ABSTRACT
As the sessile organisms, plants evolve different strategies to survive in adverse environmental conditions. The elaborate regulation of shoot branching is an important strategy for plant morphological adaptation to various environments, while the regulation of reactive oxygen species (ROS), salicylic acid (SA) and jasmonic acid (JA) is pivotal for plant responses to biotic and abiotic stresses. Recently, we have demonstrated that Arabidopsis EXB1, a WRKY transcription factor, is a positive regulator of shoot branching as a cover story in Plant Cell. Here we show that WRKY23, an EXB1 close member, has a redundant role in control of shoot branching. We further show that EXB1 is induced by H2O2, ABA or mannitol treatments, suggesting that EXB1 may also play roles in plant responses to abiotic stresses. RNA-sequencing (RNA-seq) analysis using 4EnhpEXB1-EXB1GR inducible line indicates that the genes involved in oxidative stress, oxidation reduction, SA or JA signaling pathway are regulated by EXB1 induction in a short time. We suggest that EXB1/WRKY71 transcription factor may play pivotal roles in plant adaptation to environments by both morphological and physiological ways.
KEYWORDS: Abiotic stresses, EXB1, plant adaptation, reactive oxygen species, shoot branching, WRKY transcription factor
Unlike animals, plants cannot escape from adverse environmental conditions by relocation. Plants evolve different strategies to adapt to various environmental changes. Control of shoot branching is one of the excellent strategies in response to environmental stimuli by forming proper plant morphology,1 while the regulation of producing reactive oxygen species (ROS), salicylic acid (SA) and jasmonic acid (JA) is another important strategy to adapt to environments by changing the physiological status of plant cells.2 Branches are derived from the axillary meristems (AMs) in leaf axils. AMs first develop into axillary buds by producing a few leaves. Some axillary buds can further develop into shoot branches, while other axillary buds just stay dormant. Thus, the initiation of AMs and axillary bud outgrowth are the 2 important steps that determine the final number of plant branches.3,4
The initiation of AMs and axillary bud outgrowth were regulated elaborately. The REGULATOR OF AXILLARY MERISTEMS 1 (RAX1) is a key regulator in control of AM initiation. RAX1 functions redundantly with RAX2 and RAX3.5,6 RAX1, RAX2 and RAX3 encode closely related R2R3 MYB transcription factors. The function of RAX genes in AM formation appears to be conserved throughout plant kingdom. BLIND (BL) is orthologous to RAX genes and bl mutants display defects of AM initiation in tomato and pepper.7,8 By screening an activation tagging Arabidopsis mutant collection we obtained a dominant mutant exb1-D which producing many more branches than the wild-type control. We demonstrate that EXB1 positively regulates shoot branching by affecting both axillary meristem initiation and the bud outgrowth. Furthermore, we reveal that EXB1 facilitate AM initiation by directly regulating RAX genes at the transcriptional level and also promote bud outgrowth by repressing auxin pathway.3
EXB1 encodes a WRKY transcription factor previously named WRKY71.3 WRKY family is one of the largest transcription factor families and there are 74 members in Arabidopsis genome.9 WRKY28 (EXB2), WRKY8 (EXB3), WRKY48 (EXB4) and WRKY57 (EXB5) are closely related to EXB1 in the phylogenetic tree and are proved to be redundant to EXB1. WRKY23 is also located in the EXB1 clade of the WRKY phylogenetic tree (Fig. 1A). WRKY23 has been identified to regulate auxin signaling in the downstream of IAA14, AUXIN RESPONSE FACTOR 7 (ARF7) and ARF19.10,11 WRKY23 has also been found to regulate plant embryo and root development by affecting auxin distribution through the control of flavonol biosynthesis.10,12 However no findings have demonstrated the roles of WRKY23 in shoot branching. To reveal the functions of WRKY23 in shoot branching, we first generated the construct 4EnhEXB1p-WRKY23 by using the 4 CaMV 35S enhancers and 2700-bp-long EXB1 promoter (4EnhEXB1p) to overexpress WRKY23 gene (Fig. 1B). The 4EnhEXB1p-WRKY23 transgenic plants produced many more branches than the wild-type control (Fig. 1B). The WRKY23 overexpression plants even can produced thousands of branches in a single plant, indicating that WRKY23 had high capacity to promote plant shoot branching (Fig. 1B).
Figure 1.
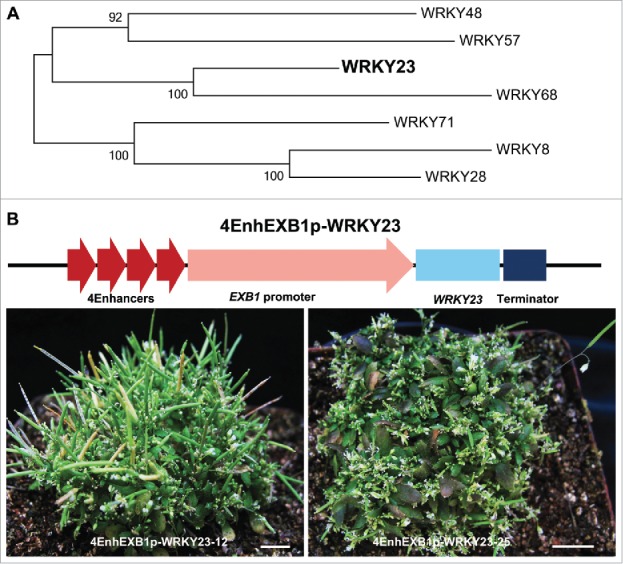
WRKY23 Had Redundant Function with EXB1. (A) The phylogenetic tree of EXB1/WRKY71, WRKY8, WRKY28, WRKY48, WRKY57, WRKY23 and WRKY68 was generated based on the full-length protein sequences using Neighbor-Joining method by MEGA 6. (B) Top, the schematic representation of 4EnhEXB1p-WRKY23 construct; Bottom, the phenotypes of 2 independent lines of 4EnhEXB1p-WRKY23 transgenic plants.
WRKY transcription factors have been reported to participate in responses to various biotic or abiotic stresses.9 In order to test whether EXB1 might also be involved in the regulation of plant responses to abiotic stresses, we first test the expression of EXB1 using quantitative RT-PCR after treatment of wild-type plants with 5 mM and 10 mM H2O2, 100 mM ABA or 300 mM mannitol. The results showed that EXB1 was induced rapidly after the treatment with H2O2, ABA or mannitol (Fig. 2), suggesting that EXB1 might also regulate plant adaptation to adverse environmental conditions physiologically.
Figure 2.
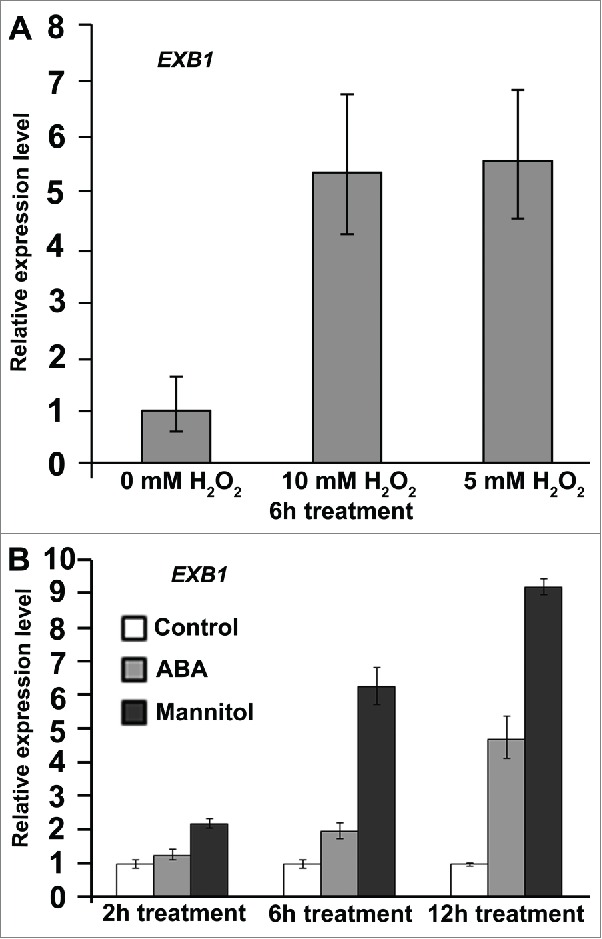
EXB1 Was Induced by H2O2, ABA or Mannitol Rapidly. (A) The relative expression level of EXB1 in wild-type plants at 6 hours after treated with 10 mM or 5 mM H2O2; (B) The relative expression level of EXB1 in wild-type plants at 2 hours, 6 hours and 12 hours after treated with 100 mM ABA or 300 mM mannitol. The expression level of EXB1 in mock-treated plants was set to 1.0. The error bars represent the SD of 3 replicates.
To understand the molecular mechanism by which EXB1 regulates shoot branching and plant responses to stresses, we fused the coding sequence of EXB1 to the sequence encoding the steroid-binding domain of the rat glucocorticoid receptor (GR)13 and put the fusion under the 4EnhEXB1p to generate the construct 4EnhpEXB1-EXB1GR. 4EnhpEXB1-EXB1GR-13 transgenic plants were treated with dexamethasone (DEX) to induce EXB1 translocation into nucleus.3 The samples were used to perform RNA-sequencing (RNA-seq) analysis. The results showed that 3114 genes were differentially expressed (fold change ≥ 2, P < 0.05) after the treatment with 30 μM for 4 hours. Among these genes, the expression level of RAX genes was rapidly increased after EXB1 induction, and the transcripts of genes involved in auxin pathways were also significantly altered.3
Our further analysis showed that the expression levels of genes related to the plant responses to oxidative stress (Table 1), SA signaling pathway (Table 2), and JA signaling pathway (Table 3) were also altered significantly (fold ≥ 2 or ≤ 0.5, p≤ 0.05) after EXB1 induction. These results further suggested that EXB1 may be involved to plant physiological responses to different stresses. Previous studies have demonstrated that WRKY8, WRKY28, WRKY48 and WRKY57 also participate in plant responses to biotic or abiotic stresses.14-17 A recent published article has demonstrated that WRKY71, WRKY8 or WRKY28 can accelerate flowering in Arabidopsis as well,18 suggesting that EXB1 might increase the possibility of successful reproduction under environmental stresses by speeding up the life cycle. These findings suggested that EXB1/WRKY71 family transcription factors may facilitate plant adaptation morphologically and physiologically through promoting shoot branching,3 and speeding up the life cycle18 in response to different environmental cues.
Table 1.
The transcript alteration of genes in response to oxidative stress by EXB1 induction.
| Gene ID | Gene product | Value of mock-treated plants | Value of DEX-treated plants | Log2(fold change) | p_value |
|---|---|---|---|---|---|
| AT1G14540 | PER4 | 2.06828 | 4.63742 | 1.2 | 0.0231123 |
| AT1G21520 | AT1G21520 | 6.35587 | 83.2197 | 3.7 | 0 |
| AT5G19890 | AT5G19890 | 0.11012 | 1.27155 | 3.5 | 0.0000828 |
| AT1G16420 | MC8 | 0.09403 | 0.9566 | 3.3 | 0.000208394 |
| AT1G52560 | AT1G52560 | 0.09277 | 0.64057 | 2.8 | 0.0229653 |
| AT4G08780 | AT4G08780 | 7.62037 | 41.3088 | 2.4 | 2.47E-11 |
| AT4G08770 | Prx37 | 19.5044 | 103.508 | 2.4 | 2.22E-16 |
| AT5G05340 | PRX52 | 6.88871 | 20.509 | 1.6 | 0.00000491 |
| AT2G38390 | AT2G38390 | 6.05359 | 17.4199 | 1.5 | 0.0000504 |
| AT4G36430 | AT4G36430 | 4.42661 | 12.6194 | 1.5 | 0.000131116 |
| AT5G06720 | ATPA2 | 3.27123 | 8.0539 | 1.3 | 0.00369748 |
| AT5G39610 | ATNAC6 | 1.89284 | 4.38157 | 1.2 | 0.0198265 |
| AT4G23190 | CRK11 | 7.61116 | 17.0319 | 1.2 | 0.000103328 |
| AT2G18150 | AT2G18150 | 6.50389 | 13.8397 | 1.1 | 0.00219204 |
| AT3G01420 | DIOX1 | 8.68423 | 18.2481 | 1.1 | 0.00033264 |
| AT3G49120 | ATPCB | 206.322 | 429.775 | 1.1 | 0.000609777 |
| AT2G37130 | AT2G37130 | 70.8261 | 144.86 | 1.0 | 0.000295334 |
| AT3G49960 | AT3G49960 | 1.00424 | 0.16773 | −2.6 | 0.00166279 |
| AT1G30870 | AT1G30870 | 0.48317 | 0.09791 | −2.3 | 0.0283957 |
| AT5G05410 | DREB2 | 16.8563 | 3.79822 | −2.1 | 1.14E-08 |
| AT5G67400 | RHS19 | 0.70408 | 0.16062 | −2.1 | 0.0112083 |
| AT1G61120 | GES | 0.23429 | 0.06097 | −1.9 | 0.0280802 |
| AT4G26010 | AT4G26010 | 1.1279 | 0.32219 | −1.8 | 0.0156379 |
| AT5G37770 | TCH2 | 217.941 | 68.1689 | −1.7 | 1.68E-09 |
| AT1G05240 | AT1G05240 | 0.8244 | 0.27342 | −1.6 | 0.0446952 |
| AT1G05250 | AT1G05250 | 0.74497 | 0.24794 | −1.6 | 0.0491903 |
| AT2G41480 | AT2G41480 | 1.8151 | 0.60604 | −1.6 | 0.0198361 |
| AT4G21830 | ATMSRB7 | 2.48263 | 0.89057 | −1.5 | 0.0258255 |
| AT3G12580 | HSP70 | 2.38277 | 0.91385 | −1.4 | 0.00509735 |
| AT4G11290 | AT4G11290 | 25.1435 | 11.3171 | −1.2 | 0.000426591 |
| AT1G44970 | AT1G44970 | 14.5406 | 6.60913 | −1.1 | 0.00129019 |
| AT4G16270 | AT4G16270 | 6.41129 | 2.93899 | −1.1 | 0.0154319 |
Table 2.
The transcript alteration of genes in SA signaling pathway by EXB1 induction.
| Gene ID | Gene product | Value of mock-treated plants | Value of DEX-treated plants | Log2(fold change) | p_value |
|---|---|---|---|---|---|
| AT4G25560 | AtMYB18 | 0.0591953 | 0.551923 | 3.2 | 0.0288016 |
| AT5G54230 | MYB49 | 0.476331 | 2.56793 | 2.4 | 0.000347191 |
| AT2G14560 | LURP1 | 50.5996 | 205.256 | 2.0 | 2.07E-09 |
| AT5G22570 | WRKY38 | 3.43386 | 13.3867 | 2.0 | 0.0000101 |
| AT3G48920 | AtMYB45 | 2.04465 | 6.3486 | 1.6 | 0.00366468 |
| AT3G49690 | RAX3 | 0.720147 | 2.11352 | 1.6 | 0.0116923 |
| AT5G54610 | ANK | 4.91594 | 12.0211 | 1.3 | 0.00113844 |
| AT4G23170 | EP1 | 22.9788 | 49.4802 | 1.1 | 0.000166857 |
| AT3G01420 | DIOX1 | 8.68423 | 18.2481 | 1.1 | 0.00033264 |
| AT5G44420 | PDF1.2 | 39.1423 | 81.5765 | 1.1 | 0.00130954 |
| AT2G14580 | PRB1 | 9.5015 | 19.4753 | 1.0 | 0.00725166 |
| AT2G36890 | MYB38 | 4.35598 | 8.51135 | 1.0 | 0.0331253 |
| AT5G13320 | GDG1 | 2.22105 | 4.30273 | 1.0 | 0.0286926 |
Table 3.
The transcript alteration of genes in JA signaling pathway by EXB1 induction.
| Gene ID | Gene product | Value of mock-treated plants | Value of DEX-treated plants | Log2(fold change) | p_value |
|---|---|---|---|---|---|
| AT2G34600 | TIFY5B | 9.53484 | 1.03625 | −3.2 | 0.000000345 |
| AT1G72520 | LOX4 | 15.1331 | 1.64327 | −3.2 | 0 |
| AT4G11280 | ACS6 | 104.144 | 12.0035 | −3.1 | 0 |
| AT1G19640 | JMT | 1.42277 | 0.161271 | −3.1 | 0.0000465 |
| AT1G32640 | RD22BP1 | 104.287 | 16.5214 | −2.7 | 0 |
| AT5G42650 | AOS | 69.7379 | 11.2917 | −2.6 | 0 |
| AT1G17380 | JAZ5 | 23.6471 | 4.03 | −2.6 | 2.88E-11 |
| AT1G17420 | LOX3 | 24.9445 | 4.26703 | −2.5 | 0 |
| AT3G23250 | ATMYB15 | 21.882 | 4.00251 | −2.5 | 1.91E-10 |
| AT3G50060 | MYB77 | 82.2341 | 19.4365 | −2.1 | 8.29E-13 |
| AT1G28480 | GRX480 | 6.93256 | 1.65924 | −2.1 | 0.00078703 |
| AT3G15210 | ATERF-4 | 92.9679 | 24.0022 | −2 | 1.56E-11 |
| AT2G02990 | RNS1 | 5.97335 | 1.49915 | −2 | 0.00042616 |
| AT3G25780 | AOC3 | 75.5138 | 19.3517 | −2 | 2.32E-11 |
| AT1G61120 | GES | 0.234291 | 0.060973 | −1.9 | 0.0280802 |
| AT1G19180 | JAZ1 | 116.687 | 31.28 | −1.9 | 3.58E-11 |
| AT4G23600 | JR2 | 11.4154 | 3.30733 | −1.8 | 0.0000113 |
| AT4G37260 | MYB73 | 88.5454 | 27.6989 | −1.7 | 0.0000105 |
| AT2G24850 | TAT3 | 6.55538 | 2.32079 | −1.5 | 0.00067809 |
| AT3G52400 | SYP122 | 61.575 | 21.9425 | −1.5 | 0.00000012 |
| AT1G20510 | OPCL1 | 70.4878 | 24.4484 | −1.5 | 8.29E-08 |
| AT1G74430 | MYB95 | 12.0119 | 4.60133 | −1.4 | 0.00257587 |
| AT5G13930 | TT4 | 83.3234 | 31.2281 | −1.4 | 0.000000296 |
| AT5G24780 | VSP1 | 62.3915 | 23.2181 | −1.4 | 0.00000055 |
| AT1G54040 | TASTY | 2.71165 | 1.07254 | −1.3 | 0.0194946 |
| AT5G13220 | TIFY9 | 18.6439 | 7.46426 | −1.3 | 0.00469042 |
| AT4G05100 | AtMYB74 | 0.759768 | 0.322459 | −1.2 | 0.116597 |
| AT5G07690 | MYB29 | 39.86 | 17.8935 | −1.2 | 0.0000415 |
| AT5G60890 | ATMYB34 | 9.95635 | 4.55748 | −1.1 | 0.0041261 |
| AT5G61420 | AtMYB28 | 81.5909 | 36.9272 | −1.1 | 0.00026045 |
| AT5G64900 | PROPEP1 | 10.7115 | 4.92772 | −1.1 | 0.0226213 |
| AT5G24770 | VSP2 | 47.7935 | 21.5581 | −1.1 | 0.0000757 |
| AT3G45140 | LOX2 | 53.0825 | 26.9621 | −1 | 0.00040705 |
| AT3G17860 | TIFY6B | 7.76129 | 3.90539 | −1 | 0.0239613 |
| AT2G06050 | OPR3 | 58.2752 | 29.0943 | −1 | 0.00092024 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31270321), by the National Key Basic Research Program of People's Republic of China (Grant No. 973-2012CB944801) and by National Transformation Science and Technology Program (Grant No. 2014ZX08009003-003).
References
- 1.Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Cell Biol 2011; 12:211-21; PMID:21427763; http://dx.doi.org/ 10.1038/nrm3088 [DOI] [PubMed] [Google Scholar]
- 2.Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002; 14:S165-S83; PMID:12045276; http://dx.doi.org/ 10.1105/tpc.000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, et al.. The WRKY transcription factor WRKY71/ EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 2015; 27:3112-27; PMID:26578700; http://dx.doi.org/ 10.1105/tpc.15.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McSteen P, Leyser O. Shoot branching. Annu Rev Plant Biol 2005; 56:353-74; PMID:15862100; http://dx.doi.org/ 10.1146/annurev.arplant.56.032604.144122 [DOI] [PubMed] [Google Scholar]
- 5.Muller D, Schmitz G, Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006; 18:586-97; PMID:16461581; http://dx.doi.org/ 10.1105/tpc.105.038745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller T, Abbott J, Moritz T, Doerner P. Arabidopsis regulator of axillary meristems1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006; 18:598-611; PMID:16473968; http://dx.doi.org/ 10.1105/tpc.105.038588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz G, Tillmann E, Carriero F, Fiore C, Cellini F, Theres K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc Natl Acad Sci USA 2002; 99:1064-9; PMID:11805344; http://dx.doi.org/ 10.1073/pnas.022516199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeifetz D, David-Schwartz R, Borovsky Y, Paran I. CaBLIND regulates axillary meristem initiation and transition to flowering in pepper. Planta 2011; 234:1227-36; PMID:21773792; http://dx.doi.org/ 10.1007/s00425-011-1479-8 [DOI] [PubMed] [Google Scholar]
- 9.Bakshi M, Oelmuller R. WRKY transcription factors: Jack of many trades in plants. Plant Signal Behav 2014; 9:e27700; PMID:24492469; http://dx.doi.org/ 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunewald W, De Smet I, Lewis DR, Lofke C, Jansen L, Goeminne G, et al.. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci USA 2012; 109:1554-9; PMID:22307611; http://dx.doi.org/ 10.1073/pnas.1121134109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grunewald W, Karimi M, Wieczorek K, Van de Cappelle E, Wischnitzki E, Grundler F, Inzé D, Beeckman T, Gheysen G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol 2008; 148:358-68; PMID:18599655; http://dx.doi.org/ 10.1104/pp.108.119131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grunewald W, De Smet I, De Rybel B, Robert HS, van de Cotte B, Willemsen V, Grundler F, Inzé D, Beeckman T, Gheysen G. Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep 2013; 14:1136-42; PMID:24157946; http://dx.doi.org/ 10.1038/embor.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 1997; 11:605-12; PMID:9107046; http://dx.doi.org/ 10.1046/j.1365-313X.1997.11030605.x [DOI] [PubMed] [Google Scholar]
- 14.Chen L, Zhang L, Li D, Wang F, Yu D. WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA 2013; 110:E1963-71; PMID:23650359; http://dx.doi.org/ 10.1073/pnas.1221347110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 2013; 74:730-45; PMID:23451802; http://dx.doi.org/ 10.1111/tpj.12159 [DOI] [PubMed] [Google Scholar]
- 16.Wu LT, Zhong GM, Wang JM, Li XF, Song X, Yang Y. Arabidopsis WRKY28 transcription factor is required for resistance to necrotrophic pathogen, Botrytis cinerea. Afr J Microbiol Res 2011; 5:5481-8; http://dx.doi.org/ 10.5897/AJMR11.781 [DOI] [Google Scholar]
- 17.Xing DH, Lai ZB, Zheng ZY, Vinod KM, Fan BF, Chen ZX. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 2008; 1:459-70; PMID:19825553; http://dx.doi.org/ 10.1093/mp/ssn020 [DOI] [PubMed] [Google Scholar]
- 18.Yu Y, Liu Z, Wang L, Kim SG, Seo PJ, Qiao M, Wang N, Li S, Cao X, Park CM, et al.. WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J 2015; PMID:26643131; http://dx.doi.org/ 10.1111/tpj.13092 [DOI] [PubMed] [Google Scholar]


