ABSTRACT
DNA is usually known as the molecule that carries the instructions necessary for cell functioning and genetic inheritance. A recent discovery reported a new functional role for extracellular DNA. After fragmentation, either by natural or artificial decomposition, small DNA molecules (between ∼50 and ∼2000 bp) exert a species specific inhibitory effect on individuals of the same species. Evidence shows that such effect occurs for a wide range of organisms, suggesting a general biological process. In this paper we explore the possible molecular mechanisms behind those findings and discuss the ecological implications, specifically those related to plant species coexistence.
KEYWORDS: Autotoxicity, extracellular DNA, litter decomposition, plant-soil negative feedback, species interactions
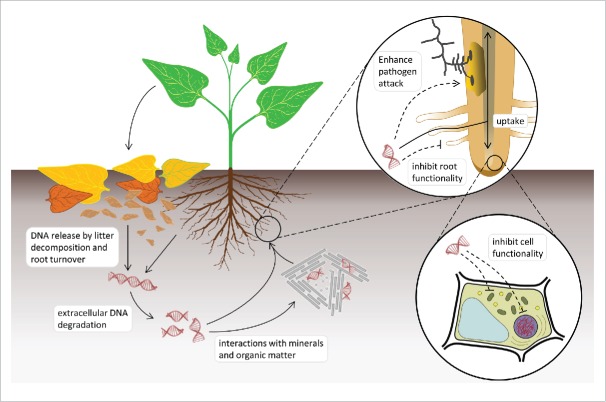
In two recent papers, Mazzoleni et al.1,2 reported on the discovery that fragmented extracellular self-DNA (i.e. DNA originating from conspecifics) produces species-specific inhibitory effects in plants. Mazzoleni et al.2 described experimental observations of autotoxicity, i.e., species-specific inhibition on seedling root growth of several species by their own decomposed litter (Fig. 1). They also showed that the application of activated carbon, known to selectively adsorb allelopathic organic compounds,3 was not effective in removing autotoxicity. Based on this evidence and previous theoretical modeling work,4,5 the hypothesis of the involvement of nucleic acids to explain the species-specific effect was formulated. DNA extraction from litter materials showed a substantial accumulation of extracellular DNA during the decomposition process. The substrates showing autotoxic effects were characterized by a high content of DNA fragments in a size range between ∼50 and ∼2000 bp. Subsequent in-depth chemical analysis by 13C NMR indeed revealed that signals related to DNA were the only ones negatively correlated with root growth on conspecific litter. Laboratory experiments confirmed the inhibitory effect of purified conspecific DNA on seed germination and root growth only when the treatment were performed using fragments of the abovementioned size range. The occurrence of the self-DNA inhibitory effect was further generalized testing several taxa such as bacteria, protozoa, algae, fungi, and insects.1
Figure 1.
Schematic representation of self-DNA soil dynamics and interactions with plant functionality.
The reported phenomenological observations raised basic questions on the underlying cellular and molecular mechanisms needing thorough investigation. Here we present pieces of information to be considered to delineate the overall frame for such investigation. First, it has been widely demonstrated that exDNA can be absorbed by living cells in both prokaryotes and eukaryotes. Microorganisms are well known to be genetically transformed by uptake of free DNA present in the environment.6,7 Recently, it has been shown that also plants are able to incorporate large organic molecules, including proteins and DNA, into roots, where fluorescently labeled DNA of 25 bp was taken up and detected inside different cells.8 Moreover, cultured mammalian cells can absorb and integrate exDNA added to the culture medium.9 In similar experiments, up to 40% of the administered exDNA was transported to the nuclei within 2-4 h10 and eventually even integrated into the genome of the guest cell.11
It is known that cells developed several defense mechanisms to protect from heterologous exDNA. The mechanisms can go from i) degradation or excretion of foreign DNA, ii) excision and loss of previously integrated DNA from the host genome, and iii) targeted inactivation of foreign molecules by specific modifications like methylation.12 These mechanisms may be primordial, like the restriction modification system involving endonucleases to prevent infection and interspecies DNA transmission in bacteria. Indeed, bacteria degrade exogenous DNA by sequence-specific restriction enzymes that cleave the foreign molecule while protecting their own genome by methylation of native DNA in a sort of primitive immune system.13 Moreover, RNA interference is an alternative mechanism to detect and react to the presence of exogenous nucleic acids (DNA/RNA) in a cell.14
More sophisticated mechanisms of defense from heterologous DNA imply the use of specific clearance from exDNA uptake in a cell, like those mediated by APOBEC dependent restriction in vertebrates.15 In such cases, foreign DNA/RNA molecules can be sensed by Toll-like receptor (TLR)-dependent and TLR-independent mechanisms16 in both cytoplasmic and endosomal compartments, signaling the induction and the production of interferons and pro-inflammatory cytokines and chemokines. Though several double-stranded DNA (dsDNA) sensors have been identified, once interferon is produced, it stimulates the transcription of many genes whose products orchestrate a wide variety of innate immune responses.
Even though the presence of defense mechanisms from extracellular heterologous DNA/RNA has been reported, nothing is known on the ability of the cells to recognize extracellular self-DNA and induce a specific response against it. In plants, it is possible to assume that a first sensing of exogenous self-DNA may occur at the level of the DNA based NET-like mantles located at the external membrane surface, since it was shown that treatments with DNases destroy the resistance to pathogen infection.17,18 However, beyond interfering with the presence of possible extracellular defensing DNA nets, the uptake of self or homologous exDNA could impact cell functionality at different levels. It can be speculated that the possible mechanisms might be ascribed to gene expression interference based on sequence-specific recognition involving RNA/DNA interactions,19 or the direct interaction with the genome structure based on mechanisms like the Small Fragment Homologous Replacement (SFHR).20 Once within the cell, Small DNA Fragments (SDFs) trigger the exchange between their sequences and the genomic DNA21 likely by a mechanism in which the fragment recognizes and anneals to its homologous target, promoting the formation of a D-loop structure. This hybrid structure could activate the endogenous machinery involved in DNA repair and, by homologous recombination, allow the SDF to be integrated into the genomic DNA.22
On these bases, we propose the hypothesis that a mixture of random self-DNA fragments, such as those produced by the degradation of the nuclear genome during decomposition of litter and soil organic matter, may cause either complete interference or inhibition of the whole cell functionality. It is fundamental to further investigate possible mechanisms of recognition, intracellular transport and response to such exposure (Fig. 1).
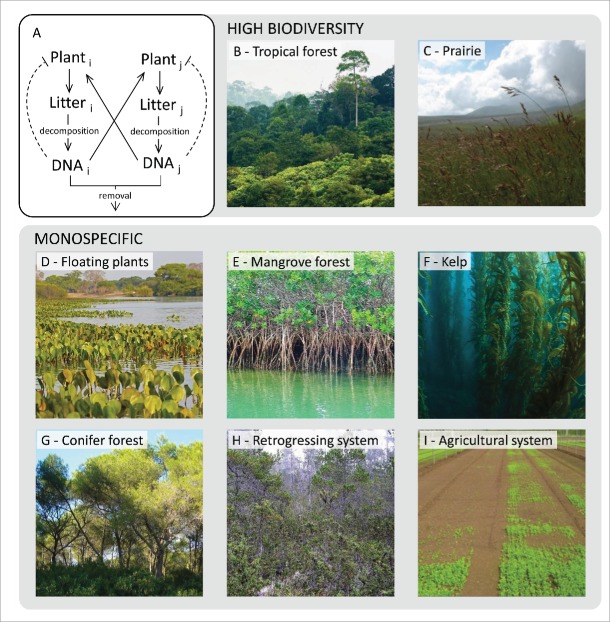
Beyond the underlying mechanisms, extracellular self-DNA inhibitory effect can bear significant implications at ecosystem level where biodiversity may be related to either accumulation or removal of DNA in the soil (Fig. 2A). A mathematical model presented by Mazzoleni et al.4 described this concept in terms of accumulation of autotoxicity, anticipating, in theoretical terms, the discovery of self-DNA inhibition. According to this model, in terrestrial ecosystems, high biodiversity levels are found in all conditions where fragments of DNA produced by litter decomposition may accumulate in the soil (Fig. 2B, C). In sharp contrast with terrestrial plants where “tierra firma” tropical forests are the most diverse ecosystems,23 aquatic tropical communities show low diversity. In general, monospecific stands occur both in salt and freshwater condition irrespectively of the latitudinal level. This is the case of floating plants (Fig. 2D; e.g. Eichornia crassipes, Lemna spp., Pistia spp. etc.), perennial species in wetlands and marshes (e.g., Phragmites australis, Spartina spp, Typha spp. etc.), gallery (e.g., Mora spp, Tabebuia spp. etc) and mangrove forests (Fig. 2E; e.g. Avicennia spp, Nypa fruticans, Rhizophora spp. etc), and also seagrass (e.g., Posidonia spp, Thalassia spp., Zoostera spp. etc), seaweed and kelp forests (Fig. 2F; e.g. Fucus spp, Laminaria spp., Macrocystis pyrifera etc),. While the reasons of this pattern remain enigmatic in the light of previous theory,24 this is highly coherent with the autotoxicity hypothesis due to self-DNA inhibition, considering that DNA molecules are water soluble thus easily removed by flushing water. Interestingly, monospecific stands can also be found in terrestrial ecosystems only when the accumulation of DNA in the soil is reduced either by slow decomposition (e.g., boreal forests) or by its degradation due to acidic soil conditions and/or burning (Fig. 2G; e.g. conifers and eucalypts forests). Several studies reported that over time scales of thousands to millions of years, and in the absence of rejuvenating disturbances that re-start primary successions, key ecosystem functions such as productivity, decomposition, and nutrient cycling undergo substantial declines, the so-called ecosystem retrogression.25 Ecosystem retrogression has been related to a progressive reduction of soil fertility, especially of phosphorus availability.26 Most notable, Turner et al.27 reported a significant accumulation of total DNA in soil in the Franz Josef glacial chronosequence, New Zealand. In such conditions, with accumulation of DNA coupled with a low species pool, the proposed model of DNA autotoxicity indeed predicts ecosystem retrogression (Fig. 2H). Finally, monospecific crops (Fig. 2I) typically show the emergence of “soil sickness,” i.e. reduced establishment and yield after several cultivation cycles, which can be recovered only by traditional agricultural practices such as either crop rotation or burning and flooding, likely to remove accumulated DNA.
Figure 2.
Ecological consequences of plant soil negative feedback mediated by self-DNA. (A) Diagram of the dynamics of DNA release in soil (solid lines) and species specific inhibitory effects (dashed lines). (B-C) Examples of high biodiversity ecosystems where the accumulation of exDNA from litter decomposition favors species coexistence. D-I) Examples of monospecific ecosystems: the occurrence of monodominance can be ascribed to the removal of self-DNA by either water (D-F) or fast degradation due to either acidic soil conditions or burning (G); the absence of competing species coupled with self-DNA accumulation leads to a decrease in the fitness of both natural (H) and agricultural (I) systems. Authors of the images can be found in the acknowledgments section.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Authors of the photographs in Fig. 2: B) sittitap/Shutterstock; C) Giuliano Bonanomi; D) Roberto Tetsuo Okamura/Shutterstock; E) Snapper 68/Shutterstock; F) Ethan Daniels/Shutterstock; G) Fabrizio Cartenì; H) Jason Sturner, distributed under a CC-BY 2.0 license; I) Giuliano Bonanomi.
References
- 1.Mazzoleni S, Carteni F, Bonanomi G, Senatore M, Termolino P, Giannino F, Incerti G, Rietkerk M, Lanzotti V, Chiusano ML. Inhibitory effects of extracellular self-DNA: a general biological process? New Phytol 2015; 206:127-32; PMID:25628124; http://dx.doi.org/ 10.1111/nph.13306 [DOI] [PubMed] [Google Scholar]
- 2.Mazzoleni S, Bonanomi G, Incerti G, Chiusano M, Termolino P, Mingo A, Senatore M, Giannino F, Cartenì F, Rietkerk M, et al.. Inhibitory and toxic effects of extracellular self-DNA in litter: a mechanism for negative plant-soil feedbacks? New Phytol 2015; 205:1195-210; PMID:25354164; http://dx.doi.org/ 10.1111/nph.13121 [DOI] [PubMed] [Google Scholar]
- 3.Hille M, den Ouden J. Charcoal and activated carbon as adsorbate of phytotoxic compounds - a comparative study. Oikos 2005; 108:202-7; http://dx.doi.org/ 10.1111/j.0030-1299.2005.13482.x [DOI] [Google Scholar]
- 4.Mazzoleni S, Bonanomi G, Giannino F, Incerti G, Dekker SC, Rietkerk M. Modelling the effects of litter decomposition on tree diversity patterns. Ecol Modell 2010; 221:2784-92; http://dx.doi.org/ 10.1016/j.ecolmodel.2010.08.007 [DOI] [Google Scholar]
- 5.Mazzoleni S, Bonanomi G, Giannino F, Rietkerk MG, Dekker SC, Zucconi F. Is plant biodiversity driven by decomposition processes? An emerging new theory on plant diversity. Community Ecol 2007; 8:103-9; http://dx.doi.org/ 10.1556/ComEc.8.2007.1.12 [DOI] [Google Scholar]
- 6.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol 1999; 53:217-44; PMID:10547691; http://dx.doi.org/ 10.1146/annurev.micro.53.1.217 [DOI] [PubMed] [Google Scholar]
- 7.Nevoigt E, Fassbender A, Stahl U. Cells of the yeast Saccharomyces cerevisiae are transformable by DNA under non-artificial conditions. Yeast 2000; 16:1107-10; PMID:10953082; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 8.Paungfoo-Lonhienne C, Lonhienne TGA, Mudge SR, Schenk PM, Christie M, Carroll BJ, Schmidt S. DNA is taken up by root hairs and pollen, and stimulates root and pollen tube growth. Plant Physiol 2010; 153:799-805; PMID:20388669; http://dx.doi.org/ 10.1104/pp.110.154963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groneberg J, Brown DT, Doerfler W. Uptake and fate of the DNA of adenovirus type 2 in KB cells. Virology 1975; 64:115-31; PMID:1114705; http://dx.doi.org/ 10.1016/0042-6822(75)90084-7 [DOI] [PubMed] [Google Scholar]
- 10.Wienhues U, Hosokawa K, Höveler A, Siegmann B, Doerfler W. A novel method for transfection and expression of reconstituted DNA-protein complexes in eukaryotic cells. DNA 1987; 6:81-9; PMID:3829890; http://dx.doi.org/ 10.1089/dna.1987.6.81 [DOI] [PubMed] [Google Scholar]
- 11.Doerfler W, Orend G, Schubbert R, Fechteler K, Heller H, Wilgenbus P, Schröer J. On the insertion of foreign DNA into mammalian genomes: Mechanism and consequences. Gene 1995; 157:241-5; PMID:7607499; http://dx.doi.org/ 10.1016/0378-1119(95)00080-P [DOI] [PubMed] [Google Scholar]
- 12.Doerfler W. Patterns of DNA methylation–evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol Chem Hoppe Seyler 1991; 372:557-64; PMID:1958315; http://dx.doi.org/ 10.1515/bchm3.1991.372.2.557 [DOI] [PubMed] [Google Scholar]
- 13.Dussoix D, Arber W. Host specificity of DNA produced by Escherichia coli*: II. Control over acceptance of DNA from infecting phage λ. J Mol Biol 1962; 5:37-49; PMID:13888713; http://dx.doi.org/ 10.1016/S0022-2836(62)80059-X [DOI] [PubMed] [Google Scholar]
- 14.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 15.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol 2010; 17:222-9; PMID:20062055; http://dx.doi.org/ 10.1038/nsmb.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner H, Bauer S. All is not Toll: new pathways in DNA recognition. J Exp Med 2006; 203:265-8; PMID:16446382; http://dx.doi.org/ 10.1084/jem.20052191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunawardena U, Rodriguez M, Straney D, Romeo JT, VanEtten HD, Hawes MC. Tissue-specific localization of pea root infection by Nectria haematococca. Mechanisms and consequences. Plant Physiol 2005; 137:1363-74; PMID:15778461; http://dx.doi.org/ 10.1104/pp.104.056366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen F, White GJ, VanEtten HD, Xiong Z, Hawes MC. Extracellular DNA is required for root tip resistance to fungal infection. Plant Physiol 2009; 151:820-9; PMID:19700564; http://dx.doi.org/ 10.1104/pp.109.142067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009; 457:413-20; PMID:19158787; http://dx.doi.org/ 10.1038/nature07756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruenert D, Bruscia E, Novelli G, Colosimo A, Dallapiccola B, Sangiuolo F, Goncz K. Sequence-specific modification of genomic DNA by small DNA fragments. J Clin Invest 2003; 112:637-41; PMID:12952908; http://dx.doi.org/ 10.1172/JCI19773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leclerc X, Danos O, Scherman D, Kichler A. A comparison of synthetic oligodeoxynucleotides, DNA fragments and AAV-1 for targeted episomal and chromosomal gene repair. BMC Biotechnol 2009; 9:35; PMID:19379497; http://dx.doi.org/ 10.1186/1472-6750-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruenert DC. Opportunities and challenges in targeting genes for therapy. Gene Ther 1999; 6:1347-8; PMID:10467357; http://dx.doi.org/ 10.1038/sj.gt.3301011 [DOI] [PubMed] [Google Scholar]
- 23.Givnish TJ. On the causes of gradients in tropical tree diversity. J Ecol 1999; 87:193-210; http://dx.doi.org/ 10.1046/j.1365-2745.1999.00333.x [DOI] [Google Scholar]
- 24.Willig MR, Kaufman DM, Stevens RD. Latitudinal Gradients of Biodiversity: Pattern, Process, Scale, and Synthesis. Annu Rev Ecol Evol Syst 2003; 34:273-309; http://dx.doi.org/ 10.1146/annurev.ecolsys.34.012103.144032 [DOI] [Google Scholar]
- 25.Peltzer DA, Wardle DA, Allison VJ, Baisden WT, Bardgett RD, Chadwick OA, Condron LM, Parfitt RL, Porder S, Richardson SJ, et al.. Understanding ecosystem retrogression. Ecol Monogr 2010; 80:509-29; http://dx.doi.org/ 10.1890/09-1552.1 [DOI] [Google Scholar]
- 26.Wardle DA, Walker LR, Bardgett RD. Ecosystem properties and Forest Decline in Contrasting Long-Term Chronosequences. Science 2004; 305:509-13; PMID:15205475; http://dx.doi.org/ 10.1126/science.1098778 [DOI] [PubMed] [Google Scholar]
- 27.Turner BL, Condron LM, Richardson SJ, Peltzer DA, Allison VJ. Soil organic phosphorus transformations during pedogenesis. Ecosystems 2007; 10:1166-81; http://dx.doi.org/ 10.1007/s10021-007-9086-z [DOI] [Google Scholar]