ABSTRACT
The majority of the vascular flowering plants form symbiotic associations with fungi from the phylum Glomeromycota through which both partners gain access to nutrients, either mineral nutrients in the case of the plant, or carbon, in the case of the fungus.1 The association develops in the roots and requires substantial remodeling of the root cortical cells where branched fungal hyphae, called arbuscules, are housed in a new membrane-bound apoplastic compartment.2 Nutrient exchange between the symbionts occurs over this interface and its development and maintenance is critical for symbiosis. Previously, we showed that DELLA proteins, which are well known as repressors of gibberellic acid signaling, also regulate development of AM symbiosis and are necessary to enable arbuscule development.3 Furthermore, constitutive overexpression of a dominant DELLA protein (della1-Δ18) is sufficient to induce transcripts of several AM symbiosis-induced genes, even in the absence of the fungal symbiont.4 Here we further extend this approach and identify AM symbiosis genes that respond transcriptionally to constitutive expression of a dominant DELLA protein and also genes that do respond to this treatment. Additionally, we demonstrate that DELLAs interact with REQUIRED FOR ARBUSCULE DEVELOPMENT 1 (RAD1) which further extends our knowledge of GRAS factor complexes that have the potential to regulate gene expression during AM symbiosis.
KEYWORDS: arbuscular mycorrhizal symbiosis, dominant DELLA protein, gibberellic acid, transcription factors
Development of arbuscular mycorrhizal (AM) symbiosis involves signal exchange between the symbionts and a highly conserved plant symbiosis signaling pathway plays a central role in controlling fungal growth into the root and the development of the symbiotic interface in the cortical cells.5,6 The gene expression changes that underlie symbiotic development have been documented through detailed transcript profiling 7,8 and some transcriptional regulators have been identified, some of which are downstream components of the symbiosis signaling pathway.4,9-11
Development of AM symbiosis is also influenced by plant hormones and treatment of roots with gibberellic acid (GA) leads either to alterations in fungal entry into the root or abolishes arbuscule development. The precise outcome depends on the concentration of GA applied.12 Genes encoding enzymes of GA biosynthesis and metabolism are differentially regulated during symbiosis 7,13-17 and associated with the colonized regions of the cortex.18 Furthermore, both bioactive and inactive GAs are present in mycorrhizal roots. All of these data suggest a role for GAs in regulating aspects of symbiotic development.18,19
DELLA proteins are members of a specific sub-group of the GRAS family of transcriptional regulators that possess a unique domain, the DELLA domain, located toward the N-terminal end of the protein.20 DELLAs function as repressors of GA signaling and their mechanism of action has been elucidated through extensive studies in Arabidopsis. Briefly, in the presence of GA, the GA receptor (GID) interacts with the DELLAs through the DELLA domain and this subsequently results in the degradation of DELLA proteins and consequently relieves their repressive effect. DELLAs interact with, or alter the activities of a wide array of transcriptional regulators and in this way, GA signaling has broad impacts on plant growth and development (reviewed in 21,22).
Recent data from several groups revealed that DELLAs are positive regulators of AM symbiosis and in M. truncatula and pea, DELLA double mutants largely fail to develop arbuscules,3,23 while in rice, a single della mutant, slr1, shows a block in development of symbiosis at the root surface.24 These phenotypes are consistent with the earlier data that showed suppression of colonization following GA treatment. Furthermore, in M. truncatula constitutive expression of a dominant DELLA protein, that lacks the DELLA domain (della1-Δ18), enables arbuscules to form, even in the presence of GA.3 These data indicate that in the legumes, DELLAs and repression of GA signaling are needed to enable arbuscule development. Additionally, constitutive expression of della1-Δ18 in various symbiosis signaling mutants, revealed a link between DELLA and the symbiosis signaling pathway and suggests that DELLAs act downstream of a central transcription factor, CYCLOPS.3 Protein interaction studies carried out in Nicotiana benthamiana leaves revealed that the rice DELLA protein, SLENDER RICE 1 (SLR1), interacts with one additional member of the GRAS transcription factor family, DELLA INTERACTING PROTEIN 1 (DIP1). Since DIP1 interacts with the GRAS transcription factor REDUCED ARBUSCULAR MYCORRHIZA1 (RAM1), it was proposed that DELLAs potentially function in a GRAS protein complex to regulate gene expression for AM symbiosis.24 In line with this hypothesis, and using the same transient N. benthamiana expression system, we found that the GRAS transcription factor REQUIRED FOR ARBUSCULE DEVELOPMENT 1 (RAD1) interacts with DELLA2 from M. truncatula (Fig. 1). RAD1 also interacts with RAM1 and influences development of AM symbiosis.4,10 Thus DELLAs interact with at least two GRAS factors, DIP and RAD1, both of which have the ability to interact with RAM1.
Figure 1.
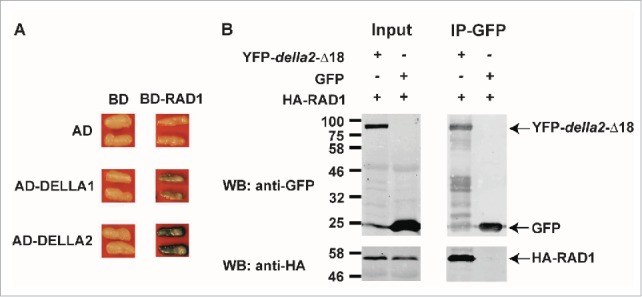
M. truncatula DELLA proteins interact with RAD1. (A) Interaction between DELLA1/2 and RAD1 in a yeast-two hybrid assay. AD, LexA-activation domain; BD, LexA-binding domain. (B) HA-tagged RAD1 co-immunoprecipitates with YFP-tagged della2-Δ18. To increase sensitivity a GA-insensitive mutant protein of DELLA2 (della2-Δ18) was used. No HA-RAD1 protein was detected following immunoprecipitation with GFP. Proteins were transiently co-expressed via agro-infiltration in N. benthamiana leaves. The methods were as described previously.33.
In the light of these studies, a major question is which downstream AM symbiosis genes are influenced by DELLA activity? Recent experiments in M. truncatula roots revealed that constitutive overexpression of a dominant DELLA protein (della1-Δ18) is sufficient to induce transcripts of the major symbiosis transcriptional regulator RAM1, and also of several other AM symbiosis-induced genes some of which are themselves regulated by RAM1.4
Here we extend this approach to search for additional symbiosis-regulated genes that are potentially direct, or indirect, transcriptional targets of DELLA activity. Transcript levels of a selection of genes whose expression has been shown previously to increase during symbiosis were evaluated in roots constitutively expressing della1-Δ18 in the absence of the fungal symbiont. Through this analysis, we identified 10 genes whose transcript levels are increased by expression of della1-Δ18 (Fig. 2), including the AM marker genes BCP1 and SCP1, which are highly expressed in arbuscule-containing cells and the adjacent cortical cells.7,25,26 Likewise transcripts for a Kunitz protease inhibitor, KPI106,27 an ortholog of a Lotus japonicus AM-induced subtilase SbtM1 18 and Vapyrin 28-30 are all increased in roots expressing della1-Δ18. In contrast, in L. japonicus roots in which symbiosis signaling was constitutively activated through expression of a gain-of-function CCaMK, both SbtM1 and Vapyrin transcripts were reduced by constitutive expression of a dominant DELLA construct.18 The M. truncatula and L. japonicus data are difficult to reconcile but as shown by Takeda et al.,18 GA signaling can have both positive and negative effects on the symbiosis and GA levels need to be tightly controlled to enable normal development of symbiosis.
Figure 2.
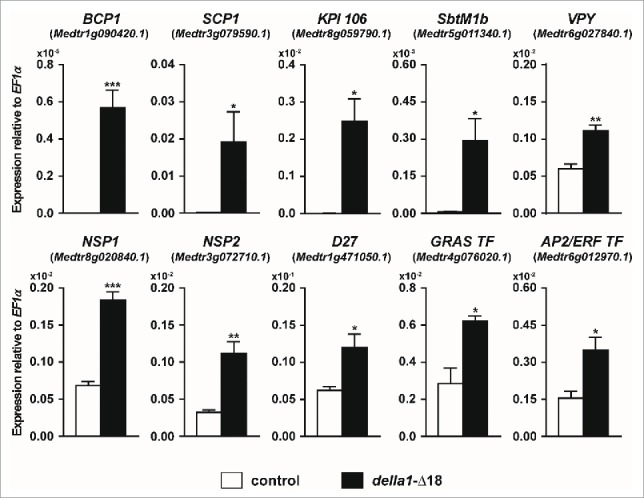
Overexpression of della1-Δ18 increases transcript levels of genes involved in AM symbiosis in the absence of the AM fungus. Transcript levels in non-colonized transgenic roots of M. truncatula expressing either a P35S:GFP vector control (control) or P35S:della1-Δ18 (della1-Δ18) at 35 d post planting, assayed by qRT-PCR. Data are averages ± SE (N ≥ 3 biological replicates). * P ≤ 0.05, ** P ≤ 0.01, ***P ≤ 0.001; t-test. ID numbers are shown in brackets and were retrieved from the M. truncatula genome version 4.0. The methods were as described previously.33
Expression of several transcription factors, including NSP1 and NSP2, two GRAS factors that influence development of symbiosis in a quantitative manner, were also induced by della1-Δ18, along with one of their downstream target genes, D27.31 These data are consistent with the earlier report that NSP1 and NSP2 transcripts are significantly reduced in a della1,della2 double mutant.3 D27 is required for strigolactone biosynthesis, thus cross-talk between GA and strigolactone signaling during symbiosis may be mediated in part through DELLAs, NSP1 and NSP2. Several other transcription factors are also induced by della1-Δ18 including an additional GRAS factor and a member of the AP2/ERF family. Currently the roles of these transcription factors in AM symbiosis are unknown.
Not all AM symbiosis-induced genes can be induced by overexpression of della1-Δ18. During AM symbiosis, GLP1 (Medtr4g052770.1), GST1 (Medtr5g076900.1) and LEC5 (Medtr5g031030.1) are highly expressed in cortex cells containing arbuscules and also in their non-colonized neighbors,7 while MtPT4 (Medtr1g028600.1) is induced specifically expressed in arbuscule-containing cells.32 We did not observe significant increases in the transcript levels of these genes in roots constitutively expressing della1-Δ18. Thus our results reveal that a subset of AM symbiosis-associated gene expression can be modulated by DELLA proteins. While this is not an exhaustive analysis and also does not distinguish between direct and indirect effects of DELLA expression, it provides a first indication of DELLA's potential sphere of influence on transcript levels during symbiosis and clues about DELLA-dependent and DELLA-independent gene expression.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Financial support was provided by the National Science Foundation Plant Genome Grant IOS 1127155 and The US. Dept. of Energy, Office of Science, Office of Biological and Environmental Research (BER), Grant No. DE FG02-08ER64628.
References
- 1.Smith SE, Read DJ. Mycorrhizal Symbiosis. San Diego, CA: Academic Press, Inc., 2008 [Google Scholar]
- 2.Harrison MJ. Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 2012; 15:691-8; PMID:23036821; http://dx.doi.org/ 10.1016/j.pbi.2012.08.010 [DOI] [PubMed] [Google Scholar]
- 3.Floss DS, Levy JG, Levesque-Tremblay V, Pumplin N, Harrison MJ. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 2013; 110:E5025-34; PMID:24297892; http://dx.doi.org/ 10.1073/pnas.1308973110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park H, Floss DS, Levesque-Tremblay V, Bravo A, Harrison M J. Hyphal branching during arbuscule development requires Reduced Arbuscular Mycorrhiza 1. Plant Physiol 2015; 169:1-15; PMID:26342109; http://dx.doi.org/ 10.1104/pp.15.01242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldroyd GED. Speak, friend, and enter:signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 2013; 11:252-63; PMID:23493145; http://dx.doi.org/ 10.1038/nrmicro2990 [DOI] [PubMed] [Google Scholar]
- 6.Gutjahr C, Parniske M. Cell and Developmental Biology of Arbuscular Mycorrhiza Symbiosis In: Schekman R, ed. Annual Review of Cell and Developmental Biol, 2013; 29:593-617. [DOI] [PubMed] [Google Scholar]
- 7.Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Kuster H. Laser Microdissection Unravels Cell-Type-Specific Transcription in Arbuscular Mycorrhizal Roots, Including CAAT-Box Transcription Factor Gene Expression Correlating with Fungal Contact and Spread. Plant Physiol 2011; 157:2023-43; PMID:22034628; http://dx.doi.org/ 10.1104/pp.111.186635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camps C, Jardinaud MF, Rengel D, Carrere S, Herve C, Debelle F, Gamas P, Bensmihen S, Gough C. Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol 2015; 208:224-40; PMID:25919491; http://dx.doi.org/ 10.1111/nph.13427 [DOI] [PubMed] [Google Scholar]
- 9.Gobbato E, Marsh JF, Vernie T, Wang E, Maillet F, Kim J, Miller JB, Sun J, Bano SA, Ratet P, et al.. A GRAS-Type Transcription Factor with a Specific Function in Mycorrhizal Signaling. Curr Biol 2012; 22:2236-41; PMID:23122845; http://dx.doi.org/ 10.1016/j.cub.2012.09.044 [DOI] [PubMed] [Google Scholar]
- 10.Xue L, Cui H, Buer B, Vijayakumar V, Delaux P-M, Junkermann S, Bucher M. Network of GRAS Transcription Factors Involved in the Control of Arbuscule Development in Lotus japonicus. Plant Physiol 2015; 167:854-71; PMID:25560877; http://dx.doi.org/ 10.1104/pp.114.255430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rich M, Schorderet M, Bapaume L, Falquet L, Morel P, Vandenbussche M, Reinhardt D. A Petunia GRAS transcription factor controls symbiotic gene expression and fungal morphogenesis in arbuscular mycorrhiza. Plant Physiol 2015; 168(3):788-97; PMID:25971550; http://dx.doi.org/ 10.1104/pp.15.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Ghachtouli N, Martin-Tanguy J, Paynot M, Gianinazzi S. First report of the inhibition of arbuscular mycorrhizal infection of Pisum sativum by specific and irreversible inhibition of polyamine biosynthesis or by gibberellic acid treatment. FEBS Lett 1996; 385:189-92; PMID:8647248; http://dx.doi.org/ 10.1016/0014-5793(96)00379-1 [DOI] [PubMed] [Google Scholar]
- 13.Gomez SK, Javot H, Deewatthanawong P, Torrez-Jerez I, Tang Y, Blancaflor EB, Udvardi MK, Harrison MJ. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol 2009; 9:1-19; PMID:19123941; http://dx.doi.org/ 10.1186/1471-2229-9-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 2009; 182:200-12; PMID:19192192; http://dx.doi.org/ 10.1111/j.1469-8137.2008.02725.x [DOI] [PubMed] [Google Scholar]
- 15.Manthey K, Krajinski F, Hohnjec N, Firnhaber C, Pühler A, Perlick AM, Küster H. Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbioses. Mol Plant-Microbe Interact 2004; 17:1063-77; PMID:15497399; http://dx.doi.org/ 10.1094/MPMI.2004.17.10.1063 [DOI] [PubMed] [Google Scholar]
- 16.Ortu G, Balestrini R, Pereira PA, Becker JD, Kuester H, Bonfante P. Plant Genes Related to Gibberellin Biosynthesis and Signaling Are Differentially Regulated during the Early Stages of AM Fungal Interactions. Mol Plant 2012; 5:951-4; PMID:22451647; http://dx.doi.org/ 10.1093/mp/sss027 [DOI] [PubMed] [Google Scholar]
- 17.Garrido JMG, Morcillo RJL, Rodriguez JAM, Bole JAO. Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol Plant-Microbe Interact 2010; 23:651-64; PMID:20367473; http://dx.doi.org/ 10.1094/MPMI-23-5-0651 [DOI] [PubMed] [Google Scholar]
- 18.Takeda N, Handa Y, Tsuzuki S, Kojima M, Sakakibara H, Kawaguchi M. Gibberellins Interfere with Symbiosis Signaling and Gene Expression and Alter Colonization by Arbuscular Mycorrhizal Fungi in Lotus japonicus. Plant Physiol 2015; 167:545-U442; PMID:25527715; http://dx.doi.org/ 10.1104/pp.114.247700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaul-Keinan O, Gadkar V, Ginzbert I, Grünzweig J, Chet I, Elad Y, Wininger S, Belausov E, Eshed Y, Artzmon N, et al.. Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomu intraradices. New Phytol 2002; 154:501-7; http://dx.doi.org/ 10.1046/j.1469-8137.2002.00388.x [DOI] [PubMed] [Google Scholar]
- 20.Harberd NP. Botany: Relieving DELLA restraint. Science 2003; 299:1853-4; PMID:12649470; http://dx.doi.org/ 10.1126/science.1083217 [DOI] [PubMed] [Google Scholar]
- 21.Sun TP. Gibberellin-GID1-DELLA: A Pivotal Regulatory Module for Plant Growth and Development. Plant Physiol 2010; 154:567-70; PMID:20921186; http://dx.doi.org/ 10.1104/pp.110.161554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daviere JM, Achard P. Gibberellin signaling in plants. Development 2013; 140:1147-51; PMID:23444347; http://dx.doi.org/ 10.1242/dev.087650 [DOI] [PubMed] [Google Scholar]
- 23.Foo E, Ross JJ, Jones WT, Reid JB. Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann Bot 2013; 111(5):769-79; PMID:23508650; http://dx.doi.org/ 10.1093/aob/mct041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu N, Luo D, Zhang X, Liu J, Wang W, Jin Y, Dong W, Liu J, Liu H, Yang W, et al.. A DELLA protein complex controls the arbuscular mycorrhizal symbiosis in plants. Cell Res 2014; 24:130-3; PMID:24343576; http://dx.doi.org/ 10.1038/cr.2013.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H. Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol 2005; 137:1283-301; PMID:15778460; http://dx.doi.org/ 10.1104/pp.104.056572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Blaylock L, Endre G, Cho J, Town CD, VandenBosch K, Harrison MJ. Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of the arbuscular mycorrhizal symbiosis. Plant Cell 2003; 15:2106-23; PMID:12953114; http://dx.doi.org/ 10.1105/tpc.014183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rech SS, Heidt S, Requena N. A tandem Kunitz protease inhibitor (KPI106)-serine carboxypeptidase (SCP1) controls mycorrhiza establishment and arbuscule development in Medicago truncatula. Plant J 2013; 75:711-25; PMID:23662629; http://dx.doi.org/ 10.1111/tpj.12242 [DOI] [PubMed] [Google Scholar]
- 28.Pumplin N, Mondo SJ, Topp S, Starker CG, Gantt JS, Harrison MJ. Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J 2010; 61:482-94; PMID:19912567; http://dx.doi.org/ 10.1111/j.1365-313X.2009.04072.x [DOI] [PubMed] [Google Scholar]
- 29.Murray JD, Muni RRD, Torres-Jerez I, Tang YH, Allen S, Andriankaja M, Li GM, Laxmi A, Cheng XF, Wen JQ, et al.. Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J 2011; 65:244-52; PMID:21223389; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04415.x [DOI] [PubMed] [Google Scholar]
- 30.Feddermann N, Muni RRD, Zeier T, Stuurman J, Ercolin F, Schorderet M, Reinhardt D. The PAM1 gene of petunia, required for intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi, encodes a homologue of VAPYRIN. Plant J 2010; 64:470-81; PMID:20804456; http://dx.doi.org/ 10.1111/j.1365-313X.2010.04341.x [DOI] [PubMed] [Google Scholar]
- 31.Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al.. Strigolactone Biosynthesis in Medicago truncatula and Rice Requires the Symbiotic GRAS-Type Transcription Factors NSP1 and NSP2. Plant Cell 2011; 23:3853-65; PMID:22039214; http://dx.doi.org/ 10.1105/tpc.111.089771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison MJ, Dewbre GR, Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002; 14:2413-29; PMID:12368495; http://dx.doi.org/ 10.1105/tpc.004861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang XC, Pumplin N, Ivanov S, Harrison MJ. EXO70I Is Required for Development of a Sub-domain of the Periarbuscular Membrane during Arbuscular Mycorrhizal Symbiosis. Curr Biol 2015; 25:2189-95; PMID:26234213; http://dx.doi.org/ 10.1016/j.cub.2015.06.075 [DOI] [PubMed] [Google Scholar]


