Abstract
Epigenetic regulation controls multiple aspects of the plant development. The N-terminal tail of histone can be differently modified to regulate various chromatin activities. One of them, the trimethylation of histone H3 lysine 27 (H3K27me3) confers a repressive chromatin state with gene silencing. H3K27me3 is dynamically deposited and removed throughout development. While components of the H3K27me3 writer, Polycomb repressive complex 2 (PRC2), have been reported for almost 2 decades, it is only recently that JUMONJI (JMJ) proteins are reported as H3K27me3 demethylases, affirming the dynamic nature of histone modifications. This review highlights recent progress in plant epigenetic research, focusing on the H3K27me3 demethylases.
Keywords: Arabidopsis, demethylation, epigenetics, flowering time, H3K27me3, histone, methylation
Histone Methylation and Demethylation
The basic subunit of a chromatin is the nucleosome, which consists of DNA wrapped around the core histone octamer (2 molcules of each of the histone H2A, H2B, H3, and H4). The amino (N)-terminal tails of these histone proteins protrude out of the nucleosome unit and are subjected to extensive post-translational modifications, including but not limited to methylation, acetylation, phosphorylation, and ubiquitination. Some of the histone modifications (e.g. acetylation, phosphorylation) are proposed to function in changing the overall charge of the histone, thereby controlling the degree of condensation of the chromatin fiber.1 However, more evidences are suggesting that histone modifications (e.g., methylation, acetylation) may serve as binding platforms to recruit other protein complexes onto the chromatin.2 The histone code hypothesis propose that these histone marks may work in a sequential or cooperative manner to regulate downstream gene activities.1
Histone methylation is one of the better-studied histone modifications, which generally occur on the lysine (K) and arginine (R) residues on the N-terminal histone tails. Depending on the location of the amino acid residue and its degree of methylation, it can confer an activating or repressing chromatin status.2 This histone modification on lysine residues is mainly catalyzed by histone methyltransferases (HMTases) containing the SET (for Suppressor of variegation 3–9, Enhancer of zeste, and Trithorax) domain. In Arabidopsis there are 37 of these HMTases which could add 1, 2 or 3 methyl group to a lysine residue, serving as ‘writer’ proteins.3 Located at the globular domain of histone H3, the surface-exposed H3K79 residue can also be methylated by the Dot1 family of methyltransferases, a class of non-SET domain-containing HMTases.4 However, the Arabidopsis genome has no apparent H3K79 methylation enrichment and does not contain Dot1-like homolog, and it has been suggested that the H3K36me3 marks in Arabidopsis may assume the similar role of H3K79me3 as a transcription elongation mark in animals.5 Besides the lysine-specific HMTases, the Arabidopsis genome contains another small family of S-adenosylmethionine (SAM) binding-domain proteins that catalyzes the methylation on arginine residues, collectively known as the Protein Arginine Methyltransferases (PRMTs).6 The PRMTs can catalyze the monomethylation, asymmetric dimethylation or symmetric dimethylation of arginine of histone H3 and H4.
Antagonizing the effect of the HMTases are the histone demethylases, which function as ‘eraser’ proteins of these histone codes. Unlike acetylation, which dynamism had been shown through the discovery of histone acetyltransferases and deacetylases, histone methylation was previously thought to be irreversible due to the lack of histone demethylases. The human Peptidyl arginine deiminase 4 (PADI4) was the first enzyme shown to be able to antagonize histone arginine methylation.7,8 However, there are arguments whether to consider PADI4 a histone demethylase because it catalyzes the conversion of methyl-arginine to citrulline instead of an unmodified arginine. A protein-protein BLAST search suggests that the Arabidopsis genome contains no PADI homolog. The existence of other proteins to carry out a similar enzymatic citrullination reaction is uncertain.
The dynamics of histone methylation was finally rectified when the demethylation of lysine 4 of histone H3 (H3K4) was first demonstrated by human Lysine specific demethylase 1 (LSD1) in Hela cells.9 In Arabidopsis, there are 4 LSD1 homologues, and like the human LSD1, some of them can demethylate modified mono-/dimethylated H3K4.10,11 Another class of histone demethylase, the JUMONJI (JMJ) proteins, can target all mono-/di-/trimethylated (me1/me2/me3) lysine residues. There are 21 JMJ histone demethylases in Arabidopsis which can be categorised into 5 groups based on their overall domain architectures and the similarities of the JmjC domain sequences (Table 1). These groups are named KDM5/JARID1, KDM4/JHDM3/JMJD2, KDM3/JHDM2, JMJD6, and JmjC domain-only groups.12,13 In this review, we focused on recent publications on the Arabidopsis H3K27me3 demethylases from two groups of JMJ proteins, namely the KDM4/JHDM3/JMJD2 and JmjC domain-only group, after briefing on H3K27me3 deposition and Polycomb group (PcG) activities (Fig. 1).
Figure 1.
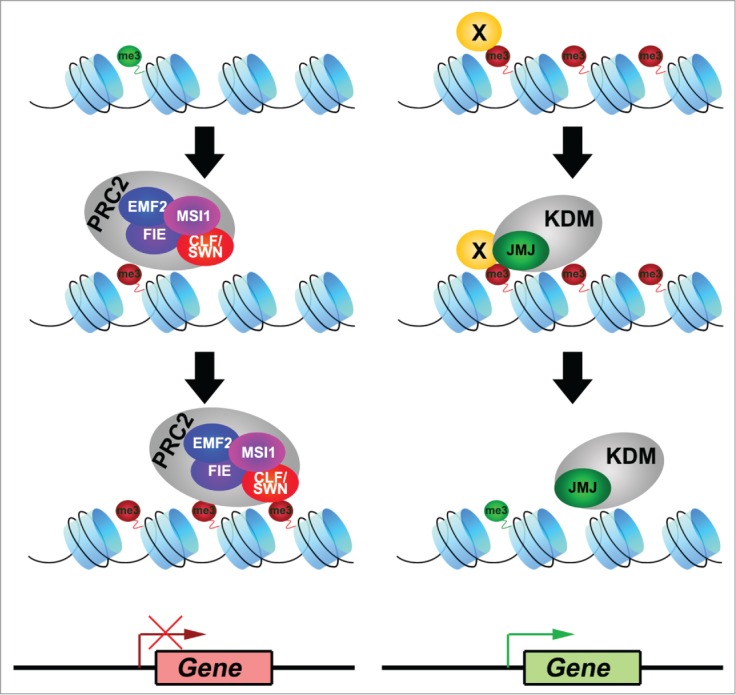
Antagonistic action of PRC2 and JMJ on gene regulation. Chromatin marked with active histone modification H3K4me3/H3K36me3 (green me3 circle) is in a permissive state of transcription. Recruitment of the Polycomb repressive complex 2 (PRC2) occurs possibly via the polycomb response elements (PREs). PRC2 complex containing core components of EMBRYONIC FLOWER 2 (EMF2), MULTICOPY SUPRESSOR OF IRA 1 (MSI1), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), CURLY LEAF (CLF) or SWINGER (SWN) mediates the deposition and spreading of the repressive histone modification H3K27me3 (red me3 circle) across the region. This results in a repressive chromatin state and silences gene expression at the region. Functioning antagonistically, histone lysine demethylase (KDM) complex containing JUMONJI (JMJ) proteins, such as EARLY FLOWERING 6 (ELF6), RELATIVE OF EARLY FLOWERING 6 (REF6), JMJ30 or JMJ32, is recruited by unknown factors (factor X) to its target sites. The catalytic JMJ demethylases remove the H3K27me3 marks from the region. Coupled with the deposition of activating histone modifications, genes at the region can resume active transcription. These antagonistic actions of PRC2 and JMJ regulate the expression of genes including, but not limited to, the flowering regulators FLOWERING LOCUS C (FLC), FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), to govern the flowering time of Arabidopsis.
Actions of PRC1 and PRC2 on Gene Repression
The H3K27me3 histone modification is correlated with gene repression and is deposited by the PcG proteins.2 In the Arabidopsis genome, at least 25% of the genes are targeted by H3K27me3 in seedlings, and these epigenetic marks are dynamically regulated during the plant's growth and development.5,14,15 The PcG proteins were first described in Drosophila and 2 of the most well-characterized PcG complexes are polycomb repressive complex 1 (PRC1) and PRC2. The Drosophila PRC2 complex contains 4 main components (Extra sex comb (Esc), Enhancer of zeste (E(z)), Suppressor of zeste 12 (Su(z)12), and p55) and catalyzes the trimethylation of H3K27 via the catalytic E(z) HMTase.16 In addition, the Drosophila PRC1 complex, which consists of main subunits Polycomb (Pc), Polyhomeotic (Ph), and the Ring finger proteins Posterior sex comb (Psc) and dRING/Sex combs extra (Sce), catalyzes the monoubiquitination of lysine 119 of histone H2A (H2AK119ub/H2Aub) through the RING ubiquitin ligase.16
In animals, the prevailing hierarchical model of PcG action has been well accepted, in which the PRC2 is recruited by the cis-acting Polycomb response elements (PREs) and deposits the H3K27me3 mark onto specific location to establish the repressive chromatin state (Fig. 1).16 PRC1 is then recruited to the region due to the specificity of its chromodomain (of Pc subunit) toward H3K27me3 to monoubiquitinate H2AK119 and further reinforces the stable repressive status.16 However, recent findings revealed that H3K27me3 is dispensable for PRC1 recruitment at Drosophila PREs.17 Moreover, there are genes targeted by H3K27me3 but lack PRC1 occupancy,18 and vice versa PRC1-mediated H2A monoubiquitination can occur independent of PRC2 activity, as in X-inactivation mediated by Xist RNA.19 Thus, it appears that gene repression may occur via the hierarchical or cooperative action of H3K27me3 and H2AK119ub marks or independently by either of the histone modifications.
The Arabidopsis genome contains homologs of all 4 core PRC2 components, and they are functionally conserved in which they mediate gene repression via the deposition of H3K27me3 marks on the chromatin (Fig. 1, reviewed in2,20). On the other hand, plants were once thought not to contain PRC1 complex due to the lack of some of the core PRC1 components, such as the H3K27me3-binding Pc protein. However, it was later shown that the homolog of the animal H3K9me2/3-binding Heterochromatin protein 1 (HP1) in Arabidopsis, LIKE HETEROCHROMATIN PROTEIN 1/TERMINAL FLOWER 2 (LHP1/TFL2) acts as the H3K27me3-binding protein and recruit putative the PRC1-like complex.21 Furthermore, recent studies identified 5 PRC1 RING finger proteins in Arabidopsis: AtRING1A/B (homologs of Drosophila Sce) and AtBMI1A/B/C (homologs of Psc) (reviewed in22,23). TFL2, AtRING1A/B,24 and AtBMI1A/B/C25,26 interact to form a PRC1-like complex in Arabidopsis and catalyze the deposition of H2AK119ub.25 Loss-of-function of atring1a/b and atbmi1a/b causes dedifferentiation of vegetative tissues into callus-like structures, which mimics the phenotype of clf swn27 and vrn2 emf228 double mutants, indicating that AtRING and AtBMI1 genes function in a similar pathway as PRC2 complex and are required to maintain the differentiated state of somatic cells.
It is noteworthy that a recent study showed that the triple mutant of the PRC1 components atbmi1a/b/c shows dramatic decrease of H3K27me3 on seed maturation genes, whereas the double mutant of the PRC2 components clf swn increased H2AK119ub, highlighting the direct or indirect regulation of each other's activity.29 Moreover, genome-wide H2AK119ub level was not significantly affected in the clf swn mutant, suggesting that PRC1 targeting can occur through pathways independent of PRC2 in plants.29 The Drosophila non-canonical PRC1 complex, called the dRing-associated factors (dRAF) complex, comprises Sce (Arabidopsis homologs: AtRING1A/B), Psc (Arabidopsis homologs: AtBMI1A/B/C), and KDM2, but lacks 2 components of the canonical PRC1 complex, Pc and Ph. Interestingly, mutation of Sce or Psc significantly decreases H2AK119ub levels, whereas depletion of Pc or Ph yield no effect on global H2AK119ub levels.30 Thus, it is suggested that, compared to the canonical PRC1 complex, the plant PRC1-like complex may more resemble the dRAF complex, and may be recruited to the chromatin independently of H3K27me3 marks to catalyze the H2AK119 monoubiquitination.22
KDM3/JHDM3/JMJD2 Group H3K27me3 Demethylases Regulate Flowering Time
The KDM4/JHDM3/JMJD2 group of Arabidopsis JMJ proteins contain 3 members: AT5G04240/JMJ11/EARLY FLOWERING 6 (ELF6), AT3G48430/JMJ12/RELATIVE OF EARLY FLOWERING 6 (REF6) and AT5G46910/JMJ13. ELF6 and REF6 were genetically identified to have opposite functions in flowering time regulation.31 Although the HMTase of H3K27me3 in Arabidopsis has been reported for almost 2 decades, the discovery of its demethylase has not been as successful. This is mainly because the metazoan H3K27me3 demethylases, Ubiquitously transcribed tetratricopeptide repeat X (UTX) and JMJD3,32 do not have a homolog in plants. It is only recently that ELF633 and REF634 are reported as the H3K27me3 demethylase in Arabidopsis.
REF6 is the first plant H3K27me2/3 demethylase reported.34 REF6 is originally shown to function as a repressor of FLOWERING LOCUS C (FLC), the expression of which is increased in the ref6 mutant.31,35 However, overexpression of REF6 causes drastic decrease of global H3K27me2/3 levels and produces phenotypes which resemble PRC2 mutants (i.e. early-flowering, curled leaf, embryonic flowering, etc).34 The early-flowering phenotype is attributed to an FLC-independent up-regulation of the floral integrator FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1).31,34 Moreover, the REF6 protein decreased H3K27me2/3 levels both in vitro and in vivo, and ref6 mutant showed H3K27me3 hypermethylation in several hundred endogenous genes, confirming its role as an H3K27 demethylase.34 Although REF6 contains C2H2 zinc finger domains at its C-terminal as a putative DNA-binding domain, it likely function as a complex like the metazoan UTX,36 and requires other transcription factors to be recruited to its target genes. Recently, it is shown that the nuclear factor Y (NF-Y) complex interacts and recruits REF6 to the SOC1 locus to remove the repressive H3K27me3 marks, thus promoting SOC1 expression.37
ELF6 was initially screened to repress the floral integrators in the photoperiod pathway, whereby mutation of elf6 results in increased FT and SOC1 expressions and an early-flowering phenotype.31 It is later reported that ELF6 also participates in the reprogramming of the epigenetic state of the floral repressor FLC.33 FLC is epigenetically silenced during vernalization (prolonged exposure to cold) to promote flowering upon returning to a warm condition,38,39 and is reactivated during reproduction to ensure proper floral behavior in the next generation.40 A hypomorphic mutant allele of elf6–5 causes defect in the epigenetic resetting of FLC in which the H3K27me3 level at the locus is maintained at a higher level in the next generation when the parental plant is vernalized.33 Furthermore, ELF6 is shown to possess H3K27me2/3 demethylation activity in vivo.33
The KDM3/JHDM3/JMJD2 group contains one more member, JMJ13, which has no functional report thus far. The JMJ13 protein contains all conserved cofactor-binding amino acids, suggesting that it is enzymatically active.12 However, its potential histone demethylation capability remains to be elucidated.
JmjC Domain-Only Group Contribute to Circadian and Thermosensory Regulations
The JmjC domain-only group of Arabidopsis JMJ proteins consists of 3 members, AT3G20810/JMJ30, AT5G19840/JMJ31, AT3G45880/JMJ32. Although AT5G63080/JMJ20 also does not have other recognizable protein domains besides a JmjC domain, its JmjC domain sequence is more similar to members of the JMJD6 group (AT5G06550/JMJ22 and AT1G78280/JMJ21). Furthermore, JMJ20 and JMJ22 are reported to function redundantly in controlling seed germination and catalyze histone arginine demethylation.41
Of the 3, JMJ30 has been previously described as functioning in the circadian systems.42,43 In the central circadian clock loop, TIMING OF CAB1 EXPRESSION 1 (TOC1) promotes the expression of morning-phased clock genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY); CCA1 and LHY in turn repress TOC1 in the evening, completing the feedback loop.44 Functioning in a similar manner as TOC1 in the central clock loop, JMJ30 acts in concert with TOC1 to promote CCA1 and LHY expression;43 subsequently, CCA1 and LHY directly bind JMJ30 and repress its expression.42 Moreover, the jmj30 mutant has shorter circadian period,42,43 and decreased CCA1 and LHY expression,43 asserting its role in circadian regulation.
We recently showed that JMJ30 and its homolog JMJ32 contribute to the thermosensory pathway of flowering control by delaying the H3K27me3-mediated repression of FLC at higher temperatures.45 Double mutant of jmj30 jmj32 produces an early-flowering phenotype when grown at elevated temperatures. Conversely, overexpression of JMJ30 results in a FLC-dependent late-flowering phenotype, with reduced H3K27me3 levels at the FLC locus, increased FLC mRNA expression and reduced FT and SOC1 expressions.42,45 Furthermore, JMJ30 is able to remove H3K27me3 in vitro and in vivo, affirming its role as an H3K27 demethylase. Although JMJ30 is diurnally expressed, the JMJ30 protein is stabilized at higher temperature which leads to its increased accumulation. JMJ30 directly binds to the FLC locus, and its heat-stabilized activity likely maintain a permissive chromatin state at the locus.45 The JMJ30/JMJ32-mediated FLC derepression thus constitutes a parallel mechanism in regulating floral transition with the other thermosensory pathways.46
This JMJ group contains another member, JMJ31, which may have lost its demethylase activity, since it has a different variant in one of its conserved key α-ketoglutarate (α-KG) binding residues.12,47 Intriguingly, the first JMJ protein isolated in mouse, Jarid2/Jumonji, also has the same amino acid substitution.48 The mouse Jarid2/Jumonji protein regulates multiple developmental processes including cardiac, haematopoietic and hepatic development.49 Despite being enzymatically inactive, Jarid2/Jumonji interacts with other histone modifier to transcriptionally regulate its target genes.49,50 Hence, it would be interesting to study the potential function of JMJ31 in modulating chromatin status of genes in Arabidopsis.
Substrate Specificity of JMJ Protein
More than a quarter of the Arabidopsis genes are targeted by H3K27me3. However, the fact that the phenotypes of elf6, ref6, jmj30, jmj32 single mutants, elf6 ref6, jmj30 jmj32 double mutants and ref6 jmj30 jmj32 triple mutant are weaker than those of the metazoan H3K27 demethylase mutant hints that there might be more H3K27me3 demethylases waiting to be uncovered, or simply because plants have greater developmental plasticity or have more robust and flexible histone modification feedback.45,51 Thus it might be interesting to observe the phenotype of the quadruple mutant of all currently known H3K27me3 demethylases, elf6 ref6 jmj30 jmj32, or a quintuple mutant of all potentially functional members of the KDM3/JHDM3/JMJD2 group and JmjC domain-only group, elf6 ref6 jmj13 jmj30 jmj32.
JMJ proteins from the same group are implicated to target similar histone modifications.12,47 Despite residing in a different group with considerable diversifications in terms of domain architecture and JmjC domain sequence, members from the KDM3/JHDM3/JMJD2 group and JmjC domain-only group both target H3K27me3 marks. Such phenomenon is also observed in the mammalian genome as members from both the KDM2/JHDM1 group and KDM3/JHDM3/JMJD2 group are capable of H3K36 demethylation.52 Crystal structures of REF6, ELF6, JMJ30, and JMJ32, with their cognate substrate (H3K27me3), will be helpful to determine their substrate specificity-determining amino acid residues.
Interestingly, the animal KDM3/JHDM3/JMJD2 group members target methylated H3K9 and H3K36,47 and other previous studies on ELF6 and REF6 also reported similar substrate preferences.35,51 One study shows that ELF6 and REF6 participate in the brassinosteroid (BR) signaling cascade through the transcription factor BRI1-EMS-SUPPRESSOR 1 (BES1).51 BES1 recruits ELF6 and REF6 to the BR target genes (e.g. TOUCH4 (TCH4)) and remove the H3K9 methylation to increase its expression, and thus BR-responsive phenotypes such as reduced cell elongation was observed in elf6 and ref6 mutants.51 Another study reports that recombinant chimeric REF6–6×His is able to demethylate H3K4me2/3 and H3K36me2/3.35 Moreover, REF6 can directly bind to the FLC locus and remove the activating histone modifications H3K4 and H3K36 methylation.35 In addition, FLC expression is strongly up-regulated, rather than down-regulated, in the ref6 mutant,31,34,35 indicating that this increased FLC expression may be the main cause of the late-flowering phenotype in ref6 mutant. This is supported by the observation that crossing of ref6 flc double mutant rescued the ref6 late-flowering phenotype to a wild-type level.31 Similar to REF6, JMJ30 is also recently reported to be able to demethylate H3K36me2/3 in vitro.53 A MYB transcription factor EARLY FLOWERING MYB PROTEIN (EFM) recruits JMJ30 to the FT locus and increases H3K36me2 levels at the region.53
The conflicting results among these reports suggest that JMJ proteins might have context dependent substrate specificity. Indeed, the metazoan JHDM3/JMJD2 group proteins such as JMJD2A show dual substrate specificities for 2 antagonizing H3K9 and H3K36 methylation marks, a repressive and activating mark respectively.47 These differences may also be due to indirect effects of histone modification crosstalks, as activating and repressing histone marks frequently show opposing dynamics.54,55 Furthermore, other JMJ proteins may retain minimal degree of substrate-flexibility and undertake non-endogenous function to compensate an extreme situation where one type of histone demethylation is compromised, as seen in the compensatory mechanism in DNA methylation.56 Similar compensatory mechanisms are evident in transcription factor activity. The carpel and fruit development gene FRUITFULL (FUL) can compensate the loss of the 2 floral meristem identity genes apetala1 (ap1) and cauliflower (cal) to mediate flower initiation, such that triple mutant of ful ap1 cal causes an even more severe phenotype than ap1 cal and produces leafy shoots in place of flowers.57 Similarly, the carpel-/fruit-specific MADS-box genes SHATTERPROOF1 (SHP1) and SHP2 also assume a non-endogenous role of AGAMOUS (AG)-compensatory activity in carpel formation in the apetala2 (ap2) ag double mutant.58 These partial compensations may be evolutionary remnants of the MADS-box and JMJ genes which retain some functional plasticity.
Conclusions and Perspectives
Post-translational histone modifications at their N-terminal tails regulate chromatin activity in distinct biological settings. Having common histone modifications such as H3K27me3 will allow group of genes functioning in the same developmental process to be regulated in a similar manner, e.g., seed maturation genes are repressed during vegetative growth and only activated during seed development.59,60 Moreover, H3K27 methylation acting in concert with other histone modifications will produce new ‘histone code’, such as the poised chromatin state of bivalent H3K27me3 and H3K4me3 marks.61,62 Although methylated histone marks may be diluted with cell cycle progression,63 having active histone demethylase such as JMJ and LSD1 proteins will allow faster response to endogenous and environmental cues.
We are beginning to understand the enzymatic activity of JMJ proteins, yet we still do not fully understand their biological roles. Many Arabidopsis genes are targeted by histone modifications but jmj mutants mostly produce flowering phenotypes.31,45,64-67 Although hundreds of genes are mis-regulated in the jmj mutants, only few of the target genes are studied in detail. Thus, genome wide ChIP-seq of JMJ proteins coupled with the expression data will aid in identifying their direct targets. Moreover, the mechanism of recruitment of these histone modifiers to specific target is of interest. A screen of JMJ-interacting partners may identify their recruiting transcription factors and also potentially uncover their roles in a larger complex. Further genetic, biochemical, and molecular analysis would increase our understanding of the functions and mechanisms of histone methylation in epigenetic regulation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Table 1.
Arabidopsis JMJ histone demethylases and their substrate specificity.
| Group | Name | Locus | Substratea | Methodsb | Reference |
|---|---|---|---|---|---|
| KDM5/JARID1 | JMJ15/MEE27 | AT2G34880 | H3K4me1/2/3 | IV | 6 |
| JMJ18 | AT1G30810 | H3K4me2/3 | Ch, IT, HI | 64 | |
| JMJ14 | AT4G20400 | H3K4me1/2/3 | Ch, IT, IV | 65–67 | |
| JMJ16 | AT1G08620 | NR | — | — | |
| JMJ19 | AT2G38950 | NR | — | — | |
| JMJ17 | AT1G63490 | NR | — | — | |
| KDM4/JHDM3/JMJD2 | JMJ12/REF6 | AT3G48430 | H3K9me3 | Ch | 51 |
| H3K4me2/3H3K36me2/3 | Ch, IT | 35 | |||
| H3K27me2/3 | Ch, IT, IV, HI | 34 | |||
| JMJ11/ELF6 | AT5G04240 | H3K9me3 | Ch | 51 | |
| H3K27me2/3 | Ch, IT, IV | 33 | |||
| JMJ13 | AT5G46910 | NR | — | — | |
| KDM3/JHDM2 | JMJ26 | AT1G11950 | NR | — | — |
| JMJ29 | AT1G62310 | NR | — | — | |
| JMJ25/IBM1 | AT3G07610 | H3K9me1/2 | Ch, IV | 68 | |
| JMJ27 | AT4G00990 | NR | — | — | |
| JMJ24 | AT1G09060 | NR | — | — | |
| JMJ28 | AT4G21430 | NR | — | — | |
| JMJD6 | JMJ22 | AT5G06550 | H3R2me2H4R3me1/2s | Ch | 41 |
| JMJ21 | AT1G78280 | NR | — | — | |
| JMJ20 | AT5G63080 | H3R2me2H4R3me1/2s | Ch, IT | 41 | |
| JmjC domain-only | JMJ30 | AT3G20810 | H3K27me2/3 | Ch, IT, IV | 45 |
| H3K36me2/3 | Ch, IT | 53 | |||
| JMJ31 | AT5G19840 | NR | — | — | |
| JMJ32 | AT3G45880 | H3K27me3 | Ch, IV | 45 |
NR, No report].
Ch, chromatin immunoprecipitation (ChIP); IT, in vitro demethylation assay; IV, in vivo demethylaion assay; HI, total histone immunoblot].
Funding
This work was supported by research grants to T.I. from Temasek Life Sciences Laboratory (TLL), and the National Research Foundation, Prime Minister's Office, Singapore under its Competitive Research Program (CRP Award No. NRF-CRP001–108). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41-5; PMID:10638745 [DOI] [PubMed] [Google Scholar]
- 2.Gan ES, Huang J, Ito T. Functional roles of histone modification, chromatin remodeling and microRNAs in Arabidopsis flower development. Int Rev Cell Mol Biol 2013; 305:115-61; PMID:23890381 [DOI] [PubMed] [Google Scholar]
- 3.Thorstensen T, Grini PE, Aalen RB. SET domain proteins in plant development. Biochim Biophys Acta 2011; 1809:407-20; PMID:21664308 [DOI] [PubMed] [Google Scholar]
- 4.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 2002; 12:1052-8; PMID:12123582 [DOI] [PubMed] [Google Scholar]
- 5.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al.. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J 2011; 30:1928-38; PMID:21487388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Ann Rev Plant Biol 2010; 61:395-420; PMID:20192747; http://dx.doi.org/ 10.1146/annurev.arplant.043008.091939 [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al.. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004; 306:279-83; PMID:15345777 [DOI] [PubMed] [Google Scholar]
- 8.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, et al.. Histone deimination antagonizes arginine methylation. Cell 2004; 118:545-53; PMID:15339660 [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119:941-53; PMID:15620353 [DOI] [PubMed] [Google Scholar]
- 10.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science 2003; 302:1751-4; PMID:14593187 [DOI] [PubMed] [Google Scholar]
- 11.Jiang D, Yang W, He Y, Amasino RM. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007; 19:2975-87; PMID:17921315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu F, Li G, Cui X, Liu C, Wang XJ, Cao X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 2008; 50:886-96; PMID:18713399 [DOI] [PubMed] [Google Scholar]
- 13.Hong EH, Jeong YM, Ryu JY, Amasino RM, Noh B, Noh YS. Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol Cells 2009; 27:481-90; PMID:19390830 [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 2007; 5:e129; PMID:17439305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. Dynamic regulation of H3K 27 trimethylation during Arabidopsis differentiation. PLoS Genet 2011; 7:e1002040; PMID:21490956; http://dx.doi.org/ 10.1371/journal.pgen.1002040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343-9; PMID:21248841; http://dx.doi.org/ 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev 2009; 19:150-8; PMID:19345089; http://dx.doi.org/ 10.1016/j.gde.2009.03.001 [DOI] [PubMed] [Google Scholar]
- 18.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al.. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008; 4:e1000242; PMID:18974828; http://dx.doi.org/ 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 2006; 25:3110-22; PMID:16763550; http://dx.doi.org/ 10.1038/sj.emboj.7601187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bemer M, Grossniklaus U. Dynamic regulation of Polycomb group activity during plant development. Curr Opin Plant Biol 2012; 15:523-9; PMID:22999383; http://dx.doi.org/ 10.1016/j.pbi.2012.09.006 [DOI] [PubMed] [Google Scholar]
- 21.Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 2007; 3:e86; PMID:17542647; http://dx.doi.org/ 10.1371/journal.pgen.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calonje M. PRC1 marks the difference in plant PcG repression. Mol Plant 2014; 7:459-71; PMID:24177684; http://dx.doi.org/ 10.1093/mp/sst150 [DOI] [PubMed] [Google Scholar]
- 23.Molitor A, Shen WH. The polycomb complex PRC1: composition and function in plants. J Genet Genom 2013; 40:231-8; PMID:23706298; http://dx.doi.org/ 10.1016/j.jgg.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 24.Chen D, Molitor A, Liu C, Shen WH. The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res 2010; 20:1332-44; PMID:21060339; http://dx.doi.org/ 10.1038/cr.2010.151 [DOI] [PubMed] [Google Scholar]
- 25.Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M. Keeping cell identity in Arabidopsis requires PRC1 RING-Finger homologs that catalyze H2A monoubiquitination. Curr Biol 2010; 20:1853-9; PMID:20933424; http://dx.doi.org/ 10.1016/j.cub.2010.09.046 [DOI] [PubMed] [Google Scholar]
- 26.Li W, Wang Z, Li J, Yang H, Cui S, Wang X, Ma L. Overexpression of AtBMI1C, a polycomb group protein gene, accelerates flowering in Arabidopsis. PLoS One 2011; 6:e21364; PMID:21701597; http://dx.doi.org/ 10.1371/journal.pone.0021364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 2004; 131:5263-76; PMID:15456723; http://dx.doi.org/ 10.1242/dev.01400 [DOI] [PubMed] [Google Scholar]
- 28.Schubert D, Clarenz O, Goodrich J. Epigenetic control of plant development by Polycomb-group proteins. Curr Opin Plant Biol 2005; 8:553-61; PMID:16043386; http://dx.doi.org/ 10.1016/j.pbi.2005.07.005 [DOI] [PubMed] [Google Scholar]
- 29.Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol 2013; 23:1324-9; PMID:23810531; http://dx.doi.org/ 10.1016/j.cub.2013.05.050 [DOI] [PubMed] [Google Scholar]
- 30.Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 2008; 22:2799-810; PMID:18923078; http://dx.doi.org/ 10.1101/gad.484208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M, Jung KJ, Doyle MR, Amasino RM, Noh YS. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004; 16:2601-13; PMID:15377760; http://dx.doi.org/ 10.1105/tpc.104.025353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007; 449:731-4; PMID:17713478; http://dx.doi.org/ 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- 33.Crevillen P, Yang H, Cui X, Greeff C, Trick M, Qiu Q, Cao X, Dean C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 2014; 515:587-90; PMID:25219852; http://dx.doi.org/ 10.1038/nature13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 2011; 43:715-9; PMID:21642989; http://dx.doi.org/ 10.1038/ng.854 [DOI] [PubMed] [Google Scholar]
- 35.Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS. Growth habit determination by the balance of histone methylation activities in Arabidopsis. EMBO J 2010; 29:3208-15; PMID:20711170; http://dx.doi.org/ 10.1038/emboj.2010.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007; 318:447-50; PMID:17761849; http://dx.doi.org/ 10.1126/science.1149042 [DOI] [PubMed] [Google Scholar]
- 37.Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun 2014; 5:4601; PMID:25105952; http://dx.doi.org/ 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- 38.Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004; 427:164-7; PMID:14712277; http://dx.doi.org/ 10.1038/nature02269 [DOI] [PubMed] [Google Scholar]
- 39.Sung S, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004; 427:159-64; PMID:14712276; http://dx.doi.org/ 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- 40.Choi J, Hyun Y, Kang MJ, In Yun H, Yun JY, Lister C, Dean C, Amasino RM, Noh B, Noh YS, et al.. Resetting and regulation of Flowering Locus C expression during Arabidopsis reproductive development. Plant J 2009; 57:918-31; PMID:19121105; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03776.x [DOI] [PubMed] [Google Scholar]
- 41.Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS. Control of seed germination by light-induced histone arginine demethylation activity. Dev Cell 2012; 22:736-48; PMID:22483719; http://dx.doi.org/ 10.1016/j.devcel.2012.01.024 [DOI] [PubMed] [Google Scholar]
- 42.Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol 2011; 155:906-15; PMID:21139085; http://dx.doi.org/ 10.1104/pp.110.167015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones MA, Covington MF, DiTacchio L, Vollmers C, Panda S, Harmer SL. Jumonji domain protein JMJD5 functions in both the plant and human circadian systems. Proc Natl Acad Sci USA 2010; 107:21623-8; PMID:21115819; http://dx.doi.org/ 10.1073/pnas.1014204108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Curr Biol 2012; 22:R648-57; PMID:22917516; http://dx.doi.org/ 10.1016/j.cub.2012.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gan ES, Xu Y, Wong JY, Goh JG, Sun B, Wee WY, Huang J, Ito T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat Commun 2014; 5:5098; PMID:25267112; http://dx.doi.org/ 10.1038/ncomms6098 [DOI] [PubMed] [Google Scholar]
- 46.Capovilla G, Schmid M, Pose D. Control of flowering by ambient temperature. J Exp Bot 2015; 66:59-69; PMID:25326628; http://dx.doi.org/ 10.1093/jxb/eru416 [DOI] [PubMed] [Google Scholar]
- 47.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715-27; PMID:16983801; http://dx.doi.org/ 10.1038/nrg1945 [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev 1995; 9:1211-22; PMID:7758946; http://dx.doi.org/ 10.1101/gad.9.10.1211 [DOI] [PubMed] [Google Scholar]
- 49.Mysliwiec MR, Carlson CD, Tietjen J, Hung H, Ansari AZ, Lee Y. Jarid2 (Jumonji, AT rich interactive domain 2) regulates NOTCH1 expression via histone modification in the developing heart. J Biol Chem 2012; 287:1235-41; PMID:22110129; http://dx.doi.org/ 10.1074/jbc.M111.315945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 2010; 464:306-10; PMID:20075857; http://dx.doi.org/ 10.1038/nature08788 [DOI] [PubMed] [Google Scholar]
- 51.Yu X, Li L, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci USA 2008; 105:7618-23; PMID:18467490; http://dx.doi.org/ 10.1073/pnas.0802254105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng SS, Kavanagh KL, McDonough MA, Butler D, Pilka ES, Lienard BM, Bray JE, Savitsky P, Gileadi O, von Delft F, et al.. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 2007; 448:87-91; PMID:17589501; http://dx.doi.org/ 10.1038/nature05971 [DOI] [PubMed] [Google Scholar]
- 53.Yan Y, Shen L, Chen Y, Bao S, Thong Z, Yu H. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev Cell 2014; 30:437-48; PMID:25132385; http://dx.doi.org/ 10.1016/j.devcel.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Howard M, Dean C. Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 2014; 24:1793-7; PMID:25065750; http://dx.doi.org/ 10.1016/j.cub.2014.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carles CC, Fletcher JC. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev 2009; 23:2723-8; PMID:19952107; http://dx.doi.org/ 10.1101/gad.1812609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al.. A role for RNAi in the selective correction of DNA methylation defects. Science 2009; 323:1600-4; PMID:19179494; http://dx.doi.org/ 10.1126/science.1165313 [DOI] [PubMed] [Google Scholar]
- 57.Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000; 127:725-34; PMID:10648231 [DOI] [PubMed] [Google Scholar]
- 58.Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003; 424:85-8; PMID:12840762; http://dx.doi.org/ 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- 59.Kim SY, Lee J, Eshed-Williams L, Zilberman D, Sung ZR. EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet 2012; 8:e1002512; PMID:22457632; http://dx.doi.org/ 10.1371/journal.pgen.1002512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, Goodrich J, Renou JP, Grini PE, Colot V, et al.. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet 2011; 7:e1002014; PMID:21423668; http://dx.doi.org/ 10.1371/journal.pgen.1002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saleh A, Al-Abdallat A, Ndamukong I, Alvarez-Venegas R, Avramova Z. The Arabidopsis homologs of trithorax (ATX1) and enhancer of zeste (CLF) establish ‘bivalent chromatin marks’ at the silent AGAMOUS locus. Nucl Acids Res 2007; 35:6290-6; PMID:17881378; http://dx.doi.org/ 10.1093/nar/gkm464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lesch BJ, Page DC. Poised chromatin in the mammalian germ line. Development 2014; 141:3619-26; PMID:25249456; http://dx.doi.org/ 10.1242/dev.113027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun B, Looi LS, Guo S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 2014; 343:1248559; PMID:24482483; http://dx.doi.org/ 10.1126/science.1248559 [DOI] [PubMed] [Google Scholar]
- 64.Yang H, Han Z, Cao Y, Fan D, Li H, Mo H, Feng Y, Liu L, Wang Z, Yue Y, et al.. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet 2012; 8:e1002664; PMID:22536163; http://dx.doi.org/ 10.1371/journal.pgen.1002664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeong JH, Song HR, Ko JH, Jeong YM, Kwon YE, Seol JH, Amasino RM, Noh B, Noh YS. Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS One 2009; 4:e8033; PMID:19946624; http://dx.doi.org/ 10.1371/journal.pone.0008033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu F, Cui X, Zhang S, Liu C, Cao X. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 2010; 20:387-90; PMID:20177424; http://dx.doi.org/ 10.1038/cr.2010.27 [DOI] [PubMed] [Google Scholar]
- 67.Yang W, Jiang D, Jiang J, He Y. A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 2010; 62:663-73; PMID:20202164; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04182.x [DOI] [PubMed] [Google Scholar]
- 68.Inagaki S, Miura-Kamio A, Nakamura Y, Lu F, Cui X, Cao X, Kimura H, Saze H, Kakutani T. Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J 2010; 29:3496-506; PMID:20834229; http://dx.doi.org/ 10.1038/emboj.2010.227 [DOI] [PMC free article] [PubMed] [Google Scholar]


