Abstract
Adaptation to stress entails a repertoire of molecular pathways that remodel the proteome, thereby promoting selective translation of pro-survival proteins. Yet, translation of other proteins, especially those which are harmful for stress adaptation is, on the contrary, transiently suppressed through mRNA decay or storage. Proteome remodeling under stress is intimately associated with the cytoplasmic ribonucleoprotein (RNP) complexes called stress granules (SGs) and processing bodies (PBs). The molecular composition and regulation of SGs and PBs in plants remain largely unknown. Recently, we identified the Arabidopsis Tudor Staphylococcal Nuclease (TSN, Tudor-SN or SND1) as a SG- and PB-associated protein required for mRNA decapping under stress conditions. Here we show that SGs localize in close proximity to PBs within plant cells that enable the exchange of molecular components. Furthermore, we provide a meta-analysis of mRNA degradome of TSN-deficient plants suggesting that TSN might inhibit the degradation of mRNAs which are involved in stress adaptation. Our results establish TSN as a versatile mRNA regulator during stress.
Keywords: mRNA decapping, processing bodies (PBs), ribonucleoprotein (RNP) complexes, stress granules (SGs), Tudor Staphylococcal Nuclease
Adaptation to stress depends on the availability of energy resources.1 Stress drives cells to an energy crisis whereupon they have to reduce energy expenditure in order to survive. To this end, eukaryotic cells compartmentalize specific mRNAs and proteins in cytoplasmic ribonucleoprotein (RNP) structures known as stress granules (SGs) and processing bodies (PBs).2 In these structures mRNA molecules are stored, degraded or kept silent in order to prevent energy expenditure on producing useless, surplus or even harmful proteins under stress conditions.3
Numerous components of SGs and PBs have been identified in yeast and animal models. SGs typically contain poly(A)+ RNA, translation initiation factors (eIFs), poly(A) binding protein (PABP) and ribosomal proteins.4 In contrast, PBs contain a suit of proteins involved in mRNA decay and translational repression, including subunits of decapping and exosome complexes, deadenylases, and RNA-binding proteins.5 Although both types of RNP complexes are present under stress conditions, they serve distinct functions. While SGs are thought to play a role in sequestering, stabilizing and storing mRNAs and translation factors,6,7 the main function of PBs is attributed to translational repression and mRNA decay, in accordance with their composition.8,9
Plants are sessile organisms subjected to and able to cope with a vast array of biotic and abiotic stresses throughout their lifespans. Recent studies have provided useful insight into the cell biology and biochemistry of SGs and PBs in plants. Sorenson and Bailey-Serres have found that Ubp1c, a SG-nucleating RNA-binding protein, is a component of the machinery that reprograms post-transcriptional gene expression during hypoxia.10 More recently, we characterized the processes of SG assembly and disassembly in plants under heat stress and established Tudor Staphylococcal Nuclease (TSN) as a structural and functional component of both SGs and PBs, essential for mRNA catabolism under stress.11
Here we extend our previous study and show that (i) SGs localize in a close proximity to PBs in plant cells, and (ii) TSN may positively regulate stability of mRNAs involved in stress adaptation pathways.
Although SGs and PBs have distinct functions and composition, several studies performed in animal and yeast models pointed to the existence of a strong physical link between them. It was demonstrated that both complexes are likely to exchange mRNA and proteins between themselves and with polysomes.4 Several observations supported the idea that similar RNP shuttle is also present in plants. First, some proteins were found to localize in both SGs and PBs, such as CCCH tandem zinc finger proteins TZF1, TZF4, TZF5 and TZF6.11,12 Second, treatment with cycloheximide, which causes trapping of mRNA in polysomes by inhibiting the translocation step during the elongation phase in protein synthesis, abolished SG and PB assembly.11,13 By performing a co-localization study using SG- and PB-associated proteins eIF4e and DCP1, respectively, we have revealed that these RNP complexes are situated in close proximity to each other in Arabidopsis root cells under heat stress (Fig. 1, arrowheads) .
Figure 1.
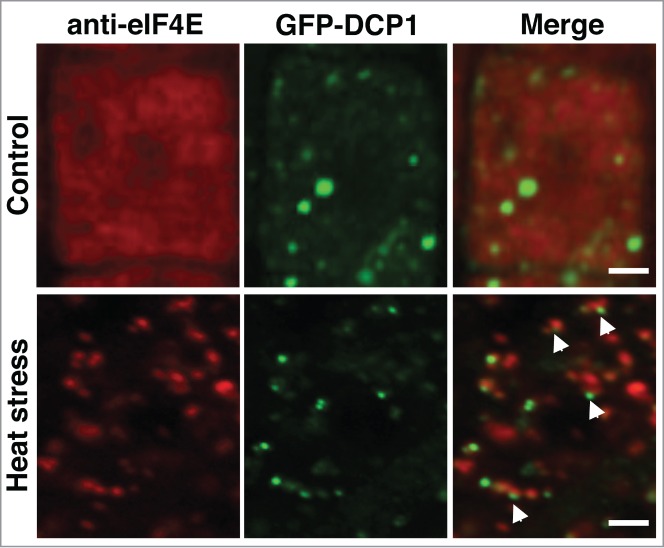
Colocalization analysis of GFP-DCP1 and eIF4E proteins. 5-day-old Col Arabidopsis seedlings expressing GFP-DCP1 (green) were heat stressed at 39°C for 40 min and then immunostained with anti-elF4E (red) as described previously.27 Root cells were examined by confocal laser microscopy. Control seedlings were grown at 23°C. Note a close proximity between eIF4E-SG and DCP1-PB denoted by arrowheads. Scale bars, 2 μm.
Shuttling of cytoplasmic mRNAs between polysomes, PBs and SGs has been well documented in stressed mammalian cells.14 Yet, the mechanisms and directionality of mRNA movement between PBs and SGs remain unresolved. A longstanding notion is that mRNAs stalled at a step of translation initiation are kept in SGs for storage, or directed to PBs for degradation. However, several studies have found that mRNA decay enzymes such as XRN1 can be also found in SGs, suggesting that mRNA degradation may also take place in these foci.8,14 Likewise, we showed that TSN is localized in both SGs and PBs and that enzymatically active tandem repeat of 4 N-terminally situated SN domains confers this localization.11
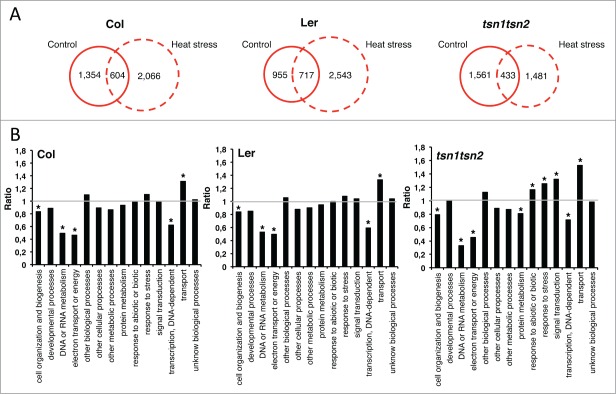
TSN is an evolutionarily conserved protein present in almost all eukaryotes, with the notable exception of Saccharomyces cerevisiae.15 The domain composition of TSN is also conserved, invariably comprising a tandem repeat of 4 non-canonical SN domains followed by a Tudor and a C-terminal partial SN domain.16 In animals, TSN functions in several gene expression pathways in both the nucleus and the cytoplasm, including regulation of transcription,17-20 pre-mRNA splicing,21 and RNA silencing.22,23 Arabidopsis genome has 2 partly redundant TSN genes (TSN1 and TSN2) which were shown to confer stress tolerance through the stabilization of mRNAs encoding secreted proteins 24 and gibberellin 20-oxidase 3, a key enzyme in gibberellin biosynthesis.25 Unlike animal TSN, in Arabidopsis TSN1 and TSN2 are exclusively cytoplasmic.11,26 However, the role of TSN in plants seems to be extended to also encompass an opposite process, viz. mRNA decay, under stress. In our recent work, the purification of uncapped mRNA molecules, intermediates of the 5′–3′ decay pathway, from Columbia-0 (Col), Landsberg erecta (Ler) and tsn1tsn2 double knock-out plants grown under control conditions or heat stress and subsequent cDNA arrays analysis revealed that TSN is essential for mRNA decapping under stress.11 While heat-stressed Col and Ler plants exhibited a pronounced increase in the accumulation of uncapped mRNA, this increase was abrogated in TSN-deficient plants (Fig. 2A). This analysis has also detected large changes in the pattern of uncapped transcripts caused by heat stress .
Figure 2.
Analysis of global mRNA decapping pattern in Arabidopsis under heat stress as affected by TSN deficiency. (A) Quantitative analysis of transcripts enriched in uncapped form in control (23°C) and heat stress (39°C for 40 min) conditions in Col, Ler and tsn1tsn2 plants. (B) GO analysis (term “Biological Process”) of transcripts enriched in uncapped form in Col, Ler and tsn1tsn2 plants under heat stress. The charts display the ratios between the percentages of mRNAs belonging to a particular GO term that show significant enrichment in uncapped form under heat stress and under control conditions. Asterisks indicate significant differences at P < 0.05, Fisher's exact test. To identify enrichment of uncapped transcripts, significance analysis was performed using TIGR MultiExperiment Viewer modelu of TM4 software.28 A further description of the microarray experiment can be found elsewhere.29 GOSlim annotation developed by TAIR was used to organize sets of genes into broad ontology categories.30
In order to determine which categories of transcripts are enriched in the uncapped form (i.e. subject to degradation) during heat stress, we grouped them into Gene Ontology (GO) Biological Process (Fig. 2B). A comparison between the heat stress versus control condition showed that the transcripts related to categories “DNA or RNA metabolism,” “electron transport or energy” and “transcription, DNA-dependent” are less uncapped in all genetic backgrounds, indicating that these transcripts are critical for heat stress adaptation. Noteworthy, transcripts of genes encoding components of stress response and signal transduction pathways are enriched in uncapped form in the tsn1tsn2 background but not in wild-type plants (Fig. 2B). These data are in agreement with the previously reported role of TSN in stabilizing certain mRNAs encoding secreted proteins.26
Here, we provide evidence that SGs and PBs are located in close proximity in plant cells, further supporting the notion that RNP components can shuttle between them. In addition, we show that transcripts related to cell signaling and stress adaptation are preferentially uncapped in tsn1tsn2 background. Further work is required to unravel the molecular mechanism that enables TSN to play 2 seemingly antagonistic roles in RNA metabolism under stress, i.e., facilitating global mRNA decapping and thus degradation, and on the other hand stabilizing particular mRNAs that are required for survival.
Funding
This work was supported by grants from Knut and Alice Wallenberg Foundation (to P.V.B.), the Swedish Research Council (to P.N.M. and P.V.B.), Pehrssons Fund (to P.V.B.), the Swedish Foundation for Strategic Research (to P.V.B.), and Olle Engkvist Foundation (to P.V.B.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hernandez I, Munne-Bosch S. Linking phosphorus availability with photo-oxidative stress in plants. J Exp Bot 2015; 66(10):2889-900; In press; PMID:25740928 [DOI] [PubMed] [Google Scholar]
- 2.Kedersha N, Anderson P. Regulation of translation by stress granules and processing bodies. Progr Mol Biol Transl Sci 2009; 90:155-85; PMID:20374741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijovic M, Arens C, Nyström T, Liu B, Sunnerhagen P. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet 2014; 10:e1004763; PMID:25375155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932-41; PMID:20064460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell 2008; 32:605-15; PMID:19061636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisin ger-Mathason TS, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, et al.. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol Cell 2008; 31:722-36; PMID:18775331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchan JR, Capaldi AP, Parker R. TOR-tured yeast find a new way to stand the heat. Mol Cell 2012; 47:155-7; PMID:22841000 [DOI] [PubMed] [Google Scholar]
- 8.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, et al.. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005; 169:871-84; PMID:15967811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Chua NH. Processing bodies and plant development. Curr Opin Plant Biol 2011; 14:88-93; PMID:21075046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorenson R, Bailey-Serres J. Selective mRNA sequestration by oligouridylate-binding protein 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 2014; 111:2373-8; PMID:24469793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez-Beltran E, Moschou PN, Smertenko AP, Bozhkov PV. Tudor staphylococcal nuclease links formation of stress granules and processing bodies with mRNA catabolism in Arabidopsis. Plant Cell 2015; 27(3):926-43; In press; PMID:25736060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogamuwa S, Jang JC. The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environm 2013; 36:1507-19; PMID:23421766 [DOI] [PubMed] [Google Scholar]
- 13.Motomura K, Le QT, Hamada T, Kutsuna N, Mano S, Nishimura M, Watanabe Y. Diffuse decapping enzyme DCP2 accumulates in DCP1 foci under heat stress in Arabidopsis thaliana. Plant Cell Physiol 2015; 56:107-15; PMID:25339350 [DOI] [PubMed] [Google Scholar]
- 14.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Persp Biol 2012; 4:a012286; PMID:22763747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard-Till RA, Yao MC. Tudor nuclease genes and programmed DNA rearrangements in Tetrahymena thermophila. Euk Cell 2007; 6:1795-804; PMID:17715366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA Jr, Välineva T, et al.. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 2009; 11:1347-54; PMID:19820703; http://dx.doi.org/ 10.1038/ncb1979 [DOI] [PubMed] [Google Scholar]
- 17.Leverson JD, Koskinen PJ, Orrico FC, Rainio EM, Jalkanen KJ, Dash AB, Eisenman RN, Ness SA. Pim-1 kinase and p100 cooperate to enhance c-Myb activity. Mol Cell 1998; 2:417-25; PMID:9809063; http://dx.doi.org/ 10.1016/S1097-2765(00)80141-0 [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Aittomaki S, Pesu M, Carter K, Saarinen J, Kalkkinen N, Kieff E, Silvennoinen O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J 2002; 21:4950-8; PMID:12234934; http://dx.doi.org/ 10.1093/emboj/cdf463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol 2003; 17:1805-14; PMID:12819296; http://dx.doi.org/ 10.1210/me.2002-0256 [DOI] [PubMed] [Google Scholar]
- 20.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol 1995; 15:4735-44; PMID:7651391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Valineva T, Hong J, Bu T, Yao Z, Jensen ON, Frilander MJ, Silvennoinen O. Transcriptional co-activator protein p100 interacts with snRNP proteins and facilitates the assembly of the spliceosome. Nucl Acid Res 2007; 35:4485-94; PMID:17576664; http://dx.doi.org/ 10.1093/nar/gkm470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH. A micrococcal nuclease homologue in RNAi effector complexes. Nature 2003; 425:411-4; PMID:14508492; http://dx.doi.org/ 10.1038/nature01956 [DOI] [PubMed] [Google Scholar]
- 23.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol 2005; 12:489-96; PMID:15895094; http://dx.doi.org/ 10.1038/nsmb936 [DOI] [PubMed] [Google Scholar]
- 24.Frey NFD, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C, Bianchi MW. RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 2010; 22:1575-91; PMID:20484005; http://dx.doi.org/ 10.1105/tpc.109.070680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan CX, Yan ZY, Wang YZ, Yan XY, Han YZ. Tudor-SN, a component of stress granules, regulates growth under salt stress by modulating GA20ox3 mRNA levels in Arabidopsis. J Exp Bot 2014; 65:5933-44; PMID:25205572; http://dx.doi.org/ 10.1093/jxb/eru334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.dit Frey NF, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C, Bianchi MW. The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 2010; 22:1575-91; PMID:20484005; http://dx.doi.org/ 10.1105/tpc.109.070680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moschou PN, Smertenko AP, Minina EA, Fukada K, Savenkov EI, Robert S, Hussey PJ, Bozhkov PV. The caspase-related protease separase (extra spindle poles) regulates cell polarity and cytokinesis in Arabidopsis. Plant Cell 2013; 25:2171-86; PMID:23898031; http://dx.doi.org/ 10.1105/tpc.113.113043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al.. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 2003; 34:374-8; PMID:12613259 [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez-Beltran E. Genome-wide analysis of uncapped mRNAs under heat stress in Arabidopsis. Genomics Data 2015; September: 7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al.. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucl Acid Res 2003; 31:224-8; PMID:12519987; http://dx.doi.org/ 10.1093/nar/gkg076 [DOI] [PMC free article] [PubMed] [Google Scholar]