In flowering plants, floral determinacy requires termination of stem cells in the center of floral meristems (FMs), and in the axils of outer floral organs, such as sepals. In the center of the FM, AGAMOUS together with other factors represses WUSCHEL expression to terminate stem cell activities.1 In the outermost whorl, APETALA1 (AP1) terminates sepal axil stem cell activities to suppress axillary secondary flower formation.2-4 Recently, we reported that low cytokinin levels was required in AP1-expressing cells to suppress sepal axillary secondary flower formation. Furthermore, we showed that the MADS-box transcription factor encoded by floral homeotic gene AP1 regulated cytokinins homeostasis by directly activating the cytokinin degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3), and suppressing the cytokinin biosynthetic gene LONELY GUY1 (LOG1). Altogether, our experimental data provided a mechanistic explanation for how axillary meristem activity is suppressed in FMs.
The indeterminate growth inflorescence meristem (IM) produces floral meristems (FMs) during reproductive stage. In contract, determinate FMs produce flowers of a particular size and form by termination of stem cell divisions in the meristem. AP1 activates floral organ identity genes to promote FM formation together with LEAFY and CAULIFLOWER.3,5-7 After FM formation, AP1 functions as a class A gene of the ABC model for floral organ identity determination to specify outer whorls of floral organs petals and sepals.8 Inflorescence-like phenotypes in ap1 flowers (Fig. 1) indicated that AP1 also prevents the formation of flowers in the axils of sepals to establish determinate FMs.2,4 But how AP1 terminates sepal axil stem cell activities to suppress axillary flowers formation?
Figure 1.

Sepal axil secondary flower phenotype in ap1 mutants. Vertical view of a wild type flower in (A), lateral view of a wild type flower in (B), and top view of a wild type inflorescence in (C). Vertical view of an ap1-1 flower in (D), lateral view of an ap1-1 flower in (E), and top view of an ap1-1 inflorescence in (F). Vertical view of an ap1-4 flower in (G), lateral view of an ap1-4 flower in (H), and top view of an ap1-4 inflorescence in (I). Note ectopic sepal axil secondary flowers in ap1 mutants.
We have recently shown that elevated cytokinin signaling is required for leaf axil axillary meristem formation.9 In flowers, we further showed that ectopic cytokinins promoted sepal axil stem cell activities to induce secondary flowers.9,10 When we elevated cytokinin levels in flowers by expressing the Arabidopsis adenosine phosphate-isopentenytransferase 8 (IPT8) gene from the AP1 promoter in wild-type plants, we observed sepal axil secondary flowers that phenocopied ap1 mutants. To understand if AP1 maintain flower determinacy by regulating cytokinin homeostasis and/or signaling, we tested cytokinin signaling and cytokinin levels in wild-type and ap1 flowers. To this end, pTCS::GFP-ER, a synthetic cytokinin signaling reporter, was introduced into the ap1-1 background. We observed an elevated GFP signal in ap1-1 flowers than in wild-type sibling flowers around the stages when secondary flowers initiate. Furthermore, we measured endogenous levels of 4 types of cytokinins, specifically zeatin riboside 5′-monophosphate, zeatin riboside, isopentenyladenine, and isopentenyladenine riboside, in young inflorescences. All four quantified cytokinins increased in ap1 mutants. Conversely, disruption of the cytokinin signaling pathway by introducing cytokinin receptor mutations into the ap1-1 mutant rescued the ectopic secondary flower phenotype.
Because AP1 encodes a MADS domain transcription factor, it is possible that AP1 directly or indirectly regulates cytokinin biosynthesis and/or degradation. A scouting of recent genome-wide binding data for AP1 and AP1 domain-specific expression data showed that a cytokinin biosynthetic gene LONELY GUY1 (LOG1), and a cytokinin degradation gene CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3) are both good AP1 downstream candidates.11,12 Using chromatin immunoprecipitation assay and transient transfection assay in protoplasts, we found that AP1 directly bound to both LOG1 and CKX3 promoter regions containing CArG motifs, which are canonical MADS domain transcription factors. Chemical inducible AP1-glucocorticoid-receptor fusion also confirmed that AP1 regulation of LOG1 and CKX3 expression did not require protein synthesis, and showed that AP1 suppressed LOG1 expression but activated CKX3 expression. We further altered LOG1 and CKX3 expression in an ap1 background to short-circuit AP1 regulation. To this end, we suppressed LOG1 expression in the AP1 expressing domain using an LOG1-specific artificial miRNA, or over expressed CKX3 under the AP1 promoter in the ap1-4 mutant. In transgenic ap1-4 mutant lines containing either an pAP1::amiR-LOG1 or an pAP1::CKX3 transgene, we found partial rescue of the sepal axil secondary flower phenotype (Fig. 2). Thus, the AP1 regulation of LOG1 and CKX3 expression is relevant to sepal axil secondary flower formation.
Figure 2.
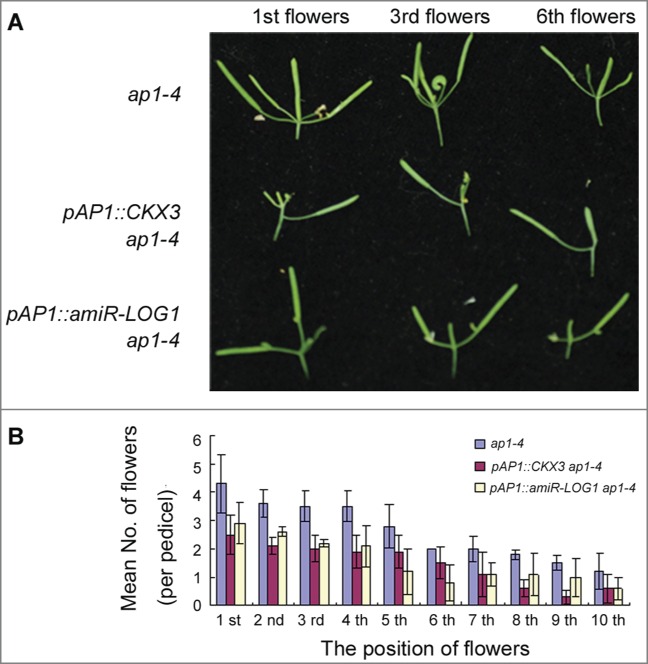
Partial rescue of the ap1 axil flower phenotype by local manipulation of CKX3 and LOG1 expression. (A) Siliques phenotypes of ap1-4, pAP1::CKX3 ap1-4 and silencing pAP1::LOG1 ap1-4. Siliques were taken at the same position of inflorescence. (B) Mean number of flowers (per pedicel) of ap1-4, pAP1::CKX3 ap1-4 and pAP1::amiR-LOG1 ap1-4 plants. Blue represents wild-type, purple represents pAP1::CKX3 ap1-4 and white represents pAP1::amiR-LOG1 ap1-4.
Transformation of floral meristems into inflorescence meristem has profound impact on seeds yield. The conversion of a single flower into a dichasium or a pleiochasium inflorescence is seen in many plant species, including cauliflower and broccoli. The cytokinin-mediated AP1 regulation is likely conserved in these vegetable crops to control their inflorescence architecture.6
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1. Sun B, Looi LS, Guo S, He Z, Gan ES, Huang J, Xu Y, Wee WY, Ito T. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 2014; 343:1248559; PMID:24482483; http://dx.doi.org/ 10.1126/science.1248559 [DOI] [PubMed] [Google Scholar]
- 2. Irish VF, Sussex IM. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 1990; 2:741-53; PMID:1983792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993; 119:721-43 [Google Scholar]
- 4. Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992; 360:273-7; PMID:1359429 [DOI] [PubMed] [Google Scholar]
- 5. Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature 1995; 377:522-4; PMID:7566148 [DOI] [PubMed] [Google Scholar]
- 6. Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science 1995; 267:522-5; PMID:7824951 [DOI] [PubMed] [Google Scholar]
- 7. Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000; 127:725-34; PMID:10648231 [DOI] [PubMed] [Google Scholar]
- 8. Bowman JL, Smyth DR, Meyerowitz EM. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991; 112:1-20; PMID:1685111 [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y. The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 2014; 26:2055-67; PMID:24850849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han Y, Zhang C, Yang H, Jiao Y. Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc Natl Acad Sci U S A 2014; 111:6840-5; PMID:24753595; http://dx.doi.org/ 10.1073/pnas.1318532111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Syst Biol 2010; 6:419; PMID:20924354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaufmann K, Wellmer F, Muino JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science 2010; 328:85-9; PMID:20360106; http://dx.doi.org/ 10.1126/science.1185244 [DOI] [PubMed] [Google Scholar]


