ABSTRACT
Citrus Huanglongbing (HLB), also known as citrus greening, has been threatening the citrus industry since the early 1900's and up to this date there are no effective cures for this disease. Field observations and greenhouse controlled studies demonstrated that some citrus genotypes are more tolerant to Candidatus Liberibacter asiaticus (CLas) pathogen than others. However, the mechanisms underpinning tolerance has not been determined yet. The phloem sap composition of CLas-tolerant and sensitive citrus varieties was studied to identify metabolites that could be responsible for their tolerance to CLas. The citrus phloem sap was collected by centrifugation and was analyzed with gas chromatography-mass spectrometry after methyl chloroformate derivatization. Thirty-three metabolites were detected in the phloem sap of the studied varieties: twenty 20 amino acids, eight 8 organic acids, and five 5 fatty acids. Interestingly, the levels of most amino acids, especially those implicated in plantdefense to pathogens such as phenylalanine, tyrosine, tryptophan, lysine, and asparagine were higher in tolerant varieties. Although the level of organic acids varied between cultivars, this variation was not correlated with citrus resistance to CLas and could be cultivar specific. The fatty acids were found in trace amounts and in most cases their levels were not significantly different among varieties. Better understanding of the mechanisms underpinning citrus tolerance to CLas will help in developing economically tolerant varieties.
KEYWORDS: Amino acids, Candidatus Liberibacter asiaticus, citrus, fatty acids, gas chromatography, Huanglongbing, methyl chloroformate, organic acids
Introduction
Citrus is one of the world’s major fruit crops with an annual global production of more than 100 million metric tons.1 Citrus fruits are a good source for carbohydrates and are rich in biological compounds that are essential for human health such as vitamins, dietary fiber, minerals, carotenoids, and flavonoids.1 Unfortunately, nowadays many diseases threaten the survival of the citrus industry. Many of these diseases such as citrus stubborn and huanglongbing (HLB) are caused by phloem sap-restricted bacteria and are spread by phloem sap-piercing insects.2 Currently, citrus greening disease is considered as the most destructive disease of citrus and it is threatening the citrus industry worldwide.
Candidatus Liberibacter which causes citrus greening is a phloem-limited, gram-negative bacterium.3 Three species of Candidatus Liberibacter are associated with citrus greening: Candidatus Liberibacter asiaticus (CLas) (Asia, North America, and Brazil), Candidatus Liberibacter africanus (CLaf) (Africa), and Candidatus Liberibacter americanus (CLam) (Brazil).4 Although the citrus greening pathogen can be spread by grafting, psyllid vectors are the major transmission means in the field.5 The Asian citrus psyllid (ACP), Diaphorina citri Kuwayama transmits CLas and CLam, while the African citrus psyllid Trioza erytrea (Del Guercio) transmits CLaf.5
ACP transmits the CLas while feeding on citrus phloem sap. Citrus and close citrus relatives are the main host plants for the ACP.5 Although ACP can survive and multiply on many host plants, ACP prefers some hosts more than others. Field observations indicated that sweet orange and Murraya paniculata (L.) Jack (orange jasmine) were the most preferred host for ACP, while Poncirus trifoliata was an occasional host.5 Although there are many observations about preferred host of ACP, comparative laboratory studies are limited. For instance, Tsai & Liu (2000) tested the preference of ACP to four 4 hosts: Citrus jambhiri Lushington (rough lemon), Murraya paniculata, Citrus × paradisi Macfad. (grapefruit), and Citrus aurantium L. (sour orange).6 Grapefruit was the most preferable host, followed by the other hosts.6 In a recent study, Richardson and Hall (2013) also showed that accessions of Poncirus trifoliata and x Citroncirus spp. were more tolerant to ACP than Citrus macrophylla Wester (Alemow); more eggs were laid on C. macrophylla than P. trifoliata accessions.7 Richardson and Hall (2013) concluded that P. trifoliata may have antixenosis and antibiosis resistance to ACP.7
Field observations also indicated that some ACP hosts are more tolerant to CLas bacteria than others.8-10 To test these observations, Folimonova et al. (2009) examined the responses of many citrus varieties to Florida CLas isolates, using graft inoculation, under controlled conditions.11 Based on intensity of symptoms, plant growth, and death, citrus genotypes were divided into three 3 major groups (sensitive, moderately tolerant, and tolerant).11 The sensitive genotypes showed severe leaf chlorosis, decreased growth, and death. The moderately tolerant plants showed mild symptoms, while the tolerant varieties showed few symptoms.11 Similar results were also reported by Sagaram and Burns (2009).12 In another study, Hajivand et al., (2009) also assessed eighteen 18 Malaysian citrus species for susceptibility to CLas by graft inoculation.13 Based on the severity of symptoms, the species were categorized in four 4g groups: sensitive (72–58% severity), moderate (50–41% severity), mild (25–17% severity), and tolerant group (no symptoms).
To defend themselves against insects and pathogens attacks, plants produce a wide spectrum of defense compounds such as terpenoids, alkaloids, nonprotein amino acids, benzoxazinoids, cyanogenic glucosides, and phenolic compounds.14-17 Although the primary function of phloem is to transport nutrients from source to sink, it is also believed that the phloem is implicated in plant defense.18 The phloem contains many cells that are implicated in the synthesis, distribution, and release of large number of defense compounds such as terpenes, alkaloids, ketones, and glucosinolates.18
Although many studies have been conducted on HLB disease, the mechanisms underpinning citrus tolerance to CLas has not been determined yet. Exploring this mystery is essential for development of economically important CLas-tolerant varieties. Some citrus varieties such as P. trifoliata which are tolerant to CLas are also tolerant to citrus tristeza virus (CTV).19 Although the mechanism of P. trifoliata resistance to CTV has not been revealed yet, it seems that it is due to the failure of the CTV to spread within its host despite its ability to replicate.20 Because several antibacterial compounds have been identified in the fruit and seed of P. trifoliata, it was also suggested that the phloem sap of CLas-tolerant varieties contain some antibacterial compounds.19,21,22
Herein, we hypothesize that the phloem sap composition of CLas-tolerant varieties is different from that of sensitive varieties. To test this hypothesis, we studied the phloem sap composition of thirteen different varieties with different degrees of tolerance to CLas.
Material and methods
Plants
Citrus plants (12 months old, about 1 m height) were kept in temperature-controlled greenhouse (28–32°C) and were watered twice a week and fertilized once a week using 20-10-20 fertilizer (Allentown, PA). The thirteen different citrus varieties that were used in this study are shown in Table 1.
Table 1.
Citrus plants used in this study.
| Citrus variety | Degree of tolerance to CLas* |
|---|---|
| Valencia sweet orange (C. sinensis (L.) Osbeck) | Sensitive (1) |
| Madam Vinous sweet orange (C. sinensis (L.) Osbeck) | Sensitive (1) |
| Duncan grapefruit (C. paradisi MacFadyen) | Sensitive (1) |
| Ruby red grapefruit (C. paradisi MacFadyen) | Sensitive (1) |
| Sour orange (C. aurantium L.) | Moderately tolerant (2) |
| Volkamer lemon (C. limonia Osbeck ‘Volkameriana’) | Moderately tolerant (2) |
| C. macrophylla Wester (Alemow) | Moderately tolerant (2) |
| Palestine Sweet lime (C. aurantifolia (Christm.) | Moderately tolerant (2) |
| Mexican lime (C. aurantifolia (Christm.) | Moderately tolerant (2) |
| Carrizo citrange (X Citroncirus webberi) | Tolerant (3) |
| Severinia buxifolia | Tolerant (3) |
| Poncirus trifoliata (L.) Raf. | Inconsistent (4) |
| Citrus latipes | Inconsistent (4) |
Folimonova et al. (2009).11
Phloem sap collection
Citrus phloem sap was collected as described by Hijaz and Killiny 2014.23 Briefly, three flush shoots (2–3 week-old, about 0.5 cm diameter) were collected from each plant from three different locations (top, middle, and bottom). Five different plants were sampled from each variety. The bark was stripped into two pieces and was manually removed from the twig. The inner part of the bark was rinsed with deionized water and dried with Kimwipes® to exclude any contamination from the xylem sap. Then the bark strips were cut into about 1-cm pieces using a sterile razor blade. To collect the phloem sap, three to five pieces of the bark tissue were vertically placed in a 0.5-ml eppendorf tube. A small hole was made at the bottom of the tube using a pin and the tube was inserted into a 2-ml eppendorf tube. The sample was centrifuged at 12,000 rpm for 15 minutes at room temperature and the phloem sap collected from each plant was pooled together and stored at −80°C until analysis.
Methyl chloroformate (MCF) derivatization
The phloem sap was derivatized with methyl chloroformate (MCF) and analyzed with GC-MS as described by Hijaz and Killiny (2014). Briefly, a 20-µl aliquot of the pure phloem was transferred to a 1-ml silanized GC-MS insert and mixed with 200 μl of NaOH (1M). Then the alkaline sample was mixed with 167 μl of methanol and 34 μl of pyridine, followed by the addition of 20 μl of MCF. The sample was vigorously mixed for 30 seconds. An additional 20 μl of MCF was added and the sample was mixed for another 30 seconds. A 100-μl aliquot of chloroform was added with vigorous mixing for 10 seconds, followed by 100 μl of sodium bicarbonate (50 mM) with vigorous mixing for 10 seconds. The upper layer was discarded and about 100 μl of the organic layer was transferred to a new insert. A few mg of sodium sulfate was added to dry the organic layer, and 0.5 μl (splitless injection) was injected into the GC-MS instrument running in the full scan mode (40–600 m/z). Standards of amino acids, organic acids, and fatty acids were derivatized as mentioned above and were used to determine the concentration of each metabolite in citrus phloem sap.
GC-MS analyses
Derivatized samples and standards were analyzed using Clarus 500 GC-MS system (Perkin Elmer, Waltham, MA, USA) fitted with an ZB-5MS column (cross-linked 5% Ph Me siloxane, 30 m × 0.25 mm × 0.025 µm film thickness). The flow rate for the hydrogen carrier gas was 1 ml/min. The GC temperature program was as follows: initial temperature was held at 70°C for 1 min, and then increased to 220°C at a rate of 10°C/min, held for 1 min, increased further to 300°C at 10°C/min, held for 0 min. The injector and the detector temperatures were set at 250°C and 260°C, respectively.
MS peak identification
GC-MS chromatograms were analyzed using TurboMass software version 5.4.2 (Perkin Elmer, Waltham, MA, USA). Peaks were first identified by comparing their mass spectra with library entries (NIST mass spectra library (National Institute of Standards and Technology, Gaithersburg, MA, USA); Wiley 9th edition (John Wiley and Sons, Inc., Hoboken, NJ, USA)). Identification was further confirmed by comparing the retention time and mass spectra of the detected metabolites with those of derivatized standards.
Data processing and statistical analysis
Data was manually aligned using retention time and mass values. Principal component analysis (PCA) was performed on metabolite concentrations to discriminate among varieties using JMP version 9.0 (SAS Institute Inc.,). Post hoc pairwise comparison followed by Tukey honestly significant difference (Tukey’s HSD) test was used to compare the mean concentration of each metabolite in the different varieties. The two-way hierarchical cluster analysis (HCA), based on Ward’s method (with 95% confidence), followed by the heat-map using the concentration of individual metabolites (Table 2) were also performed to construct the similarity dendrogram and visualize differences in the phloem sap profile between the different varieties. Simple linear regression and step wise forward selection (SWFS) were also conducted to elucidate any relation between the detected metabolites and citrus resistance to CLas infection.
Table 2.
Phloem sap metabolites concentrations (mM) in different citrus varieties as analyzed by GC-MS after methylchloroformate derivatization (n ranges between 4 and 5).
| compound | Valencia | Palestine Sweet lime | Poncirus trifoliata | Volkamer lemon | Rubey red grapefruit | Severinia buxifolia | Sour orange | Mexican lime | Carrizo citrange | Duncan grapefruit | Madam Vinous | Alemow | Citrus latipes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fumaric | 0.44±0.03f | 2.75±0.43cdef | 0.74±0.17f | 4.96±1.04abcd | 5.70±1.63abc | 4.48±1.89bcde | 3.97±1.37bcdef | 6.21±1.03abc | 2.841.79cdef | 1.59±0.95ef | 7.88±2.08a | 6.55±2.99ab | 1.42±0.80ef |
| succinic acid | 0.18±0.02bc | 0.31±0.06bc | 0.14±0.01c | 0.34±0.04bc | 0.24±0.05bc | 1.06±0.37a | 0.17±0.02c | 0.31±0.08bc | 0.13±0.03c | 0.11±0.03c | 0.45±0.16b | 0.26±0.08bc | 0.21±0.04bc |
| malate | 10.25±2.05e | 42.22±11.53e | 24.497.01e | 161.77±32.35ab | 147.52±46.80ab | 165.90±42.45ab | 147.08±20.85ab | 126.96±15.98bcd | 75.93±43.35cde | 35.47±26.79e | 205.52±34.21a | 136.75±33.65abc | 47.22±20.08e |
| benzoic acid | 13.06±1.34a | 1.73±0.46defg | 0.56±0.16fg | 4.05±0.68de | 13.72±1.39a | 5.33±0.76cd | 8.56±0.78bc | 0.25±0.04g | 2.72±0.58defg | 9.36±3.45b | 13.90±2.26a | 0.25±0.08g | 1.87±0.50efg |
| glycine | 0.06±0.01g | 0.26±0.05cdefg | 0.56±0.15ab | 0.48±0.07bcd | 0.46±0.17bcd | 0.27±0.05cdefg | 0.25±0.04defg | 0.11±0.03g | 0.51±0.05bc | 0.41±0.13bcde | 0.38±0.08bcdef | 0.20±0.08efg | 0.79±0.19a |
| alanine | 0.23±0.04f | 0.46±0.07ef | 1.31±0.20bcd | 1.46±0.24abc | 1.45±0.44abc | 1.29±0.14bcd | 0.99±0.22cdef | 0.90±0.34cdef | 0.86±0.35cdef | 0.95±0.31cdef | 1.79±0.51ab | 1.04±0.17cde | 2.05±0.32a |
| valine | 0.06±0.01c | 0.32±0.09abc | 0.44±0.14ab | 0.16±0.06bc | 0.12±0.02c | 0.33±0.10abc | 0.10±0.01c | 0.12±0.02bc | 0.09±0.01c | 0.14±0.04bc | 0.20±0.08bc | 0.11±0.05bc | 0.57±0.41a |
| leucine | 0.06±0.01e | 0.24±0.06bcd | 0.54±0.15a | 0.20±0.08cde | 0.14±0.02cde | 0.40±0.09ab | 0.12±0.02cde | 0.16±0.04cde | 0.10±0.03de | 0.16±0.04cde | 0.21±0.09cde | 0.13±0.05cde | 0.26±0.04bc |
| GABA | 1.22±0.29g | 2.62±0.54efg | 6.01±2.96bcde | 6.99±1.70bc | 7.62±1.13bc | 6.75±1.64bc | 6.23±1.05bcd | 2.85±0.47efg | 3.27±0.95defg | 4.84±1.36cdef | 8.57±1.21b | 1.84±0.51fg | 16.49±2.07a |
| isoleucune | 0.11±0.01d | 0.36±0.05bcd | 0.72±0.24a | 0.60±0.13a | 0.59±0.06ab | 0.58±0.04ab | 0.52±0.08abc | 0.29±0.05cd | 0.32±0.08cd | 0.32±0.13cd | 0.73±0.08a | 0.23±0.08d | 0.22±0.08d |
| threonine | 0.10±0.02b | 0.24±0.09b | 0.38±0.19b | 0.30±0.04b | 0.24±0.05b | 0.31±0.08b | 0.18±0.02b | 0.11±0.02b | 0.18±0.02b | 0.22±0.10b | 0.26±0.06b | 0.10±0.04b | 1.46±0.59a |
| malic acid | 9.11±1.34e | 28.84±7.09cde | 15.93±3.52e | 61.71±19.50ab | 75.64±6.89a | 53.55±25.21abcd | 55.30±7.00abc | 58.30±6.10abc | 34.59±17.67bcde | 19.31±13.69e | 78.83±10.36a | 61.76±11.28ab | 27.03±7.57de |
| proline | 2.53±0.30b | 9.98±1.02ab | 12.28±3.61ab | 16.04±8.89ab | 18.73±5.86ab | 18.78±9.21ab | 17.62±4.34ab | 11.57±4.15ab | 16.50±7.27ab | 18.22±8.55ab | 20.48±4.67a | 10.52±3.85ab | 20.86±9.82a |
| asparagine | 2.69±0.17b | 8.15±1.71b | 48.64±16.36ab | 7.15±6.29b | 8.09±1.85b | 5.12±1.26b | 2.27±0.77b | 0.15±0.03b | 6.39±4.75b | 2.18±1.21b | 1.86±0.37b | 3.03±1.72b | 80.78±69.02a |
| aspartic acid | 0.67±0.06c | 1.45±0.20bc | 3.12±1.37bc | 3.80±1.03bc | 3.13±0.47bc | 2.79±1.09bc | 1.94±0.17bc | 1.30±0.06bc | 1.34±0.42bc | 1.23±0.42c | 2.50±0.51bc | 0.95±0.24c | 7.28±3.01a |
| quinic acid | 0.79±0.22bc | 11.131.78bc | 15.92±9.53bc | 19.10±5.86bc | 11.59±3.14bc | 121.30±40.65a | 5.04±1.37bc | 10.92±2.34bc | 6.30±3.08bc | 2.35±2.68c | 8.32±2.22bc | 17.89±7.16bc | 39.07±38.76b |
| citric acid | 0.64±0.06b | 2.86±0.95b | 16.30±15.40b | 13.53±6.38b | 12.55±2.55b | 59.20±24.29a | 5.74±0.91b | 5.55±0.68b | 6.89±3.87b | 2.20±3.14b | 14.78±16.10b | 7.07±3.97b | 18.88±19.39b |
| serine | 1.06±0.20de | 1.49±0.34cde | 4.68±2.05ab | 2.96±1.05bcd | 3.33±0.73bc | 3.17±1.40bcd | 1.31±0.16de | 0.69±0.04e | 1.82±0.42cde | 2.12±0.66cde | 1.99±0.53cde | 0.52±0.24e | 6.73±1.28a |
| glutamine | 1.80±2.30b | 1.13±0.19b | 3.89±1.98b | 1.91±0.63b | 1.92±0.34b | 1.41±0.66b | 0.80±0.22b | 0.24±0.01b | 1.03±0.48b | 0.91±0.36b | 0.68±0.27b | 0.49±0.26b | 11.28±9.58a |
| salicylic acid | 0.05±0.01ab | 0.04±0.00ab | 0.03±0.03ab | 0.04±0.01ab | 0.12±0.10a | 0.09±0.06ab | 0.06±0.01ab | 0.04±0.00ab | 0.02±0.01b | 0.06±0.02ab | 0.11±0.03a | 0.04±0.01ab | 0.03±0.00b |
| glutamic acid | 0.17±0.01b | 0.02±0.01cd | 0.07±0.09bcd | 0.07±0.03bcd | 0.05±0.02bcd | 0.32±0.15a | 0.02±0.01d | 0.12±0.03bcd | 0.03±0.01cd | 0.02±0.01d | 0.06±0.01bcd | 0.06±0.02bcd | 0.13±0.03bc |
| methionine | 0.01±0.00d | 0.03±0.01d | 0.08±0.01a | 0.06±0.01abc | 0.040.01bcd | 0.06±0.01ab | 0.02±0.01d | 0.03±0.01d | 0.02±0.01d | 0.01±0.00d | 0.03±0.02cd | 0.02±0.02d | 0.03±0.02d |
| cysteine | 0.00±0.00b | 0.01±0.01b | 0.03±0.02ab | 0.03±0.01b | 0.02±0.01b | 0.02±0.01b | 0.02±0.03b | 0.02±0.02b | 0.01±0.01b | 0.01±0.00b | 0.02±0.01b | 0.02±0.01b | 0.05±0.01a |
| phenyla-lanine | 0.05±0.00c | 0.13±0.02bc | 0.25±0.03a | 0.10±0.04bc | 0.08±0.01c | 0.23±0.06a | 0.06±0.01c | 0.09±0.02bc | 0.08±0.01c | 0.08±0.02c | 0.10±0.03bc | 0.11±0.03bc | 0.16±0.06b |
| myristic | ND | ND | Tr | Tr | Tr | Tr | ND | ND | ND | Tr | Tr | Tr | Tr |
| palmitic | 0.03±0.00bcd | 0.03±0.00bcd | 0.03±0.00bcd | 0.04±0.01abc | 0.04±0.00abc | 0.04±0.00abc | 0.03±0.01cd | 0.02±0.01d | 0.03±0.00bcd | 0.03±0.01bc | 0.04±0.00ab | 0.04±0.01ab | 0.05±0.00a |
| lysine | 0.10±0.01b | 0.23±0.05b | 0.33±0.07b | 0.23±0.09b | 0.34±0.15b | 0.51±0.12b | 0.12±0.02b | 0.13±0.01b | 0.09±0.03b | 0.24±0.13b | 0.29±0.15b | 0.09±0.05b | 1.27±0.51a |
| histidine | 0.04±0.02bc | 0.08±0.06bc | 0.15±0.08bc | 0.04±0.03c | 0.06±0.04bc | 0.24±0.07b | 0.01±0.01c | 0.01±0.01c | 0.01±0.01c | 0.06±0.06c | 0.05±0.04bc | 0.02±0.02c | 0.46±0.23a |
| linoelic acid | Tr | ND | ND | Tr | Tr | Tr | ND | ND | ND | Tr | Tr | Tr | Tr |
| oleic acid | Tr | Tr | Tr | Tr | Tr | Tr | Tr | ND | Tr | Tr | Tr | Tr | Tr |
| stearic | 0.02±0.00bcde | 0.02±0.00bcde | 0.03±0.00abcd | 0.03±0.00abcd | 0.03±0.00abc | 0.03±0.00abcd | 0.02±0.01de | 0.02±0.00e | 0.02±0.00cde | 0.03±0.01bcd | 0.03±0.00ab | 0.03±0.00abcd | 0.04±0.00a |
| tyrosine | 0.1±0.00bcd | 0.15±0.02a | 0.14±0.01a | 0.08±0.03bcd | 0.07±0.01cd | 0.13±0.02ab | 0.04±0.01d | 0.06±0.01d | 0.05±0.00d | 0.05±0.02d | 0.07±0.02cd | 0.03±0.01d | 0.11±0.05abc |
| tryptophan | 0.0±0.00bc | 0.04±0.02bc | 0.03±0.00bc | 0.02±0.01bc | 0.01±0.00bc | 0.12±0.06a | 0.01±0.00c | 0.04±0.01bc | 0.01±0.00bc | 0.02±0.01bc | 0.02±0.01bc | 0.02±0.01bc | 0.06±0.05b |
| amino acids | 13.6±11.08 | 26.06±27.39 | 21.12±83.66 | 55.03±42.67 | 46.42±46.47 | 44.12±42.84 | 40.59±32.61 | 20.88±18.99 | 36.24±32.70 | 51.71±32.18 | 41.30±40.27 | 11.94±19.52 | 96.87±151.04 |
| organic acids | 24.23±24.26 | 48.57±47.68 | 21.84±49.61 | 129.30±103.73 | 110.47±119.57 | 203.51±245.01 | 80.43±78.84 | 89.78±81.59 | 74.22±53.49 | 36.03±34.98 | 106.69±124.28 | 85.48±93.80 | 124.68±88.51 |
| fatty acids | 0.06±0.07 | 0.07±0.07 | 0.08±0.08 | 0.10±0.09 | 0.08±0.09 | 0.08±0.09 | 0.07±0.07 | 0.06±0.05 | 0.07±0.07 | 0.10±0.08 | 0.09±0.09 | 0.07±0.09 | 0.11±0.11 |
| total | 37.89±35.41 | 74.71±75.14 | 43.03±133.34 | 184.44±146.48 | 156.98±166.13 | 247.71±287.93 | 121.1±111.51 | 110.72±100.63 | 110.53±86.25 | 87.84±67.24 | 148.08±164.65 | 97.49±113.42 | 221.67±239.66 |
Means in the same line with the same letters are not significantly different from each other (Tukey test, p > 0.05). nd: not detected and tr: < 0.01 mM.
The identity of all compounds was confirmed using authentic standards except malate was tentatively identified using the mass spectra library.
Results
We studied the phloem sap because it is the main source of nutrients for Candidatus Liberibacter asiaticus (CLas) and the Asian citrus psyllids (ACP). In addition, we focused on the amino acid composition of the phloem sap because amino acids are precursors for most of the secondary metabolites implicated in plant defense. Methyl chloroformate (MCF) derivatization was chosen because it reacts with amino and organic acids, but does not react with sugars which are abundant in the phloem sap. Thirty-three compounds were detected in the phloem sap after MCF derivatization (Table 2). These metabolites can be classified into three groups: amino acids (20), organic acids (8), and fatty acids (5).
All of the amino acids detected in the citrus phloem sap were proteinogenic amino acids except γ-aminobutyric acid (GABA). The concentration of GABA in citrus phloem sap ranged from 1 to 16 mM (Table 2). Proline was the most predominant amino acid in all varieties except for C. latipes and P. trifoliata. The major amino acid in these two varieties was asparagine. The concentration of proline ranged from 3.0 to 20.9 mM and it showed little variation among the studied varieties. The total concentration of amino acids in the citrus phloem sap ranged from 13 to 97 mM. C. latipes was the highest in total amino acids, whereas Valencia was the lowest (Table 2). Seven organic acids (fumaric, succinic, malic, quinic, benzoic, citric, and salicylic acid) were identified in citrus phloem sap. Malic acid was the main organic acid (Table 2). Citric and quinic acid were also abundant and their concentration ranged from (1 to 18 mM) and (1 to 39 mM), respectively. Low amounts (< 0.1 mM) of myristic, palmitic, stearic, oleic, and linoleic were also detected in the citrus phloem sap.
PCA and cluster analysis indicated that the phloem sap of CLas-tolerant varieties was different from CLas-sensitive varieties
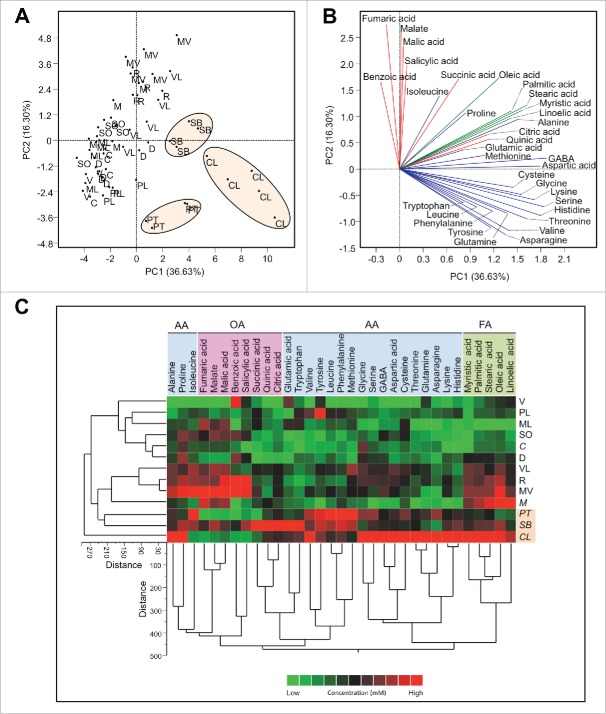
The principal component analysis (PCA) of the different citrus varieties generated using the concentration of all metabolites in Table 2 is shown in Fig. 1A. Principal component 1 and 2 explained about 52.9 % of the variation. Most of the tolerant cultivars, especially C. latipes, S. buxifolia, and P. trifoliata clustered to the right of the PCA plot. In addition, V. lemon and Palestine lime also clustered close to the previous varieties. This clustering indicated that the phloem sap composition of these varieties was different from the rest of the other varieties (sensitive). The loading plot (Fig. 1B) showed that these varieties were higher in most of the metabolites, especially amino acids (bottom of high right quadrant and top of the lower right quadrant). On the other hand, the rest of the varieties (sensitive) clustered to the left of the plot and they were low in most of the metabolites, especially amino acids. In agreement with the PCA analysis, the cluster analysis (CA) showed that C. latipes, and S. buxifolia were similar and clustered closely to each other (Fig. 1C). The differences in the metabolites of the phloem sap among the different citrus varieties are also visualized in the heat map (Fig. 1C).
Figure 1.
Principal components analysis-scatter plot (A), its associated loading plot (B) and two-way hierarchical cluster analysis and heat-map (C) showing the distribution of different citrus varieties using the concentration of detected compounds in phloem saps (n ranges from 4 to 5). Row represents variety and column compound concentrations. Cells are colored based on concentration in mM. AA: amino acids, OA: organic acids, FA: fatty acids. Varieties abbreviations: Valencia sweet orange (V), Madam Vinous sweet orange (MV), Duncan grapefruit (D), Ruby red grapefruit (R), Sour orange (SO), Volkamer lemon (VL), Alemow (M), Palestine sweet lime (PL), Mexican lime (ML), Carrizo citrange (C), Severinia buxifolia (SB), Poncirus trifoliata (PT), and Citrus latipes (CL).
The PCA generated using the concentration of amino acids also separated C. latipes, S. buxifolia, and P. trifoliata from the other varieties (Fig. 2A). As shown in the loading plot (Fig. 2B) these varieties were higher in most of the amino acids (top and lower right quadrants).
Figure 2.

Principal components analysis (PCA)-scatter plot and its associated loading plot showing the distribution of different citrus varieties using the concentrations of detected compounds (n ranges from 4 to 5). (A) PCA-scatter plot using the concentrations of the amino acid group and its PCA-loading plot (B), (C) PCA-scatter plot using the concentration of organic acid group and its PCA-loading plot (D), (E) PCA-scatter plot using the concentration of fatty acid group and its PCA-loading plot (F).Varieties abbreviations: Valencia sweet orange (V), Madam Vinous sweet orange (MV), Duncan grapefruit (D), Ruby red grapefruit (R), Sour orange (SO), Volkamer lemon (VL), Alemow (M), Palestine sweet lime (PL), Mexican lime (ML), Carrizo citrange (C), Severinia buxifolia (SB), Poncirus trifoliata (PT), and Citrus latipes (CL).
The PCA generated using the organic acids (Fig. 2C) showed that organic acid profiles of S. buxifolia and Madam vinous were different from other varieties. As shown in the loading plot (Fig. 2D), S. buxifolia, was high in quinic, citric, and succinic acid, whereas Madam vinous was high in malate. Although fatty acids are an important in defense signaling in plants, the PCA and CA generated using the concentration of fatty acids did not show any clear clustering of any of the selected varieties (Figs. 2E-F).
Tukey test confirmed PCA and CA results and revealed the differences among varieties
In agreement with the PCA result the Tukey’s test showed that tolerant cultivars were high in most of the phloem sap components, especially the amino acids. For example, C. latipes was the highest in alanine, valine, lysine, serine, glutamic acid, aspartic acid, threonine, asparagine, glutamine, GABA, palmitic, stearic, and cysteine (Table 2). In addition, it was the second highest variety in phenylalanine and the third in tyrosine (Table 2). P. trifoliata was the highest in leucine, isoleucine, phenylalanine, methionine, and cysteine and was the second highest variety in asparagine, glutamine, valine, glycine, and tyrosine (Table 2). S. buxifolia was the highest in tryptophan, leucine, succinic, citric acid, and quinic acid (Table 2). In addition, S. buxifolia was the second highest cultivar in phenylalanine, and the third highest variety in methionine, tyrosine, and valine (Table 2). Volkamer lemon was high in methionine and aspartic acid and the fourth in quinic acid (Table 2). Palestine lime was the highest in tyrosine and the third in valine and tryptophan (Table 2). Mexican lime was third in cysteine and fourth in glutamic acid (Table 2). C. macrophylla was the third in quinic acid and the fifth in phenylalanine (Table 2). C. citrange was the second highest variety in quinic acid and the third in glycine (Table 2).
Interestingly, the levels of phenylalanine, tyrosine, and tryptophan as well as their total level were correlated with citrus tolerance to CLas (Figs. 3A-C). As shown in Fig. 3A, P. trifoliata, S. buxifolia, C. latipes, Palestine lime, C. macrophylla, and Volkmer lemon were the highest in phenylalanine. Palestine lime, P. trifoliata, S. buxifolia, C. latipes, and Volkamer lemon were the highest in tyrosine (Fig. 3B). S. Buxifolia, C. latipes, Mexican lime, Palestine lime, and P. trifoliata were the highest in tryptophan (Fig. 3C). In the same manner, S. Buxifolia, P. trifoliata, C. latipes, Palestine lime, and Volkamer were the highest in the total of these three amino acids (Fig. 3D). On the other hand, most of the sensitive cultivars were low in these amino acids (Fig. 3A-C).
Figure 3.

Average concentration of phenylalanine (A), tyrosine (B), tryptophan (C), and total of phenylalanine, tyrosine, and tryptophan together in different citrus varieties (D). (n ranges from 4 to 5)., Varieties abbreviations: Valencia sweet orange (V), Madam Vinous sweet orange (MV), Duncan grapefruit (D), Ruby red grapefruit (R), Sour orange (SO), Volkamer lemon (VL), Alemow (M), Palestine sweet lime (PL), Mexican lime (ML), Carrizo citrange (C), Severinia buxifolia (SB), Poncirus trifoliata (PT), and Citrus latipes (CL).
Simple linear regression (SLR) and Stepwise forward regression (SWFR) suggest that citrus tolerance to CLas is dependent on phloem sap composition
The relationship between the phloem sap metabolites and citrus tolerance to CLas was investigated using simple and multiple linear regression (Table 3). The simple linear regression showed that most of the phloem sap amino acids were implicated in citrus resistance to CLas (Table 3). As shown in Table 3, phenylalanine was highly correlated with citrus tolerance to CLas (R2: 0.4, P-value < 0.0001). Other amino acids such as valine, leucine, tyrosine, tryptophan, histidine, lysine, cysteine, methionine, serine, and glutamine were also positively correlated with citrus tolerance to CLas (Table 3). On the other hand, organic acids were negatively correlated with citrus tolerance to CLas (Table 3), however this correlation was not strong except for benzoic acid (R2: 0.4, P-value < 0.0001). Fatty acids also did not show a strong correlation with citrus tolerance to CLas; R2 was either too small or p-value was larger than 0.05 (Table 3).
Table 3.
Simple linear regression (SLR) and stepwise forward regression (SWFR) results.
| Simple linear regression (SLR) between the concentration of phloem sap metabolites in different citrus varieties and their degrees of tolerance to CLas. | |||||
|---|---|---|---|---|---|
| compound | R2 | P-value | Regression formula | ||
| fumaric | 0.07 | 0.0330 | rank = 2.4672713 − 0.1033573*fumaric | ||
| succinic acid | 0.01 | 0.5423 | rank = 2.0159913 + 0.2544109*succinic acid | ||
| malate | 0.05 | 0.0858 | rank = 2.4228736 − 0.0033815*malate | ||
| benzoic acid | 0.40 | <0.0001 | rank = 2.846556 − 0.1247415*benzoic acid | ||
| glycine | 0.24 | <0.0001 | rank = 1.2137884 + 2.464121* L-glycine | ||
| alanine | 0.08 | 0.02008 | rank = 1.4595241 + 0.5635009* L-alanine | ||
| valine | 0.33 | <0.0001 | rank = 1.4365366 + 3.1788655* L-valine | ||
| leucine | 0.35 | <0.0001 | rank = 1.1479306 + 4.6403882* L-leucine | ||
| GABA | 0.14 | 0.0018 | rank = 1.5367921 + 0.0968826*GABA | ||
| isoleucine | 0.01 | 0.8939 | rank = 2.1275553 − 0.0894776* L-isoleucine | ||
| threonine | 0.26 | 0.0001 | rank = 1.6537338 + 1.4218318* L-threonine | ||
| malic acid | 0.06 | 0.0372 | rank = 2.5545697 − 0.0107363*malic acid | ||
| proline | 0.01 | 0.4064 | rank = 1.8546825 + 0.0159869* L-proline | ||
| asparagine | 0.30 | <0.0001 | rank = 1.8293058 + 0.0201661* L-asparagine | ||
| aspartic acid | 0.21 | <0.0001 | rank = 1.4964464 + 0.2476606* L-aspartic acid | ||
| quinic acid | 0.13 | <0.0024 | rank = 1.8535996 + 0.0121467*quinic acid | ||
| citric acid | 0.09 | 0.0153 | rank = 1.8560199 + 0.0190329*citric acid | ||
| serine | 0.29 | <0.0001 | rank = 1.3563804 + 0.3112015* L-serine | ||
| glutamine | 0.21 | <0.0001 | rank = 1.8325813 + 0.1282487* L-glutamine | ||
| salicylic acid | 0.10 | 0.0106 | rank = 2.5278381 − 7.9565749*salicylic acid | ||
| glutamic acid | 0.03 | 0.2052 | rank = 1.9431001 + 1.7149994* L-glutamic acid | ||
| methionine | 0.21 | <0.0001 | rank = 1.4207654 + 20.700626* L-methionine | ||
| cysteine | 0.26 | <0.0001 | rank = 1.481304 + 31.896956* L-cysteine | ||
| phenylalanine | 0.41 | <0.0001 | rank = 0.8664052 + 10.784315* L-phenylalanine | ||
| myristic acid | 0.06 | 0.0451 | rank = 1.2877006 + 151.58439* myristic | ||
| palmitic acid | 0.07 | 0.0343 | rank = 1.2694889 + 24.498395* palmitic | ||
| lysine | 0.24 | <0.0001 | rank = 1.6619978 + 1.4715583* L-lysine | ||
| histidine | 0.30 | <0.0001 | rank = 1.7394908 + 4.0341786*histidine | ||
| linoleic acid | 0.01 | 0.4235 | rank = 1.8792104 + 39.64309*linoleic acid | ||
| oleic acid | 0.01 | 0.3618 | rank = 1.825534 + 37.441287*oleic acid | ||
| stearic | 0.05 | 0.0804 | rank = 1.4599026 + 23.933753* stearic | ||
| tyrosine | 0.18 | 0.0003 | rank = 1.3115472 + 9.9256764* L-tyrosine | ||
| tryptophan |
0.14 |
0.0019 |
rank = 1.7886113 + 10.823357* L-tryptophan |
||
| Stepwise forward regression (SWFR) between the concentration of phloem sap metabolites in different citrus varieties and their degree of tolerance to CLas. | |||||
| Compound (step number) |
R2 |
P-value |
Mallowe’s Criterion (Cp) |
Akaike information criterion (AICc) |
Bayesian information criterion (BIC) |
| phenylalanine (1) | 0.41 | 0.0000 | 70.419 | 476.516 | 482.749 |
| benzoic acid (2) | 0.61 | 0.0000 | 28.482 | 452.017 | 460.191 |
| serine (3) | 0.70 | 0.0000 | 8.0795 | 435.048 | 445.088 |
| stearic acid (4) | 0.73 | 0.0148 | 3.9023 | 430.992 | 442.82 |
| fumaric (5) | 0.76 | 0.0039 | −1.917 | 424.284 | 437.818 |
| linoleic acid (6) | 0.78 | 0.0316 | −3.884 | 421.664 | 436.818 |
| GABA (7) | 0.79 | 0.0815 | −4.367 | 420.863 | 437.548 |
| leucine (8) | 0.80 | 0.0918 | −4.616 | 420.323 | 438.441 |
| glutamic acid (9) | 0.81 | 0.1473 | −4.235 | 420.704 | 440.156 |
Stepwise forward regression (SWFR) was also used to investigate the relation between phloem sap metabolites and varieties’ tolerance to CLas. The best model that accounts for this relation is shown in (Table 3). In agreement with the SLR, phenylalanine was also the most important component in the model and it explained about 50% of the variation in varieties’ tolerance to CLas. Benzoic acid which showed a negative correlation with citrus resistance to CLas in SLR was the second most important component in the model and it accounted about 20% of the variation (Table 3). Serine, GABA, leucine, stearic acid, linoleic acid, and glutamic acid were also significant to the model (Table 3).
Discussion
Clustering of the tolerant and sensitive cultivars into two groups indicated that the phloem sap composition of the tolerant cultivars was different from that of susceptible cultivars. This clustering also indicated that the phloem sap composition may have an influence on citrus tolerance to CLas and ACP. Our results showed that most of the CLas-tolerant cultivars were high in amino acids, especially those implicated in plant defense to pathogens such as phenylalanine, tyrosine, and tryptophan.
Phenylalanine is the end product of the shikimate pathway which also produces tyrosine and tryptophan.24 On the other hand, phenylalanine is the first precursor in phenylpropanoid cycle which is involved in plant defense, structural support, and survival.25 The phenylpropanoid pathway produces many important secondary metabolites which are involved in plant defense such as flavonoids, lignin, lignans, coumarins, and hydroxycinnamic acid conjugates.26
Induction of phenylalanine ammonia lyase (PAL), the first key enzyme of the phenylpropanoid pathway, has been observed in tobacco, soybean, cucumber, and melon after viral and pathogen infections.27-29 Induction of PAL and cinnamate 4-hydroxylase (C4H), the second enzyme in phenylpropanoid pathway, was also observed in cucumber and tomato after infection with the bacterial pathogen Pseudomonas syringae.30,31 Induction of PAL and C4H enzymes has been also observed in many insect-damaged plants.32 It is also believed that the biosynthesis of phenylalanine, the first precursor in phenylpropanoid cycle, is induced after insect damage.32
In addition, phenylalanine is implicated in the synthesis of salicylic acid (SA) which mediates plant defense against pathogens and insect attack.17,33 SA accumulation, in both infected and distal leaves after pathogens attack, is necessary to activate pathogenesis-related gene expression and induce the synthesis of defensive compounds in infected plants.33 Although SA could be synthesized from several pathways, silencing and chemical inhibition of PAL genes in plants indicated that the synthesis of SA from cinnamate (produced from phenylalanine) was the most important pathway during pathogen infection.33,34 Like salicylic acid, the phytohormone ethylene could be derived from amino acids. Ethylene controls many aspects of the plant life cycle and plays an important role in plant response to biotic and abiotic stresses.35 Ethylene can be derived from methionine via three key enzymatic reactions.35 The knockout of the methionine cycle in Arabidopsis reduced ethylene synthesis in seedlings.36
Recently, Slisz et al. (2012) showed that juice from symptomatic CLas-positive sweet orange fruit was lower in sugar, and many amino acids including alanine, arginine, isoleucine, leucine, proline, threonine, and valine compared to that obtained from healthy fruits.37 On the other hand, the concentration of phenylalanine, asparagine, and histidine were higher in CLas-positive fruits.37 The high levels of phenylanine in symptomatic fruits, was explained by the inhibition of the phenylpropanoid biosynthesis pathway by CLas.37 However, it is also possible that the increased demand for phenylalanine in the phenylpropanoid pathway may increase the synthesis of phenylalanine in CLas-infected fruits. In agreement with the latter possibility, Görlach et al. (1995) suggested that the increased demand for phenylalanine in the phenylpropanoid pathway after elicitor treatment increased de novo synthesis of phenylanine biosynthesis.38 Our results together with Slisz’s observation suggested that phenylalanine may play an important role in citrus response to CLas.
In addition to phenylalanine, tyrosine and tryptophan were also high in CLas-tolerant varieties. Like phenylalanine, tyrosine and tryptophan are also precursors for many secondary metabolites which are implicated in plant defense against biotic stress.16 Phenylalanine, tyrosine, and tryptophan are also implicated in the synthesis of plant phenolics.16 Constitutive phenolics act as feeding deterrents for herbivores, however their role in resistance against fungi, bacteria, and nematode is controversial.16 Although cultivars with high levels of constitutive phenolics were less attractive to herbivores, it is believed that the speed and the duration of de novo biosyntheses of phenolics are more important for resistance than constitutive concentrations.16 Because the CLas-tolerant cultivars contain high levels of these aromatic amino acids, these cultivars may contain high levels of phenolics or they could synthesize higher amounts of phenolics compounds rapidly after ACP and CLas attack. The high levels of constitutive phenolics or their rapid synthesis from de novo may explain why some citrus varieties are more tolerant to ACP and CLas. However, more work is needed to test the role of citrus phenolics against CLas.
CLas-tolerant varieties were higher in amino acids which are implicated in plant defense. This result was not surprising because many plant defensive compounds such as glucosinolates are derived from amino acids.39 In addition, some amino acid metabolic pathways are an important part in plant immune system.40 For example, pipecolic acid which is formed from lysine, derived from aspartic acid, has been found to induce the production of SA and camalexin in Arabidopsis leaves inoculated with bacterial pathogens.40 Although the levels of amino acids in plants are under a continual state of flux and only one time point was investigated, our results indicated that amino acids may play an important role in citrus tolerance to CLas. In addition, the induction of these amino acids in CLas-infected citrus plants (non-published data) supports our current results. Time-wise determination of amino acids levels in CLas-tolerant and susceptible varieties before and after infection with CLas could reveal more insights about the role of amino acids in citrus response against CLas.
Field observation and controlled studies showed that ACP prefers some citrus varieties more than others.5-7 However, the characteristics that make some varieties resistant to ACP are not known yet.41 Plant resistance to ACP could result from different traits such as non-preference, antibiosis, and tolerance.42 It is also possible that non-preferred hosts could be deficient in essential nutrients. However, because most of the tolerant cultivars were, in general, rich in amino acids including the essential ones, this possibility can be eliminated.
Although SLR showed that fatty acids were not implicated in citrus tolerance to CLas, SWFS showed that linoleic acid and stearic acid were necessary in the model. Fatty acids are precursors for jasmonic acid which is released in response to wounding and herbivore attack (Zeier, 2013). High levels of these fatty acids in CLas tolerant varieties may explain why they are also tolerant to ACP.
Plant defense against bacterial pathogens and insects requires the biosynthesis of metabolites with antimicrobial or toxic activities and these metabolites could be produced constitutively (phytoanticipins) and/or induced (phytoalexins) after biotic stress.40 Plant defense also depends on the performance of plant metabolism.40 Because many plant defensive compounds are derived from amino acids, the metabolism of certain amino acids impacts plant resistance to pathogens.40 Our results showed that phloem sap of CLas-tolerant varieties was higher in amino acids implicated in plant resistance to pathogens and insects. The high amounts of these amino acids may be responsible for the tolerance of these varieties to CLas and ACP; these varieties may contain higher amount of constitutive defensive compound and/or they may synthesize them in more efficient manner after infection.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank our lab members for critical reading of the manuscript.
Funding
This study was supported with grant #558 for NK received from Citrus Research and Development Foundation (CRDF).
References
- 1.Liu Y, Heying E, Tanumihardjo SA. History, global distribution, and nutritional importance of citrus fruits. Compr Rev Food Sci F 2012; 11 (6):530-545; http://dx.doi.org/ 10.1111/j.1541-4337.2012.00201.x [DOI] [Google Scholar]
- 2.Bové JM, Garnier M. Phloem- and xylem-restricted plant pathogenic bacteria. Plant Sci. 2002; 163:1083-1098; http://dx.doi.org/ 10.1016/S0168-9452(02)00276-5 [DOI] [Google Scholar]
- 3.Jagoueix S, Bové JM, Garnier M. The phloem-limited bacterium of greening disease of citrus is a member of the alpha subdivision of the Proteobacteria. Int J Syst Bacteriol 1994; 44:379-86; PMID:7520729; http://dx.doi.org/ 10.1099/00207713-44-3-379 [DOI] [PubMed] [Google Scholar]
- 4.Tatineni S, Sagaram US, Gowda S, Robertson CJ, Dawson WO, Iwanami T, Wang N. In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 2008; 98 (5):592-599; PMID:18943228; http://dx.doi.org/ 10.1094/PHYTO-98-5-0592 [DOI] [PubMed] [Google Scholar]
- 5.Halbert SE, Manjunath KL. Asian citrus psyllids (Sternorrhyncha: Psyllidae) greening disease of citrus: a literature review and assessment of risk in Florida. Fla Entomol 2004; 87 (3):330-53; http://dx.doi.org/ 10.1653/0015-4040(2004)087%5b0330:ACPSPA%5d2.0.CO;2 [DOI] [Google Scholar]
- 6.Tsai JH, Liu YH. Biology of Diaphorina citri (Homoptera: Psyllidae) on four host plants. J Econ Entomol 2000; 93 (6):1721-5; PMID:11142304; http://dx.doi.org/ 10.1603/0022-0493-93.6.1721 [DOI] [PubMed] [Google Scholar]
- 7.Richardson ML, Hall DG. Resistance of Poncirus and citrus x Poncirus germplasm to the Asian citrus psyllid. Crop Sci 2013; 53 (1):183-188; http://dx.doi.org/ 10.2135/cropsci2012.02.0091 [DOI] [Google Scholar]
- 8.Fraser LR, Singh D. Reaction of Indian citrus varieties to greening virus. Pages 365-366 in: Proc. First Int. Citrus Symp 1969; Chapman HD. (editor). Publications of Department of the University of California, Riverside. [Google Scholar]
- 9.Gonzales CI, Vinas RC, Vergara LA. Observations of 110 citrus cultivars in an area severely infested by leaf mottling. Pages 38-40 in: Proc. Fifth Conf. Int. Organ. Citrus Virol 1972. Tokyo Price W. C., ed. University of Florida Press, Gainesville. [Google Scholar]
- 10.Koizumi M, Prommintara M, Linwattana G, Kaisuwan T. Field evaluation of citrus cultivars for greening disease resistance in Thailand. Pages 274-279 in: Proc. Twelfth Conf. Int. Organ. Citrus Virol 1993. New Delhi Moreno P., da Graça J. V., and Timmer L. W., eds. International Organization of Citrus Virologists, University of California, Riverside. [Google Scholar]
- 11.Folimonova SY, Robertson CJ, Garnsey SM, Gowda S, Dawson WO. Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology 2009; 99 (12):1346-1354; PMID:19900000; http://dx.doi.org/ 10.1094/PHYTO-99-12-1346 [DOI] [PubMed] [Google Scholar]
- 12.Sagaram M, Burns JK. Leaf chlorophyll fluorescence parameters and huanglongbing. J Am Soc Hortic Sci 2009; 134 (2):194-201. [Google Scholar]
- 13.Hajivand S, Thohirah Lee A, Kamaruzaman S, Siti Nor AA, Nur Ashikin PA. Differential reaction of citrus species in Malaysia to Huanglongbing (HLB) disease using grafting method. Am J Agricul Biol Sci 2009:4 (1):32-38; http://dx.doi.org/ 10.3844/ajabssp.2009.32.38 [DOI] [Google Scholar]
- 14.Fürstenberg-Hägg J, Zagrobelny M, Bak S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013; 14 (5):10242-10297; PMID:23681010; http://dx.doi.org/10328566 10.3390/ijms140510242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazid M, Khan TA, Mohammad F. Role of secondary metabolites in defense mechanisms of plants. Biol Med J 2011; 3 (2):232-249. [Google Scholar]
- 16.Bennett RN, Wallsgrove RM. Secondary metabolites in plant defense-mechanisms. New Phytol 1994; 127 (4):617-633; http://dx.doi.org/ 10.1111/j.1469-8137.1994.tb02968.x [DOI] [PubMed] [Google Scholar]
- 17.Maffei ME. Sites of synthesis, biochemistry and functional role of plant volatiles. South African J Bot 2010; 7 (4):612-631; http://dx.doi.org/ 10.1016/j.sajb.2010.03.003 [DOI] [Google Scholar]
- 18.Hagel M, Onoyovwi A, Yeung EC, Facchini PJ. 2013. Role of phloem sap in plant defense. In phloem molecular cell biology, systemic communication, biotic interactions. GA Thompson, van Bell AJE (editors). John Wiley @ Sons, Ames, Iowa: C2013. P. 251-270; http://dx.doi.org/ 10.1002/9781118382806. [DOI] [Google Scholar]
- 19.Albrecht U, Bowman KD. Tolerance of the trifoliate citrus hybrid US-897 (Citrus reticulata Blanco x Poncirus trifoliata L. Raf.) to huanglongbing. Hort Sci 2011; 46 (1):16-22. [Google Scholar]
- 20.Albiach-Marti MR, Grosser JW, Gowda S, Mawassi M, Satyanarayana T, Garnsey SM, Dawson WO. Citrus tristeza virus replicates and forms infectious virions in protoplasts of resistant citrus relatives. Mol Breeding 2004; 14 (2):117-128. [Google Scholar]
- 21.Kim DH, Bae EA, Han MJ. Anti-Helicobacter pylori activity of the metabolites of poncirin from Poncirus trifoliata by human intestinal bacteria. Biol Pharm Bull 1999; 22 (4):422-4; PMID:10328566; http://doi.org/10.1248/bpb.22.422 [DOI] [PubMed] [Google Scholar]
- 22.Rahman A, Al-Reza SM, Yoon JI, Kang SC. In vitro inhibition of foodborne pathogens by volatile oil and organic extracts of Poncirus trifoliata Rafin. seeds. J Food Sci Agric 2009; 89 (5):876-81; http://dx.doi.org/10.1002/jsfa.3527 [Google Scholar]
- 23.Hijaz F, Killiny N. Collection and Chemical Composition of Phloem Sap from Citrus sinensis L. Osbeck (Sweet Orange). PLoS ONE 2014; 9 (7):e101830; PMID:25014027; http://dx.doi.org/ 10.1371/journal.pone.0101830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 1999; 50:473-503; PMID:15012217: http://dx.doi.org/ 10.1146/annurev.arplant.50.1.473 [DOI] [PubMed] [Google Scholar]
- 25.Vgot T. Phenylpropanoid biosynthesis. Mol Plant 2010; 3:2-20; PMID:20035037; http://dx.doi.org/ 10.1093/mp/ssp106 [DOI] [PubMed] [Google Scholar]
- 26.Fraser CM, Chapple C. The phenylpropanoid pathway in Arabidopsis. The Arabidopsis book/Am Society Plant Biol 2011; 9, e0152; http://dx.doi.org/ 10.1093/mp/ssp10610.1199/tab.0152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrini L, Rohfritsch O, Fritig B, Legrand M. Phenylalanine ammonialyase in tobacco. Molecular cloning and gene expression during the hypersensitive reaction to Tobacco Mosaic Virus and the response to a fungal elicitor. Plant Physiol. 106, 877-886; PMID:7824656; http://dx.doi.org/10./1104/pp./106./3./877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellés JM, Lopez-Gresa MP, Fayos J, Pallas V, Rodrigo I, Conejero V. Induction of cinnamate 4-hydroxylase and phenylpropanoids in virus-infected cucumber and melon plants. Plant Sci 2008; 174:524-533; http://dx.doi.org/ 10.1016/j.plantsci.2008.02.008 [DOI] [Google Scholar]
- 29.Delledonne M, Xia YJ, Dixon RA, Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature 1988; 394:585-588; PMID:9707120; http://dx.doi.org/10.1038/29087 [DOI] [PubMed] [Google Scholar]
- 30.Smith-Becker J, Marois E, Huguet EJ, Midland SL, Sims JJ, Keen NT. Accumulation of salicylic acid and 4-hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-lyase activity in petioles and stems. Plant Physiol 1998; 116:231-238; PMID:9449843; http://dx.doi.org/10./1104/pp./116./1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López-Gresa MP, Torres C, Campos L, Lison P, Rodrigo I, .Identification of defense metabolites in tomato plants infected by the bacterial pathogen Pseudomonas syringae. Environ Exp Bot 2011; 74:216-228. http://dx.doi.org/ 10.1016/j.envexpbot.2011.06.003 [DOI] [Google Scholar]
- 32.Bernards MA, Bastrup-Spohr L. Phenylpropanoid metabolism induced by wounding and insect herbivory. In Induced Plant Resistance to Herbivory. Schaller Andreas. (editor) Springer Science & Business Media; Berlin, Germany: C2008. P. 189-211. [Google Scholar]
- 33.Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defense. Nature 2002, 417 (6888):571-571; PMID:11734859; http://dx.doi.org/10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- 34.Chen Z, Zheng Z, Huang J, Lai Z, Fan B. Biosynthesis of salicylic acid in plants. Plant Signal Behav 2009; 4 (6):493-6; PMID:19816125; http://dx.doi.org/ 10.4161/psb.4.6.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot 2009; 60; 3311-3336; PMID:19567479; http://dx.doi.org/ 10.1093/jxb/erp204 [DOI] [PubMed] [Google Scholar]
- 36.Buerstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M. The role of methionine recycling for ethylene synthesis in Arabidopsis. Plant J 2007; 49 (2):238-249; PMID:17144895; http://dx.doi.org/ 10.1111/j.1365-313X.2006.02942.x [DOI] [PubMed] [Google Scholar]
- 37.Slisz AM, Breksa AP 3rd, Mishchuk DO, McCollum G, Slupsky CM. Metabolomic Analysis of Citrus Infection by 'Candidatus Liberibacter' Reveals Insight into Pathogenicity. J Proteome Res 2012; 11 (8):4223-4230; PMID:22698301; http://dx.doi.org/ 10.1021/pr300350x [DOI] [PubMed] [Google Scholar]
- 38.Görlach J, Raesecke HR, Rentsch D, Regenass M, Roy P, Zala M, Keel C, Boller T, Amrhein N, Schmid J. Temporally distinct accumulation of transcripts encoding enzymes of the prechorismate pathway in elicitor-treated, cultured tomato cells. Proc Natl Acad Sci U S A 1995; 92 (8):3166-3170; PMID:11607524; PMID:11607524; http://dx.doi.org/ 10.1073/pnas.92.8.3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sønderby IE, Geu-Flores F, Halkier BA. Biosynthesis of glucosinolates- gene discovery and beyond. Trends Plant Sci 2010; 15:283-290; PMID:20303821; http://dx.doi.org/ 10.1016/j.tplants.2010.02.005 [DOI] [PubMed] [Google Scholar]
- 40.Zeier J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell and Envir 2013; 36 (12):2085-2103; PMID:23611692; http://dx.doi.org/ 10.1111/pce.12122 [DOI] [PubMed] [Google Scholar]
- 41.Ammar E-D, Richardson ML, Abdo Z, Hall DG, Shatters RG Jr. Differences in Stylet Sheath Occurrence and the Fibrous Ring (Sclerenchyma) between x Citroncirus Plants Relatively Resistant or Susceptible to Adults of the Asian Citrus Psyllid Diaphorina citri (Hemiptera: Liviidae). PLoS ONE 2014; 9 (10):e110919; PMID:25140675; http://dx.doi.org/ 10.1371/journal.pone.0110919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Painter RH. Resistance of plants to insects. Annu Rev Entomol 1958; 3:267-290; http://dx.doi.org/ 10.1146/annurev.en.03.010158.001411 [DOI] [Google Scholar]