ABSTRACT
The cellular patterning of Arabidopsis root epidermis is a well-characterized system for study of how single-layered cells are arranged in a particular spatial order. Previously, we found that histone acetylation plays an important role in regulating epidermal differentiation by relaying positional information. To investigate the underlying mechanisms, we screened all available mutants of both HDAC and HAT families. Analyses of mutants of HDAC family members revealed that among single mutants, only HDA6, HDA18 and HDA19 exhibited ectopic H cells at the N position. Similarly, among HAT family members, only single mutants for GCN5 and HAF2 exhibited altered epidermal phenotypes, which were unexpectedly similar to the phenotypes observed in HDAC mutants. Based on these results, together with the previous findings regarding the regulatory mechanisms of HDA18 and HDA6, we proposed that homeostasis of histone acetylation is important for robustness of the regulatory network responsible for the cellular patterning of the Arabidopsis root epidermis.
KEYWORDS: Arabidopsis root epidermis, cellular patterning, histone acetyltransferase (HAT), histone deacetylase, mutant analysis
To explore whether histone acetylation is involved in morphogenetic events during plant development, we treated Arabidopsis seedlings with Trichostatin A (TSA), a specific inhibitor of histone deacetylase (HDAC). Pattern formation of root epidermal cells was dramatically disturbed upon TSA application, indicating that histone acetylation affects cellular patterning.1
The single-layered root epidermis of Arabidopsis consists of 2 types of cells, hair (H) and non-hair (N), whose arrangement depends on their spatial relationships with the underlying cortical cells.2 Genes encoding transcriptional factors, receptor-like kinases and other proteins have been identified based on their mutant phenotypes as playing roles in the regulation of cellular patterning in the Arabidopsis root epidermis (recently summarized by3,4). Successive investigation following up the TSA treatments1 uncovered that HISTONE DEACETYLASE 18 and 6 (HDA18 and HDA6) also play roles in the regulatory network,4,5 by targeting genes of both known and unknown function involved in the process, such as the unknown function kinase genes, ETC1 and GL2. It is therefore intriguing to elucidate how many HDAC and HAT genes are involved in the process.
In Arabidopsis genome, there are 18 members in the HDAC family, among which 16 have predicted intact HDAC domains, and 12 members in the HAT family.6,7 To clarify which members in the HDAC and HAT families affect root epidermal pattering, we screened all the available mutants of the 2 gene families for their genotypes and phenotypes (Tables 1, 2).
Table 1.
Mutant analysis of 16 Arabidopsis HDAC family members with 35 lines.
| Sub family | Gene | Mutant | Mutation type / T-DNA insertion site | Gene expression test | Root epidermal phenotype |
|---|---|---|---|---|---|
| RPD3 /HDA1 | HDA2 | SALK_110030 | T-DNA / 5′-UTR | Down regulated | × |
| CS3987 | RNAi | Downregulated | × | ||
| SALK_041074 | T-DNA / promoter | Normally expressed | – | ||
| HDA5 | SALK_093312 | T-DNA / exon | Not expressed | × | |
| SALK_030624 | T-DNA / 3′-UTR | Not expressed after T-DNA | × | ||
| HDA6; RPD3B | axe1-4 | point mutation | Down regulated | ✓ | |
| CS1894 | point mutation | – | ✓ | ||
| CS1895 | point mutation | – | ✓ | ||
| HDA7 | SALK_002912C | T-DNA / 5′-UTR | Not expressed | × | |
| HDA8 | CS30953 | RNAi | Not expressed | × | |
| CS30954 | RNAi | Not expressed | |||
| CS30955 | RNAi | Not expressed | |||
| SALK_010451 | T-DNA / between genes | Normally expressed | – | ||
| HDA9 | SALK_007123 | T-DNA / exon | Not expressed | × | |
| HDA14 | SALK_144995 | T-DNA / exon | Downregulated | × | |
| SALK_035491 | T-DNA / between genes | Normally expressed | – | ||
| HDA15 | SALK_004027 | T-DNA / exon | Not expressed | × | |
| HDA18 | SALK_006938 | T-DNA / exon | Not expressed after T-DNA | ✓ | |
| CS847004 | T-DNA / intron | Not expressed before T-DNA | ✓ | ||
| HDA19; RPD3A | SALK_139445 | T-DNA / exon | Not expressed | ✓ | |
| CS30925 | RNAi | Down regulated | ✓ | ||
| CS30926 | RNAi | Downregulated | – | ||
| HD2 | HDT1; HD2A | CS3969 | RNAi | Down regulated | × |
| CS3970 | RNAi | Downregulated | × | ||
| HDT2; HDA2B | CS30871 | RNAi | Not expressed | × | |
| CS30872 | RNAi | Down regulated | – | ||
| CS30873 | RNAi | Downregulated | – | ||
| SALK_035452 | T-DNA / promoter | Normally expressed | – | ||
| HDT3; HD2C | SALK_129799 | T-DNA / intron | Not expressed after T-DNA | × | |
| SALK_136925 | T-DNA / intron | Not expressed | × | ||
| SALK_002860 | T-DNA / 5′-UTR | Normally expressed | – | ||
| HDT4; HD2D | CS30956 | RNAi | Not expressed | × | |
| SALK_095273 | T-DNA / promoter | Normally expressed | – | ||
| SIR2 | SRT1 | SALK_001493 | T-DNA / exon | Not expressed after T-DNA | × |
| SRT2 | SALK_149295 | T-DNA / exon | Not expressed | × |
Bold font indicates genes with mutants showing disturbed root epidermal patterning. x indicates mutant showing normally root epidermal patterning as wild type; ✓ indicates mutant showing disturbed root epidermal patterning phenotype; – indicates phenotype not yet observed.
Table 2.
Mutant analysis of 12 Arabidopsis HAT family members with 20 lines.
| Sub family | Gene | Mutant | Mutant type / T-DNA insertion site | Gene expression test | Root epidermal phenotype |
|---|---|---|---|---|---|
| GNAT | HAG1; GCN5 | SALK_106557 | T-DNA / exon | Not expressed | ✓ |
| CS24025 | RNAi | Down regulated | ✓ | ||
| HAG2 | SALK_051832 | T-DNA / intron | Not expressed | × | |
| HAG3; HAC8 | SALK_104121 | T-DNA / 3′-UTR | Normally expressed | – | |
| CS3981 | RNAi | Downregulated | × | ||
| CS3983 | RNAi | Down regulated | × | ||
| MYST | HAG4; HAC6 | SALK_027726C | T-DNA / exon | Not expressed | × |
| HAG5 | SALK_106046C | T-DNA / intron | Not expressed | × | |
| CBP | HAC1 | SALK_122894 | T-DNA / exon | Not expressed | × |
| SALK_082118 | T-DNA / exon | Not expressed | × | ||
| HAC2 | SALK_049434 | T-DNA / 5′-UTR | Downregulated | × | |
| HAC4 | SALK_006923 | T-DNA / exon | Not expressed | × | |
| HAC5 | SALK_152684 | T-DNA / exon | Not expressed | × | |
| SALK_054116 | T-DNA / exon | Not expressed | × | ||
| SALK_075639C | T-DNA / exon | Not expressed | × | ||
| HAC12 | SALK_052490 | T-DNA / intron | Not expressed | × | |
| SALK_071102 | T-DNA / 3′-UTR | Not expressed | × | ||
| TAFII250 | HAF1 | SALK_110848C | T-DNA / exon | Not expressed | × |
| HAF2 | SALK_088103C | T-DNA / intron | Not expressed | ✓ | |
| SALK_038282C | T-DNA / intron | Not expressed | ✓ |
Bold font indicates genes with mutants showing disturbed root epidermal patterning. x indicates mutant showing normally root epidermal patterning as wild type; ✓ indicates mutant showing disturbed root epidermal patterning phenotype; – indicates phenotype not yet observed.
As shown in Table 1, we analyzed 35 mutant lines for HDAC genes available in various stocks, including T-DNA insertion, point mutation, and RNAi lines, to verify their genotypes (Table 1). Then, we examined the expression level of the respective HDAC genes in their homozygous mutants except for the point mutation lines. In total, twenty-nine lines exhibited significantly decreased expression of the corresponding genes. For these 29 mutant lines, we screened the phenotype in terms of the cellular patterns of root epidermis using paraffin sections of the root tip according to.1 Of the 16 members of HDAC gene family, only 3, i.e. hda185, hda64, and hda19 (Fig. 1 and Table 3), exhibited altered cellular patterning that converted cells at N positions to H fate.
Figure 1.
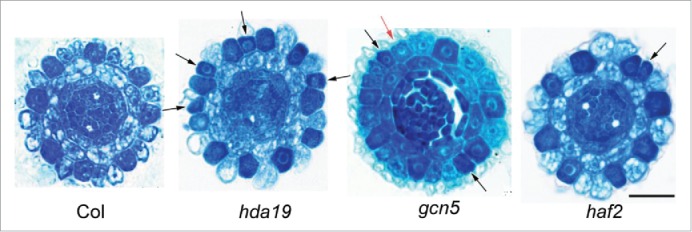
Phenotype of hdac and hat mutants for root tip epidermal patterning. Cross-sections and toluidine blue staining of root tips of 5-day-old seedlings. Black arrows indicate the ectopic darkly stained developing hair cells at N positions. Red arrow indicates the lightly stained developing non-hair cell at the H position. Bar = 20 μm.
Table 3.
Quantification of ectopic H and N cells in the root tips of 5-day-old seedlings.
| Gene | Sample | Ratio of root hair (%) | N cell at H position (%) | H cell at N position (%) | n |
|---|---|---|---|---|---|
| Col | 42.3±2.4 | 0.8d±1.4 | 2.1±2.9 | 17 | |
| HDA19 | SALK_139445 | 48.7±4.0 | 6.3±6.1b | 18.4±5.6a | 33 |
| CS30925 | 42.0±2.6 | 0±0 | 13.1±5.0a | 10 | |
| GCN5 | SALK_106557 | 47.7±7.7 | 4.8±7.2b | 19.1±8.6a | 6 |
| CS24025 | 46.1±3.7 | 0.5±1.0 | 10.1±5.9a | 20 | |
| HAF2 | SALK_088103C | 45.3±2.6 | 0.9±1.7 | 9.6±4.4a | 16 |
| SALK_0382823C | 47.6±3.7 | 3.0±5.9 | 9.2±5.9a | 41 |
Values indicate mean ± SD (a, b: differs significantly from the wild type, a: p < 0.01; b: p < 0.05; Student's t test.).
Similarly, we analyzed twenty mutant lines for HAT genes available in various stocks (Table 2) for their genotypes. Nineteen lines of HAT mutants exhibited significantly decreased expression of the corresponding genes. The phenotype screen revealed that of the 12 members of HAT gene family, only gcn5 and haf2 exhibited the altered cellular patterning, in which cells at N positions were converted to H fate (Table 3 and Fig. 1). The phenotype of gcn5 was more severe than that of haf2.
Two interesting phenomena were uncovered by this saturation mutant analysis of all members of HDAC and HAT gene families for the phenotype of altered cellular patterning of Arabidopsis root epidermis. The first was that only a few, not all, members of the HDAC/HAT gene families give rise to altered cellular patterning as single mutants. The high sequence conservation of the HADC and HAT domain suggests that all members of each gene family should have similar enzymatic activity, such as HDA6 and HDA19/AtHD1 for HDAC,8,9 except HDA10a and HDA17a, which have truncated HDAC domains.6 Thus, the sequences outside of the functional domains or interactions between the HDAC/HAT proteins and other proteins may possibly play indispensible roles in the functional specificity leading to a particular phenotype.
A second phenomenon was that the mutants of HDAC and HAT gene families exhibit similar phenotypes. Based on their enzymatic activities, the proteins of the 2 families should have opposite function in histone acetylation. The similar phenotypes observed in the mutants of the 2 gene families suggest that homeostasis of the histone acetylation status might be important in the regulation of cellular patterning of Arabidopsis root epidermis. We have shown that HDA6 and HDA18 target different genes, and the 2 members of the HAT family, GCN5 and HAF2, may target additional genes involved in the cellular patterning. The homeostasis hypothesis is supported by our investigation of the mechanism of HDA18 function. In that study, we found that both loss-of-function mutation and overexpression lines exhibited similar phenotypes and up-regulation of target genes.5
Based on the roles of HDA6 and HDA18 in cellular patterning of Arabidopsis root epidermis, we have proposed that the ultimate role of histone acetylation in this phenotype is to enhance the robustness of the transcriptional regulation network.4 Such an opinion is open to be examined as the molecular mechanisms of GCN5 and HAF2 in producing this phenotype remain to be elucidated. In addition, we currently cannot rule out the possibility of combinational involvement of other members of the HDAC and HAT families in cellular patterning.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Support for this work is from the National Natural Science Foundation of China (grant # 30570901 to SB and 31370220 to WC).
References
- 1.Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc Natl Acad Sci U S A 2005; 102:14469-74; PMID:16176989; http://doi.org/24596575 10.1073/pnas.1503143102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, et al.. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development (Cambridge, England) 1994; 120:2465-74; PMID:1246652724596575 [Google Scholar]
- 3.Schiefelbein J, Huang L, Zheng X. Regulation of epidermal cell fate in Arabidopsis roots: the importance of multiple feedback loops. Front Plant Sci 2014; 5:47; PMID:24596575; http://dx.doi.org/ 10.3389/fpls.2014.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li DX, Chen WQ, Xu ZH, Bai SN. HISTONE DEACETYLASE6-defective mutants show increased expression and acetylation of ENHANCER OF TRIPTYCHON AND CAPRICE1 and GLABRA2 with small but significant effects on root epidermis cellular pattern. Plant Physiol 2015; 168:1448-58; PMID:26143251; http://dx.doi.org/ 10.1104/pp.15.00821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C, Li LC, Chen WQ, Chen X, Xu ZH, Bai SN. HDA18 affects cell fate in arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell 2013; 25(1):257-69; PMID:23362208; http://dx.doi.org/ 10.1105/tpc.112.107045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, et al.. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 2002; 30:5036-55; PMID:12466527; http://dx.doi.org/19327164 10.1093/nar/gkf600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alinsug MV, Yu CW, Wu K. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol 2009; 9:37; PMID:19327164; http://dx.doi.org/ 10.1186/1471-2229-9-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong PM, Tian L, Chen ZJ. Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell Res 2006; 16:479-88; PMID:16699543; http://dx.doi.org/ 10.1038/sj.cr.7310059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, et al.. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 2006; 20:1283-93; PMID:16648464; http://dx.doi.org/ 10.1101/gad.1417706 [DOI] [PMC free article] [PubMed] [Google Scholar]


