The thermal cycle used is as follows: 95°C for 10 min for activation, 40 cycles of 95°C for 15 s for denaturation and 60°C for 1 min for annealing and extension. The specificity of reaction was analyzed in melting curve analysis. The amplified products were also run in 1.5% agarose gel to confirm the expected band size of the fragment. The relative transcript level of the mRNA for different tissues is determined by −ΔΔCT values in comparison with the shoot values. For stress, untreated was considered as control and fold change was calculated by a similar method.
Results
Chickpea whole genome shotgun contigs database of desi variety (ICC4958) was used to retrieve the contigs having sequences homologous to START protein. Arabidopsis START domain containing protein (AT5G54170) was used as query sequence. The contigs showing significant alignments to the query sequence and consisting of E-value up to 0.05 were selected for the identification of chickpea START proteins. All the predicted proteins from the selected contigs were analyzed for the presence of START domain using Pfam and SMART databases. Our analysis of the chickpea genome (Desi variety) resulted in the identification of 36 genes containing START domain (Table 1).
Table 1.
List of START domain proteins in chickpea
| S.NO | ID | Protein Size (aa) | Type | Start Domain Region | Chromosome Location |
|---|---|---|---|---|---|
| 1 | Contig23652 | 737 | HD-START | 257–476 | I |
| 2 | Contig15638 | 804 | HD-START | 238–458 | Scaffold |
| 3 | Contig2884 | 750 | HD-START | 261–484 | IV |
| 4 | Contig2569 | 729 | HD-START | 249–468 | VIII |
| 5 | Contig13133 | 777 | HD-START | 289–513 | VII |
| 6 | Contig22402 | 762 | HD-START | 270–493 | Scaffold |
| 7 | Contig5986 | 814 | HD-START | 326–552 | III |
| 8 | Contig14368 | 810 | HD-START | 290–514 | VIII |
| 9 | Contig12681 | 722 | HD_START | 227–450 | I |
| 10 | Contig1668 | 713 | HD_START | 235–458 | IV |
| 11 | Contig3503 | 624 | HD_START | 149–367 | I |
| 12 | Contig3505_1 | 464 | HD_START | 227–448 | I |
| 13 | Contig 3505_2 | 524 | MINIMAL_START | 44–262 | I |
| 14 | Contig1250 | 755 | HD_START | 273–494 | I |
| 15 | Contig80 | 619 | MINIMAL_START | 262–375 | II |
| 16 | Contig5081 | 372 | MINIMAL_START | 1–114 | VI |
| 17 | Contig3504 | 463 | MINIMAL_START | 35–258 | I |
| 18 | Contig12904 | 311 | MINIMAL_START | 83–153 | I |
| 19 | Contig12363 | 851 | HD_START_MEKHLA | 165–372 | VI |
| 20 | Contig4799 | 829 | HD_START_MEKHLA | 164–371 | I |
| 21 | Contig9932 | 838 | HD_START_MEKHLA | 160–367 | VI |
| 22 | Contig3514 | 844 | HD_START_MEKHLA | 165–372 | VI |
| 23 | Contig5082 | 526 | HD_START | 418–524 | VI |
| 24 | Contig10550 | 793 | HD_START_MEKHLA | 161–341 | VII |
| 25 | Contig8429 | 761 | HD_START_MEKHLA | 142–351 | V |
| 26 | Contig10484 | 452 | MINIMAL_START | 329–422 | VI |
| 27 | Contig380 | 848 | HD_START_MEKHLA | 158–366 | VI |
| 28 | Contig25266 | 493 | MINIMAL_START_TM | 182–386 | I |
| 29 | Contig632 | 437 | MINIMAL_START_TM | 155–317 | VIII |
| 30 | Contig2750 | 399 | MINIMAL_START_TM | 98–306 | IV |
| 31 | Contig5716 | 425 | MINIMAL_START_TM | 139–289 | V |
| 32 | Contig4555 | 188 | MINIMAL_START | 32–165 | VII |
| 33 | Contig54670 | 147 | MINIMAL_START | 6–123 | Scaffold |
| 34 | Contig6542 | 235 | MINIMAL_START | 42–211 | V |
| 35 | Contig16710 | 477 | MINIMAL_START | 108–271 | I |
| 36 | Contig30763 | 251 | MINIMAL_START | 77–251 | VII |
Phylogenetic analysis
The grouping of chickpea START proteins was analyzed through phylogenetic tree reconstruction. Arabidopsis, rice, chickpea and soybean START proteins were used to reconstruct the phylogenetic tree. The tree revealed that the major classes of chickpea START proteins viz. minimal START, TM-START, HD-START, and HD-START-MEKHLA were grouped into distinct clades (Fig. 1). One of the least studied groups of START proteins having transmembrane domain (TM START) was clustered in a separate group in the phylogenetic reconstruction. The TM-START clade consisted of 22 proteins; 4 of chickpea, four of rice, six of Arabidopsis, and eight of soybean. The phylogenetic clade containing only TM-START proteins of Arabidopsis, rice, chickpea and soybean can be further divided into subclades I and II (Fig. 2). The first clade with 6 TM-START proteins consists of one member from chickpea and rice and two each from Arabidopsis and soybean. The second clade consists of 16 proteins; this clade is further divided into two subclades i.e Group IIa and Group IIb. Group IIa consists of six proteins and Group IIb of 10. The four chickpea TM-START proteins were found to be closely related to soybean than Arabidopsis and rice. To understand more about the grouping of these proteins we resorted to analyzing other motifs present in these proteins.
Figure 1.
Neighbor-joining phylogeny of Arabidopsis, chickpea, rice and soybean START proteins performed in MEGA6. Colored branch indicates the separate grouping of TM-START proteins.
Figure 2.
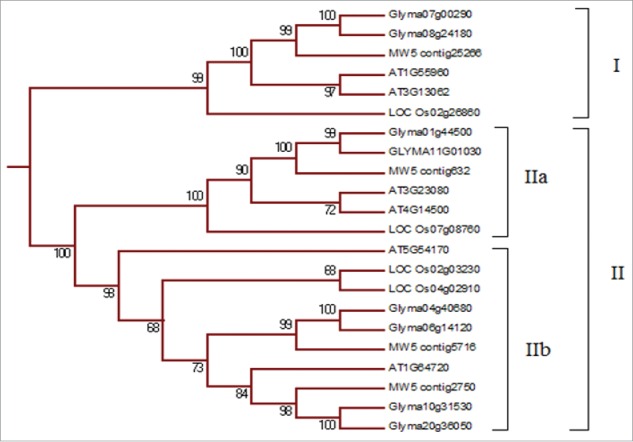
A clade consisting of TM-START proteins of Arabidopsis, chickpea, rice and soybean. From the phylogenetic tree of 149 START proteins, 22 TM-START proteins of which six are from Arabidopsis, 8 from soybean, and four each from rice and chickpea were identified.
Pfam and SMART analyses revealed the presence of domains other than START in chickpea proteins under study. The domains identified include the homeodomain (HD-START), HD-START with MEKHLA domain and transmembrane (TM) domain. We could identify 14 proteins with HD-START domain, seven proteins with HD-START-MEKHLA domain, 15 proteins with minimal START among which four proteins contained transmembrane (TM) regions. Chickpea START proteins containing TM domain were initially identified using TMHMM Server v. 2.0 and further verified using Kyte Doolittle Hydropathy Plot, and PSIPRED protein Sequence Analysis workbench. At least one transmembrane domain was predicted in all the sequences by all the three programs used (Table S1). For further analysis, we took only the START proteins having transmembrane domain as this class of proteins has not been studied in detail. The TM-START proteins of chickpea were analyzed through multiple sequence alignment, phylogeny, motif prediction, cis-element prediction, and gene expression analysis.
We included the START proteins from Arabidopsis, rice, chickpea and soybean (retrieved from TAIR, Rice Genome Annotation Project and PlantGDB) for the analysis. The soybean START proteins are provided in Supplementary Table 3. Analysis of sequence conservation of TM-START proteins was performed through multiple sequence alignment using MUSCLE program. In the START domain region of TM-START proteins, 33 amino acids were found to be invariable for the Arabidopsis, rice, chickpea and soybean (Fig. 3). Also, most striking feature in the transmembrane region in the carboxy terminal is rather than the conservation of specific amino acids in specific positions, hydrophobic groups containing amino acids like tryptophan and valine were highly conserved. Apart from these two regions, the N-terminal region of proteins also consists of conserved groups of hydrophobic regions presumed to function as transmembrane domain. Our prediction of transmembrane domains in these proteins by various programs showed the presence of at least one transmembrane domain in the amino- or C-terminal regions of the proteins.
Figure 3.
Multiple alignment of START domain of TM-START proteins from Arabidopsis, chickpea, rice and soybean performed in TCOFFEE alignment program. BOXSHADE output reveals the conservation of amino acids in these proteins. Asterisk reveals the conservation of amino acid residues that are required for the binding of head group of phosphatidylcholine.
Motif analysis
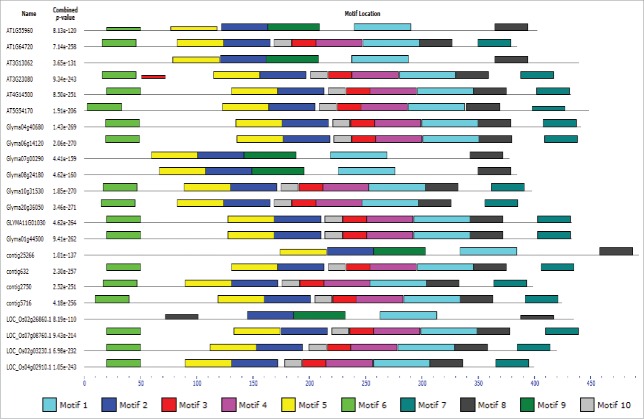
MEME motif analysis of all the TM-START domain proteins of 4 genera revealed highly conserved motifs in the START domain. Several conserved amino acids were present in the motifs of the START domain. The TM-START proteins of Arabidopsis, rice, chickpea, and soybean (22 proteins) were analyzed separately for the conservation of motifs between them. Apart from the conserved motifs in the START domain, other motifs were also conserved (Fig. 4). Motif 6 was present in the N-terminal region of 17 proteins, motif 8 and motif 7 were present in the C-terminal region of 22 and 16 proteins, repectively. The InterPro search revealed that motif 6 had a transmembrane signature sequence in the amino–terminal, whereas motif 7 and 8 had transmembrane signature sequences in the C-terminal. Motif 6 identified in the amino-terminal region was found to be present in 7 of the TM-START proteins. In this motif, the amino acid tryptophan was found to be conserved and the consensus sequence is P[LI]W[LIV][AT][VF][MFI][IF]G[VL][VL][VI]GW[SA]W[KR]P[KR]W. Motif 8 identified in the carboxy terminal region before transmembrane motif was found to have an invariable lysine residue in the TM proteins of Arabidopsis, chickpea and soybean and rice.
Figure 4.
Motifs identification in 22 TM-START proteins of Arabidopsis, rice, chickpea and soybean was performed using MEME. Motifs 6, 7 and 8 are related to transmembrane domain.
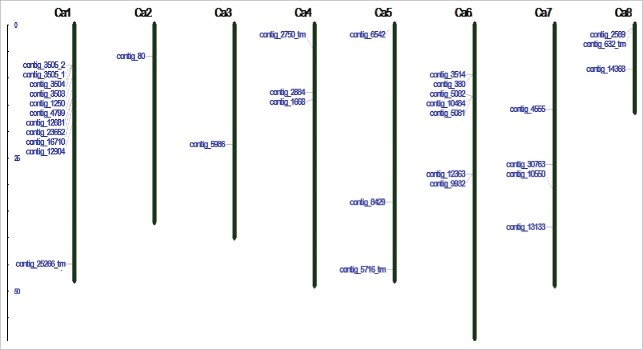
The contigs of desi chickpea genome are not anchored on to pseudomolecules of linkage groups. Thus, we used the kabuli genome17 linkage group for the chromosomal mapping of START proteins (Fig. 5), which revealed that three START proteins were in the scaffold and the other 33 were mapped on the 8 chromosomes of chickpea. Eleven START genes were present on chromosome 1 as a cluster spanning 5–20 cM distance. Seven genes were present on chromosome 6, four on chromosome 7, three each on chromosome 4, 5 and 8, and one each on chromosome 2 and 3. The four transmembrane START genes, namely contig 25266, 2750, 5716 and 632 were present in linkage groups 1, 4, 5 and 8, respectively.
Figure 5.
Chromosomal mapping of identified START proteins from chickpea. Ca1–Ca8 represents the number of chickpea chromosomes distanced by centimorgan. Three genes present in the scaffold are not mapped.
Expression analysis
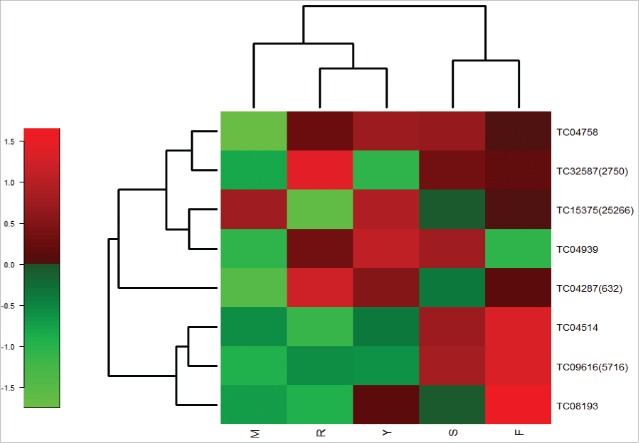
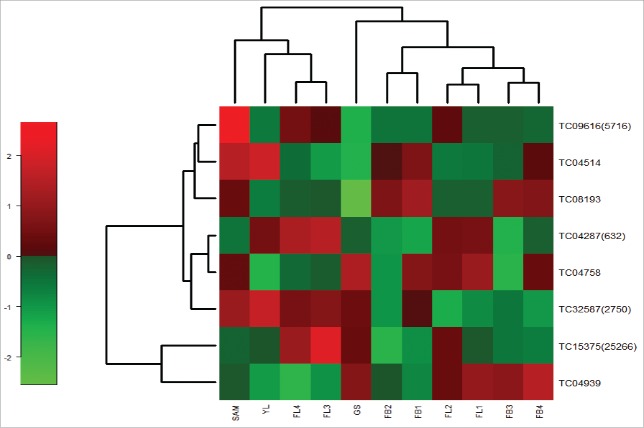
A heat map generated using normalized read counts from the chickpea transcriptome database (CTDB) (Fig. 6) revealed that the expression of all the TM-START protein genes was more pronounced in the flower tissues of chickpea at stages 3 and 4 (Fig. 7). The expression analysis of TM-START genes of Arabidopsis were also compared to identify specific patterns of expression. The expression of the Arabidopsis genes in 10 developmental stages was analyzed in Genvestigator (Fig. S1). All the Arabidopsis TM-START genes express throughout the various developmental stages. Two genes, AT4G14500 and AT5G54170, were highly induced in the senescent leaves while At3G13062 showed no expression in the senescent leaves. Similar to the observation in chickpea TM-START genes, all Arabidopsis genes were expressed in the flower stages.
Figure 6.
Heat map of chickpea START proteins generated using the normalized read counts of different tissues. CTDB was used to retrieve the read counts. Tentative consensus (TC) ID along with contig number in brackets represents TM-START proteins. S-Shoot: R-Root: F-Flower bud: Y-Young pod: M-Mature leaf.
Figure 7.
Heat map of chickpea START proteins generated using the normalized read counts of different tissues and flower stages.CTDB was used to retrieve the read counts. Tentative consensus (TC) ID along with contig number in brackets represents TM-START proteins. SAM-Shoot apical meristem: FB1-4-Flower bud stages: FL1-FL4- Flower stages:GS-Germinating seedlings:YL-Young leaf.
Expression analysis for the Arabidopsis genes was also performed for different stress conditions (Fig. S2). One of the genes, AT5G54170, was found to be induced in abiotic stresses like drought and salt stress, wherein it was found to be expressed in root epidermis. Apart from the abiotic stresses, biotic stresses like leaf miner infection, Pseudomonas infection also led to induced expression of this gene. AT1G64720, a paralog of AT5G54170, showed induction under salt-stress.
Quantitative real time-PCR analysis of TM-START protein genes
We had earlier shown that contig5716 is wound-responsive and also studied its expression in various tissues of chickpea.35 In this study we examined the expression of all the four TM-START genes in chickpea in different tissues collected at the pod development stage (Fig. 8). The analysis revealed that three of the TM-START genes (contig 25266, 2750 and 5716) had similar pattern of expression. The level of expression in roots and flowers was higher compared to shoots and pod wall. The level of expression of these genes was very low in the developing grain. Contig 632 was expressed at lower levels compared to the other TM-START genes in all the tissues.
Figure 8.
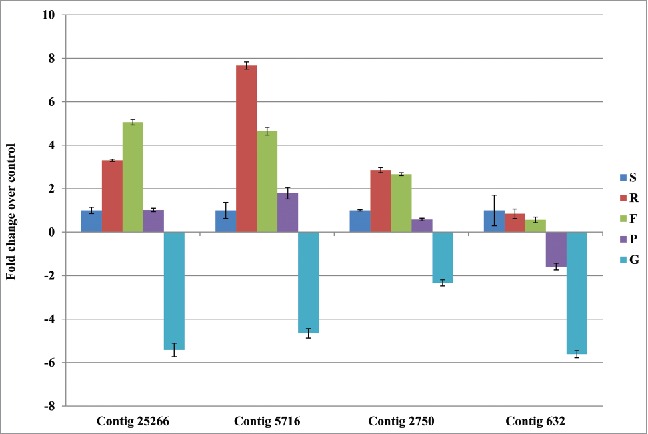
qPCR analysis of TM-START proteins of chickpea genes done in different tissues. Relative fold change values are given with respect to GADPH2 was used as reference genes. S-Shoot: R-Root: F-Flower : P-Young pod: G-Grain. The expression in shoot was taken as 1 and compared to other tissues.
These TM-START genes were also analyzed for their response to different stresses such as wound, cold, heat, drought, and salt at different time points (Fig. 9). Under cold stress, contig_632 showed up-regulation (3 fold) within 6 h of stress induction and down-regulated at 24 h of stress treatment Contig_5716 showed upregulation (2 fold) at 1 h and at later time points dropped to basal levels. Contig_2750 was downregulated (3 fold) 15 min after stress treatment. In response to drought condition, two contigs (2750 and 5716) showed slight upregulation ( 2–3 fold) within 15 minutes of stress and the expression level was maintained at a higher level up to 6–24 h of stress induction. Contrary to this, contig25266 showed varying levels of up- and down-regulation in drought stress, while contig632 showed consistent downregulation (∼8 fold) after 15 min. The pattern of expression kinetics that was observed for heat stress treatment was downregulation (3–10 fold) in early time points i.e. from 15 min to 1 h, whereas up-regulation (8–32 fold) was noticed during later time points of 6–24 h after stress treatment. Wound response showed consistent upregulation (∼8 fold) up to 1 h of contig_5716 and contig_632. At 24 h after treatment, the expression comes down to the basal level. In response to salt stress, all contigs showed similar expression profile through different time points. However, contig_2750, 5716 and 25266 showed consistent increase in expression after 15 min of incubation up to 6 h, while contig_632 showed downregulation (∼4 fold) at early stages of stress treatment and later attains the basal level.
Figure 9.
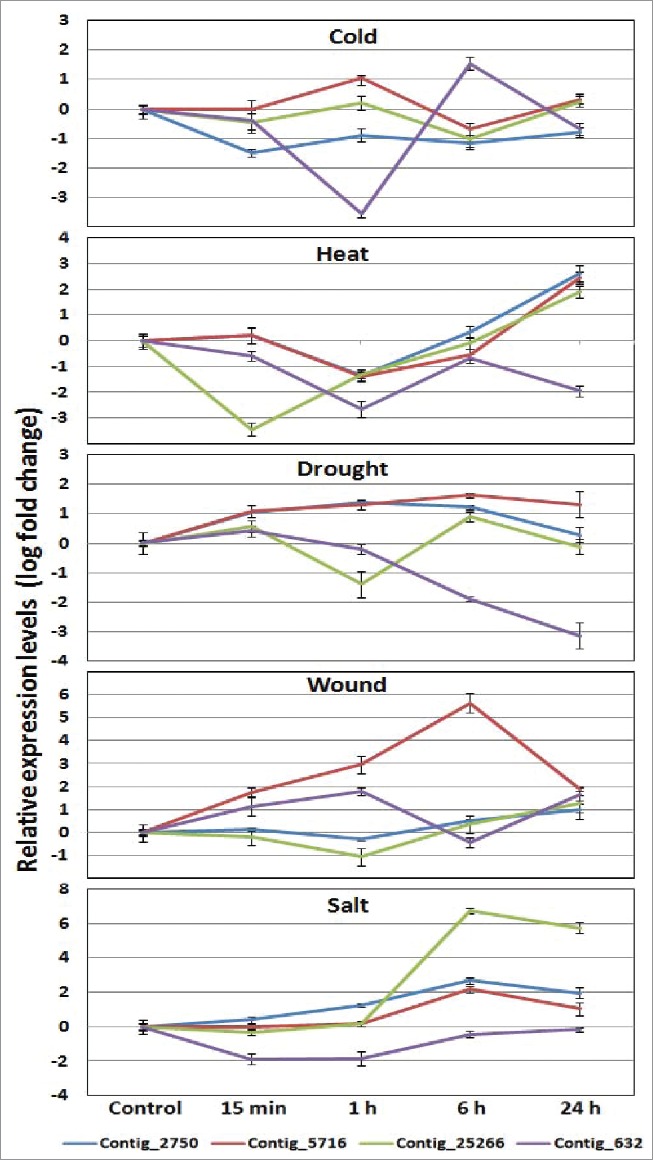
Quantitative real time PCR analysis of the 4 TM-START genes under stresses such as cold, heat, drought, wound, and salt at varying time intervals.
cis-element analysis
The upstream region (2 kb) from the predicted transcription start sites (TSS) of all chickpea START genes were taken for cis-element identification (Fig. 10). Skn-1 for endosperm specific expression, HSE (heat stress element) for binding of heat stress transcription factors, circadian for circadian control and Box 4 for light responsiveness cis-elements were identified in all four chickpea TM-START promoters. cis-elements like Gbox for light responsiveness, ARE for anti oxidant responsiveness element, Box-I for wound responsiveness, GCN-4 for endosperm expression, GATA-motif for light responsiveness, and also elements like Sp1, TCA-element, TCT motif was identified in three of the four TM-START proteins. For example, contig 2750 and contig 5716 have both w-box and Box-w1 cis-elements, which are predicted for wound responsiveness (w-box; cis-acting regulatory element involved in direct fungal elicitor stimulated transcription of defense genes and activation of genes involved in response to wounding, Box-w1; fungal elicitor responsive element).
Figure 10.
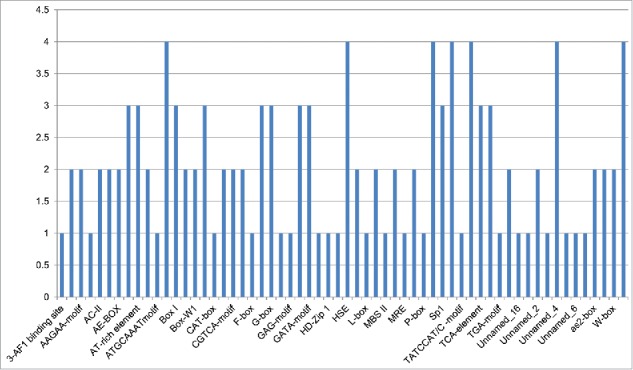
Chart showing the number of cis-elements identified in TM-START proteins of chickpea. The analysis was carried out using PlantCARE program. The name of the selected motifs are provided in the supplementary table.
Discussion
TM-START genes- members of highly diverged gene family in plants
The productivity of chickpea crop is adversely affected by several biotic and abiotic stresses. A variety of plant genes encoding for both structural, functional and transcription factors are known to play important roles in successfully managing the hostile effects posed by a wide range of stresses. Availability of the chickpea draft genome sequence prompted us to undertake identification of genes for START domain containing proteins and their expression in response to abiotic stress conditions. This is because, START domain containing proteins have been shown to function in the plant development regulation2 only. Most of the genes regulating plant development are also shown to contribute to stress management in plants We focused our study on the structural proteins i.e., Trans-Membrane START proteins (TM-START) which are only four, among the total 36 START domain containing proteins, in the chickpea nucleotide database. These were identified after mining, through using START domain as marker/query sequence and phylogenetic reconstruction with Arabidopsis, rice and soybean sequences. An interesting feature we observed through sequence comparision is that till now no DUF-1336 and PH domain containing START proteins have been identified among the 36 chickpea START domain containing proteins. However, completion of the chickpea genome might uncover a few more START domain containing proteins. Sequence alignment showed conservation of the amino acids required for the binding of glycerol-3-phosphoryl choline head group of phosphotidyl-choline3 along with conservation of hydrophobic groups at both amino- and C-terminal region presumed to be transmembrane domains.
Phylogenetic analysis showed that START domain containing proteins diverged and evolved into special classes like HD-START, HD-Zip START, START minimal protein and TM-START in the past from other organisms (non-plants) and these are specific to plants2. This, we concluded based on a previous study by Schrick et al.2 which suggests a separate clade for Arabidopsis START proteins when compared with mammalian, animalia and bacterial START proteins. In a similar finding, soybean HD-START proteins were also clustered separately in comparison to other HD proteins of soybean.36 Formation of two separate clades between monocots and dicots for TM-START proteins strongly suggests frequent/latest evolving gene family might be for functional specialization acquisition or to make plants more hardy in response to the changing environmental conditions. It was also noticed in the phylogenetic analysis that the HD-ZIP START proteins were separately clustered and consists of genes from monocots and dicots supporting the presence of TM-START in both crops species.
Expression studies show chickpea TM-START contigs to be involved in developmental and stress regulation
Higher expression of START genes in floral tissues, which is consistent with our qRT-PCR experiment, suggests a role for TM-START domain containing proteins in floral organ development. The START domain containing HD-ZIP genes in chickpea showed higher expression in young leaves, flower and pod wall tissues. The predominant expression of the TM-START genes in flower tissues of chickpea needs to be investigated in relation to flower development. However, availability of Arabidopsis flower sterility mutant and its association with serine decarboxylase suggests a strong but putative role of START domain containing proteins in floral organ development. The enzyme serine decarboxylase catalyzes formation of ethanolamine, a precursor for choline, phosphatidylethanolamine and phosphatidylcholine.37
Thus, the predominant expression of the TM-START genes in flower tissues of chickpea has to be studied in relation to flower development. But, TM-START proteins are not the only group of proteins that may be involved in the transfer of phosphatidylcholine in plants. There are other phosphotidyl choline transfer proteins which are predominantly expressed in flowers.38 Therefore, the possible function of TM-START domain proteins in flower development needs further exploration in plants. The role of phosphatidylcholine in various aspects of plant development is yet to be elucidated. Interestingly, it was recently shown that phosphatidylcholine binds florigen to induce flowering.39 To authenticate TM-START proteins role in flower induction, these genes should be expressed in the mersitem. Available expression profile indicated high levels of expression in shoot apical meristem (Fig. 9) for two chickpea TM-START (contig 2750, 5716) genes supporting their predicted role in floral organ development. Besides this, one of the Arabidopsis TM-START genes (AT5G54170) genes showed very early response to wound stress35. This gene (AT5G54170) is close to the orthologs contig_5716 and contig_2750 we identified in chickpea in phylogenetic tree. The cis-elements search in the upstream promoter sequences of these contigs indicated presence of stress-responsive elements (wound and fungal elicitor responsive, mention other elements also). Moreover our experiment (q-PCR) also supports the induction of these chickpea TM-START genes in response to wound, cold, heat, drought, and salt stresses. Furthermore, detailed time course study with variety of stresses showed strong up-regulation of contig 5716 in response to wound, drought, heat and salt stresses thereby indicating its putative role in general stress response pathway. Simultaneously, in response to heat and cold stress reversal in expression suggests it to be a putative candidate for stress-responsive pathway. TM-START proteins are known to manage transfer and binding of phosphotidyl choline molecules through membranes and directly could be correlated to stress responsiveness. One of the studies in chickpea indicated substrate phosphorylation of stress induced calcium dependent protein kinase (CaCDPK) after addition of phosphatidyl choline.40 Hence, from the study we assume that TM-START domain containing proteins work upstream of the CaCDPK pathway for stress response.
Future prospects and conclusion
Among stress related gene families, START proteins that play an important link between lipid molecules and plant development, deserve special attention. Further, methods to monitor the real time variations in the levels of phosphatidylcholine in situ are needed. The homologs of PCTP in plants, TM-START which probably bind to phosphatidylcholine have to be studied further to understand the role of lipids in signaling and plant development. In this context, we identified 36 chickpea START proteins and studied the four TM-START domain proteins through in silico and expression analysis. Our analysis revealed the predominant expression of TM-START genes in flower tissue and early induction of three TM-START genes in response to wound, salt and drought stress and delayed induction in response to heat stress. Further work, however, will provide insights into the role of these genes in plants subjected to stress and would provide impetus for further characterization of START superfamily of genes in chickpea and other legumes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
Funding
VS and PTJ acknowledge the senior research fellowship from the Indian Agricultural Research Institute. PC acknowledges the senior research fellowship from the Council of Scientific and Industrial Research, New Delhi, India. Financial support from Indian Council of Agricultural Research under the functional genomics component of National Project on Transgenic Crop (NPTC) is gratefully acknowledged.
References
- 1.Stocco DM. StAR protein and the regulation of steroid hormone biosynthesis. Ann Rev Physiol 2001; 63:193-213 [DOI] [PubMed] [Google Scholar]
- 2.Schrick K, Nguyen D, Karlowski WM, Mayer KFX. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 2004; 5:R41; PMID:15186492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol 2002; 9:507-11; PMID:12055623 [DOI] [PubMed] [Google Scholar]
- 4.Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci U S A 2002; 99:6949-54; PMID:12011453; http://dx.doi.org/ 10.1073/pnas.052140699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyster KM. The membrane and lipids as integral participants in signal transduction: lipid signal transduction for the non-lipid biochemist. Adv Physiol Educ 2007; 31:5-16; PMID:17327576; http://dx.doi.org/ 10.1152/advan.00088.2006 [DOI] [PubMed] [Google Scholar]
- 6.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature 2003; 426:803-9; PMID:14685229; http://dx.doi.org/ 10.1038/nature02188 [DOI] [PubMed] [Google Scholar]
- 7.Tabunoki H, Sugiyama H, Tanaka Y, Fujii H, Banno Y, Jouni ZE, Kobayashi M, Sato R, Maekawa H, Tsuchida K. Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae. J Biol Chem 2002; 277:32133-40; PMID:12052833; http://dx.doi.org/ 10.1074/jbc.M204507200 [DOI] [PubMed] [Google Scholar]
- 8.Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci 1999; 24:130-2; PMID:10322415; http://dx.doi.org/ 10.1016/S0968-0004(99)01362-6 [DOI] [PubMed] [Google Scholar]
- 9.Roth GE, Gierl MS, Vollborn L, Meise M, Lintermann R, Korge G. The Drosophila gene Start1: a putative cholesterol transporter and key regulator of ecdysteroid synthesis. Proc Natl Acad Sci 2004; 101:1601-6; PMID:14745013; http://dx.doi.org/ 10.1073/pnas.0308212100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venkata BP, Schrick K. START domains in lipid / sterol transfer and signaling in plants. 2006; In: Proceedings of the 17th International Symposium on Plant Lipids, Michigan State University Press. [Google Scholar]
- 11.Ito M, Sentoku N, Nishimura A, Hong S-K, Sato Y, Matsuoka M. Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J 2002; 29:497-507; PMID:11846882; http://dx.doi.org/ 10.1046/j.1365-313x.2002.01234.x [DOI] [PubMed] [Google Scholar]
- 12.Kubo H. ANTHOCYANINLESS2, a Homeobox gene affecting anthocyanin distribution and root development in arabidopsis. Plant Cell 1999; 11(7):1217-26; PMID:10402424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 1994; 8:1388-99; PMID:7926739; http://dx.doi.org/ 10.1101/gad.8.12.1388 [DOI] [PubMed] [Google Scholar]
- 14.McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 2001; 411:709-13; PMID:11395776; http://dx.doi.org/ 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- 15.Tang D, Ade J, Frye CA, Innes RW. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J 2005; 44:245-57; PMID:16212604; http://dx.doi.org/ 10.1111/j.1365-313X.2005.02523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurwitz N, Pellegrini-Calace M, Jones DT. Towards genome-scale structure prediction for transmembrane proteins. Philos Trans R Soc Lond B Biol Sci 2006; PMID:16524835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Esch JJ, Shiu S-H, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB. Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 2006; 18:3429-42; PMID:17189345; http://dx.doi.org/ 10.1105/tpc.106.044537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hothorn M, Belkhadir Y, Dreux M, Dabi T, Noel JP, Wilson IA, Chory J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011; 474:467-71; PMID:21666665; http://dx.doi.org/ 10.1038/nature10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al.. The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 2013; 25:3472-90; PMID:24045019; http://dx.doi.org/ 10.1105/tpc.113.117168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar'an B, et al.. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 2013; 31:240-6; PMID:23354103; http://dx.doi.org/ 10.1038/nbt.2491 [DOI] [PubMed] [Google Scholar]
- 21.Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, et al.. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 2013; 74:715-29; PMID:23489434 [DOI] [PubMed] [Google Scholar]
- 22.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res 2004; 32:W309-12; PMID:15215400; http://dx.doi.org/ 10.1093/nar/gkh379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 2005; 33:W451-4; PMID:15980510; http://dx.doi.org/ 10.1093/nar/gki487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol 1997; 268:78-94; PMID:9149143; http://dx.doi.org/ 10.1006/jmbi.1997.0951 [DOI] [PubMed] [Google Scholar]
- 25.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al.. The Pfam protein families database. Nucleic Acids Res 2012; 40:D290-301; PMID:22127870; http://dx.doi.org/ 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci 1998; 95:5857-64; PMID:9600884; http://dx.doi.org/ 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 2001; 305:567-80; PMID:11152613; http://dx.doi.org/ 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 28.Kyte J, Doolittle R. A simple method for displaying the hydropathic character of a protein. J Mol Biol 1982; 157:105-32; PMID:7108955; http://dx.doi.org/ 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 29.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000; 16:404-5; PMID:10869041; http://dx.doi.org/ 10.1093/bioinformatics/16.4.404 [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725-29; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202-8; PMID:19458158; http://dx.doi.org/ 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002; 30:325-7; PMID:11752327; http://dx.doi.org/ 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garg R, Patel RK, Jhanwar S, Priya P, Bhattacharjee A, Yadav G, Bhatia S, Chattopadhyay D, Tyagi AK, Jain M. Gene discovery and tissue-specific transcriptome analysis in chickpea with massively parallel pyrosequencing and web resource development. Plant Physiol 2011; 156:1661-78; PMID:21653784; http://dx.doi.org/ 10.1104/pp.111.178616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garg R, Sahoo A, Tyagi AK, Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem Biophys Res Commun 2010; 396:283-8; PMID:20399753 [DOI] [PubMed] [Google Scholar]
- 35.Satheesh V, Chidambaranathan P, Jagannadham PT, Kohli D, Jain PK, Bhat SR, Srinivasan R. A polyketide cyclase/dehydrase and lipid transport superfamily gene of Arabidopsis and its orthologue of chickpea exhibit rapid response to wounding. Ind J Genet Pl Breed 2014; 74:463-70; http://dx.doi.org/ 10.5958/0975-6906.2014.00871.2. [DOI] [Google Scholar]
- 36.Chen X, Chen Z, Zhao H, Zhao Y, Cheng B, Xiang Y. Genome-Wide analysis of soybean hd-zip gene family and expression profiling under salinity and drought treatments. PLoS One 2014; 9(2): e87156; PMID:24498296; http://dx.doi.org/ 10.3390/ijms1303317610.1371/journal.pone.0087156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon Y, Yu S-I, Lee H, Yim JH, Zhu J-K, Lee B-H. Arabidopsis serine decarboxylase mutants implicate the roles of ethanolamine in plant growth and development. Int J Mol Sci 2012; 13:3176-88; PMID:22489147; http://dx.doi.org/ 10.3390/ijms13033176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mo P, Zhu Y, Liu X, Zhang A, Yan C, Wang D. Identification of two phosphatidylinositol/phosphatidylcholine transfer protein genes that are predominately transcribed in the flowers of Arabidopsis thaliana. J Plant Physiol 2007; 164: 478-86; PMID:16697077; http://dx.doi.org/ 10.1016/j.jplph.2006.03.014 [DOI] [PubMed] [Google Scholar]
- 39.Nakamura Y, Andres F, Kanehara K, Liu YC, Dormann P, Coupland G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat Commun 2014; 5:3553; PMID:24698997; http://dx.doi.org/ 10.1038/ncomms4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixit AK, Jayabaskaran C. Phospholipid mediated activation of calcium dependent protein kinase 1 (CaCDPK1) from chickpea: a new paradigm of regulation. PLoS One 2012; 7:e51591; PMID:23284721; http://dx.doi.org/ 10.1371/journal.pone.0051591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.