Abstract
A secondary, non-linear, lateral part of ethylene signaling pathway has been anticipated and speculated before. Recently, it has been found that part of the proteomic response of Eruca sativa to silver nitrate (which is an inhibitor of ethylene signaling) is related to sulfur metabolism. Using public Arabidopsis thaliana microarray data, I show that silver nitrate mimics the signal of sulfur starvation at the transcriptome level. This, combined with data mined from literature, indicates that ethylene receptors are localized at the beginning of the response to sulfur deficiency in plants. This means that the non-linear, lateral part of ethylene signaling pathway exists and is responsible for transduction of the signal of sulfur deficit. Here, I present a model of such a pathway and anticipate it to be the starting point for more detailed analysis of the lateral part of ethylene signaling pathway and the exact mechanism of sulfur status sensing in plants.
Keywords: Arabidopsis thaliana, ethylene receptors, ethylene signaling, silver nitrate, sulfur deficiency
Introduction
Ethylene is a simple hydrocarbon gas that is known as one of plant hormones. Most of ethylene response effects are induced through linear pathway (Fig. 1) of ethylene signal transduction.1 In Arabidopsis thaliana, ethylene is sensed by ETR1 (ETHYLENE RECEPTOR1), ETR2, ERS1 (ETHYLENE RESPONSE SENSOR1), ERS2, EIN4 (ETHYLENE INSENSITIVE4). These five members of ethylene receptors family work as negative regulators upstream the CTR1 (CONSTITUTIVE TRIPLE RESPONSE1). When ethylene is absent, the receptors activate CTR1, a serine/threonine-protein kinase which directly phosphorylate the EIN2 and therefore suppresses the ethylene signal transduction.2,3 In the presence of ethylene, CTR1 stays inactive which results in EIN2 proteolytical processing. After that the C-terminal domain of EIN2 migrates to the nucleus2-4 where activates the transcription factors EIN3, EIL1 (EIN3 like1) and EIL2 5,6 and therefore initiates transcriptional cascade in response to ethylene. Noteworthy, another EIL, namely the EIL3 also known as SLIM1 (SULFUR LIMITATION1), has been shown to control the transcription of most of the genes related to plant response to sulfur deficit and not partake in response to ethylene.7
Figure 1.
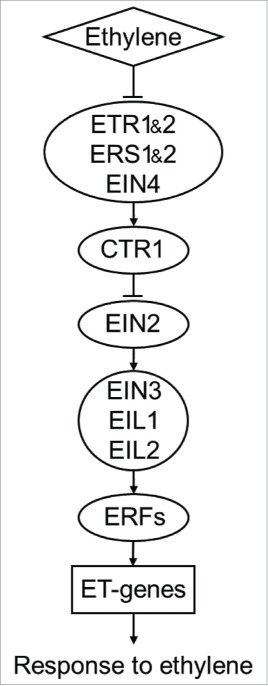
Current linear model of ethylene signaling pathway in Arabidopsis. Ethylene is sensed by 5 members of ethylene receptors family: ETR1 (ethylene receptor 1), ETR2, ERS1 (ethylene response sensor 1), ERS2, EIN4 (ethylene insensitive 4). When ethylene is absent, the receptors activate CTR1 (constitutive triple response 1), which directly phosphorylate the EIN2 and therefore suppresses the ethylene signal transduction.2,3 In the presence of ethylene, CTR1 stays inactive and EIN2 activates the transcription factors EIN3, EIL1 (EIN3 like1) and EIL2 5,6 and therefore initiates transcriptional cascade in response to ethylene.
A secondary, non-linear, lateral part of ethylene signaling pathway has been anticipated and speculated before based on overlapping and non-overlapping roles of ethylene receptors (reviewed in8) as well as on research regarding role of MAPK (mitogen-activated protein kinase) signaling cascade in ethylene signaling.9 No clue of such lateral part of ethylene signaling has been demonstrated yet apart from the strong theoretical need for the explanation of the ethylene receptor differences and controversy of MAPK cascade involvement in ethylene signaling pathway.
Ethylene is involved in many processes of plant growth and development, such as seed germination, seedling growth, leaf, root, stem and flower development, fruit ripening, organ senescence and abscission. It also plays an important role in plant response to both biotic and abiotic stresses such as drought, water-logging, flooding, wounding, mechanical impedance, salinity, and various pathogens (for reviews see: 10,11). In addition, there is strong evidence about involvement of ethylene in plant responses to nutritional stresses. Changed level of ethylene production was reported as a result of nitrogen (N), phosphorus (P), potassium (K), calcium (Ca), magnesium (Mg) and iron (Fe) deficiency.12-14
In recent decades anthropological sulfur dioxide (SO2) emission to the atmosphere has been greatly reduced.15 That caused mostly positive effects; however, the decrease in the amount of sulfur (S) in the natural environment, has become lately a limiting factor in plant production in some areas over the world.16 Therefore a strong stimulus to investigate the metabolism of S and its regulation in plants appeared. Despite the intensive research in the area, some questions remain unanswered. One of the fundamental, open problems is the nature of perception and signaling of the early stages of S deficiency.
A particularly strong link of S metabolic pathway with ethylene is stressed by the fact that ethylene biosynthesis starts from methionine, which is converted to S-adenosyl-methionine (SAM) (for recent review see: 17). Therefore, multiple attempts to link S deficit with ethylene signaling pathway have been undertaken previously, but no simple conclusion has been provided. The increase of ethylene production, during response to S deficiency, by Nicotiana tabacum 18 and Solanum lycopersicum 19 plants has been reported, but most of the ethylene responsive genes remains unaffected,18 like in microarray studies on Arabidopsis thaliana.20,21 The factor, responsible for lack of correlation for results of ethylene measurements and transcriptomics, might be the regulation of ethylene signaling. Recently, it has been shown that Nicotiana attenuata plants, which heterologously expressed the mutant A. thaliana ethylene receptor etr1–1, were impaired in sulfate uptake and S metabolism.22 Such changes, according to current knowledge, have to be mediated through EIL3. Moreover, it has been found lately that part of the proteomic response of Eruca sativa to Ag+ ions is related to S metabolism.23 Noteworthy, the Ag+ ions inhibit the hormone signaling at the sensing level hence silver nitrate (AgNO3) is frequently used to study ethylene perception.
In this work, the draft of model of secondary, lateral part of ethylene signaling is presented. The fundamental part of the model is EIL3, already known as the central transcription factor in plant response to S deficiency. Previous data suggests that signal to EIL3 might be relayed from ethylene receptors.22,23 To test such hypothesis, the response to S deficiency and AgNO3 treatment should share characteristic similarities shown in this study.
Materials and Methods
The Perturbations Tool (also described as GENE SEARCH across Perturbations) of Genevestigator 24 software was used for finding genes similarly up-regulated under chosen conditions and for visualization of the expression levels of the identified genes. Default settings was used: all base categories were selected, the base checkboxes were not shown, the threshold was set at 0.5. The methodology behind the tool utilizes the Kg parameter which measures how specific the up-regulation of a gene is in the target categories compared to the base categories. The relevant part of software documentation define the Kg parameter as:
where si,g is the meta-profile signal for category i and gene g, Tu is the set of all target categories in the upregulated group, Td is the set of all target categories in the down-regulated group, B+ is the set of all base categories with positive log-ratios (log2(si,g) > 0) and B− is the set of all base categories with negative log-ratios (log2(si,g) < 0).
Results
In this work, to find correlations in plant responses to S deficit and AgNO3 treatment, I analyzed the publicly available Arabidopsis microarray data and identified a group of 20 genes (Table A1) up-regulated in the similar manner under both conditions (Fig. 2, Table A2). Many of these genes are considered as the S deficiency markers. The 11 of 20 recognized genes could be found among 32 genes previously reported to be under the control of a transcription factor EIL3.7 Noteworthy, based on the list of the mentioned 32 genes, EIL3 is considered to be a central transcription factor responsible for plant early response to S deficiency. On the other hand, the data, for expression of the 20 identified genes in slim1 knock-out plants (Table A3), suggest that 3 more genes may (at least partially) stay under the control of EIL3, namely SIP1;2 (SMALL INTRINSIC PROTEIN1;2) (affected in both knockout lines) and LSU2 (RESPONSE TO LOW SULFUR2) as well as OPR1;2 (12-oxophytodienoate reductase1;2) (affected only in one knock-out line, slim1–1). Moreover, among the remaining 6 genes only APR2 (adenylylsulfate reductase2) and APR3 have been upregulated by S deficit in control plants, in the cited experiment. This indicates that expression of only 2 gens (APR2 and APR3), out of 20 identified in this study, for sure do not stay under EIL3 influence during plant early response to S deficiency.
Figure 2.
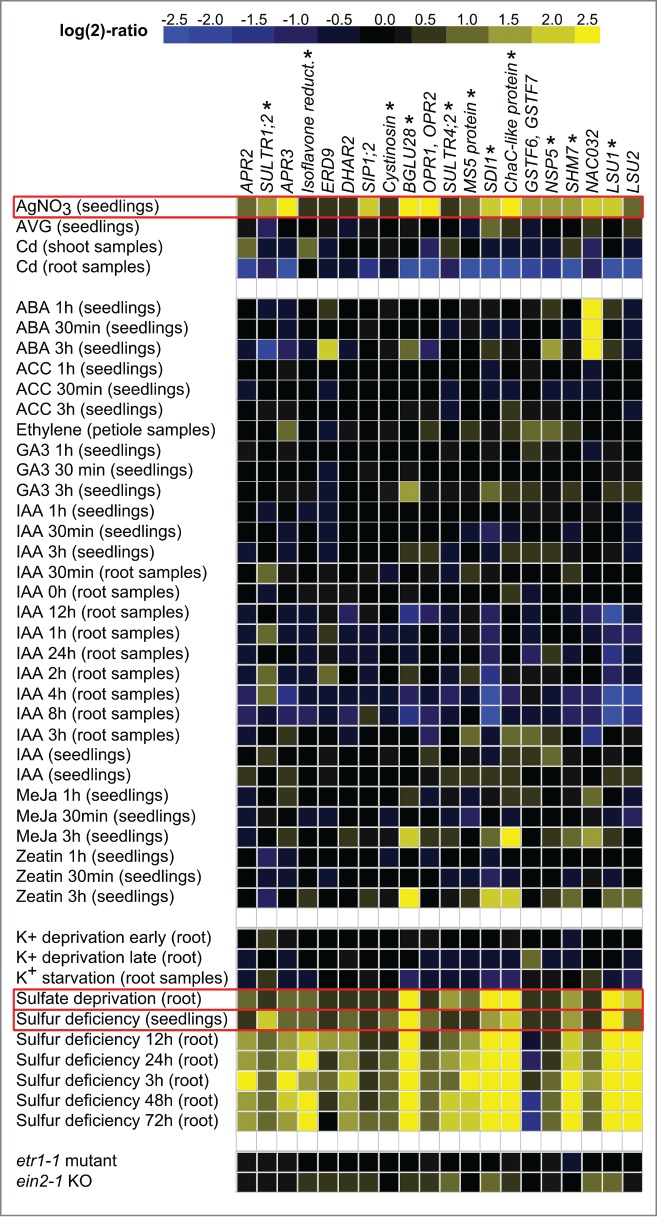
Expression of the A. thaliana genes found as similarly upregulated under S deficit and AgNO3 treatment. Similar upregulation has been found based on data from experiments marked with red boxes. Genes previously reported to be under the control of transcription factor EIL3 (also known as SLIM1) are marked with asterisks. For annotations of genes see Table A1, for numerical data see Table A2.
To determine how S deficiency is coupled with ethylene signaling, I revealed that no correlation in regulation of transcription (for the 20 genes) could be found between S deficit and treatment with ethylene, ethylene precursor 1-Aminocyclopropane-1-carboxylic acid (ACC), or inhibitor of ethylene biosynthesis Aminoethoxyvinylglycine (AVG) (Fig. 2, Table A2). Consequently, the observed correlation, in gene expression, in responses to S deficit and AgNO3 treatment, is specific to AgNO3 action.
Some new effects of AgNO3 treatment (unrelated to the ethylene signaling pathway) have been reported recently.23,25,26 Therefore, I had to examine the hypotheses that AgNO3 mimics the S deficit signal not by its action on the ethylene receptors.
Heavy metals (including Ag) induce particular plant response including production of glutathione (GSH) and heavy metal chelators, the phytochelatins (PCs). The excessive need for the S containing metabolites (such as PCs and GSH) is especially strong for cadmium (Cd).27 Therefore Cd could cause some type of S deficiency itself. The effect of Ag+ ions might be similar to Cd2+ ions as has been recently proposed.23 However, the toxic effect of heavy metals depends on the concentration and time of treatment, and the induction of transcription of the selected 20 genes could not be detected in short time experiments utilizing low concentration of Cd2+ ions (Fig. 2, Table A2).
On the other hand, even concentration as low as 5µM of AgNO3 could greatly reduce the potassium (K+) ions influx to roots.25 However, I showed that the 20 genes of interest are not induced by neither early nor late K starvation (Fig. 2, Table A2).
One more effect of AgNO3 is the increase of auxin efflux from Arabidopsis root tips.26 Such increase could result in decreased level of endogenous indole-3-acetic acid (IAA). However, microarray data related to various conditions related to IAA, reveal no influence on the expression of most of the 20 genes of interest (Fig. 2, Table A2).
In summary, the co-expression of the 20 genes under S deficit and AgNO3 treatment could not be explained by the generic toxicity of heavy metals, the decrease of K+ uptake or increase of auxin efflux. So, according to current knowledge, the mechanism of AgNO3 ability to mimic the S deficit signal have to be linked with ethylene signaling, more precisely with binding to ethylene receptors. Interestingly, Ag+ ions inhibits the ethylene perception, but promotes ethylene binding to ETR1 and ERS1.28 Besides, experiments showing that the effect of AgNO3 on the linear ethylene signaling pathway is connected with the hormone receptors have underlined the dominant role of ETR1.28 Therefore, I examined the data for the A. thaliana etr1–1 mutant which produces mutated ETR1 protein that could not relay signal in linear ethylene signaling pathway. The results signify that the correlation documented in this study is not dependent on ETR1 role in primary linear ethylene signaling pathway (Fig. 2, Table A2). In Arabidopsis 4 other ethylene receptors exist (ETR2, ERS1, ERS2 and EIN4) and could supplement the role of ETR1.8 Therefore, I investigated the microarray data for the A. thaliana ein2–1 knockout mutant of the downstream element of the pathway, EIN2.29 The observed mild induction of only a fraction of the 20 genes could not fully explain the AgNO3 or S deficit effect (Fig. 2, Table A2). This shows that the mechanism that I looked for is not dependent on the inhibition of signal transduction through the primary, linear ethylene signaling pathway.
Discussion
Data presented in this manuscript indicates that AgNO3 mimics the signal of S deficiency. The identification of 20 genes similarly up-regulated in response to S deficit and AgNO3 treatment could be explained only by EIL3 action. Therefore AgNO3 somehow influences EIL3.
Here, I propose the model of the lateral part of ethylene signaling pathway responsible for transduction of S deficit signal (Fig. 3). It is lateral as it shares the beginning with the primary, linear ethylene pathway but leads to expression of different genes at the end. It begins at the ethylene receptors level where the S status in cell should be recognized. The exact mechanism of such sensing is yet to be discovered, but the potential signaling molecule should function analogously to AgNO3. The next element is EIL3 which, in my model, plays similar role to the EIN3 in primary ethylene signaling. In contrast to other family members (namely EIN3, as well as EIL1 and EIL2 5,6), EIL3 is the member of the family that has not been shown to partake in response to ethylene, but instead modulates the transcription of most of the genes related to S deficit.7 Noteworthy, my model links EIL3 with lateral part of ethylene signaling as the changes in transcription of most of the 20 genes identified in this work could be explained by EIL3 action.
Figure 3.

Model of role of AgNO3 in primary, linear and suggested secondary, lateral ethylene signaling pathway. Secondary ethylene pathway (responsible for transduction of signal of S deficit) is presented on the left, primary on the right. The most speculative parts of pathway (including 3 distinct possibilities of signal transduction from ethylene receptors to EIL3 (also known as SLIM1) is light gray. AHP1 (histidine-containing phosphotransmitter1), ARR2 (2-component response regulator ARR2), MAPK (mitogen-activated protein kinase), MEK also known as MAPKK (MAPK kinase).
Based on current knowledge the signal from the receptors to the EIL3 might be transferred through: (I) 2-component signaling pathway,30-32 (II) MAPK signaling cascade,9,33,34 (III) unidentified molecules, (IV) directly.
The link of the 2-component signaling pathway with ethylene signaling is on the receptors level. The interaction of AHP1 (histidine-containing phosphotransmitter1) with ETR1 30 and modulation of ethylene response by ARR2 (2-component response regulator ARR2) have been reported.31,32 The possibility of existence of pathway, independent from main linear ethylene signaling, is supported by the exhibition of partial response to ethylene in ctr1 null mutants.35,36 The two-component phosphorelay has been suggested as a candidate for such lateral pathway.8 However, MAPK cascade has been shown to directly modulate EIN3 stability 9 and therefore it also could be responsible for lateral ethylene signaling. Interestingly, during plant response to S deficiency, transcription of only fraction of ethylene responsive genes is induced,18,20,21 despite increased ethylene production.18,19 Noteworthy, stabilization due to phosphorylation by MAPK of at least one form of ACC synthases, and therefore increase in ethylene biosynthesis has been shown.37-39 On the other hand, the similarity of EIN3 and EIL3 also point to MAPK cascade as it has been shown that the stability of EIN3 is regulated by phosphorylation mediated by MAPK kinases.9 Additionally, such hypothesis could finally resolve the controversy of involvement of MAPK cascade in linear ethylene signaling pathway.40,41 Many of the inconsistent results of studies on MAPKs could be explained by different S status in cells of used plants.
While further research is needed to identify the way of signal transduction to EIL3, the next step of the early response to S deficit is induction of the genes identified in this study. Noteworthy, the candidates for modulators of the response to S starvation should be found among these genes. Indeed, a regulatory role of LSUs and SULTR1;2 (SULFATE TRANSPORTER1;2) has been reported recently.18,42 Interestingly, transcription of the homologues of EIN4 and ERS1 is misregulated in Nicotiana tabacum plants with the lowered level of UP9 family (in Arabidospsis the family is called LSU) transcripts.18 Moreover, the mentioned research has underlined the complexity of the linkage between ethylene signaling and S starvation as increased ethylene production in response to S deficit has been reported, but not in the plants with lowered transcription of the UP9 family members.
One of the ways to verify the proposed model is to study the plant response to S deficiency in the plants with changed level of the ethylene receptor genes. For example overexpression of ethylene receptor genes should cause the plants inability to sense current S status in the cells. Such plants should have S deficiency symptoms even under optimal sulfate supply. Noteworthy, it has been recently shown that Nicotiana attenuata plants, which heterologously expressed the mutant A. thaliana receptor etr1–1, were impaired in sulfate uptake and S metabolism.22 The mentioned results include abnormal phenotype of 35S-etr1 seedlings under optimal sulfate supply and correspond with the model proposed in this study. Noteworthy, the data presented in the mentioned study22 in the light of the model presented in this study, clearly links ETR1 with the EIL3 action and therefore with sensing of S status. Moreover this linkage is not related with ETR1 role in primary ethylene signaling pathway as the used mutated receptor could not relay the signal of ethylene downstream the linear pathway. On the other hand, such mutation seems to not affect the ETR1 ability to properly function in the lateral part of ethylene signaling pathway. This means that data from the cited study22 could be treated as experimental validation of the model of lateral ethylene signaling pathway presented in this study. Summarizing, the data obtained using Nicotiana attenuata plants, which heterologously expressed the mutant A. thaliana receptor etr1–1 22 indicates that the correlation identified in this study could be explained by AgNO3 effect on ethylene receptors resulting in relay the signal to EIL3, which regulates most of the plant early response to S deficit.
For the first view, most of the effects showed in this study could be explained also by a hypothesis that silver ions could directly block S uptake. However, no data to confirm such theory exists in literature. On the other hand, data cited in the previous paragraph clearly confirms my model. Additionally, it has been proposed that the effect of Ag+ ions might be similar to other heavy metals.23 This means that silver ions should actually increase not decrease the S uptake. Noteworthy, data presented in this study (Fig. 2, Table A2) also indicates that low concentration of Ag+ ions should not result in S deficit.
I anticipate my model to be the starting point for more detailed analysis of the non-primary ethylene signaling pathway. Previously, the thinking of ethylene signaling pathway has been restricted only to functions linked with ethylene, but even the core elements of pathway might actually play role in more complex signaling. Utilizing this new point of view, the knowledge about structure and role of ethylene signaling pathway could be expanded. Additionally, my work explains the nature of up to now open, fundamental problem with determining the exact mechanism of S status sensing in plants. The ethylene signaling has not been seen before as the candidate to transfer S deficit signal because only the primary, linear pathway has been considered.
Acknowledgments
I thank my colleagues Lidia Borkiewicz, Katarzyna Zientara-Rytter and Ewa Krzywinska for critical comments on the letter and Agnieszka Sirko for discussions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Alonso JM, Stepanova AN. Ethylene Signaling Pathway. Sci Signal 2014; (Connections Map in the Database of Cell Signaling, as seen 23 July 2014), http://stke.sciencemag.org/cgi/cm/stkecm;CMP_13899. [Google Scholar]
- 2.Ju C, Yoon GM, Shemansky JM, Lin DY, Ying ZI, Chang J, Garrett WM, Kessenbrock M, Groth G, Tucker ML, et al.. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc Natl Acad Sci 2012; 109:19486-91; PMID:23132950; http://dx.doi.org/ 10.1073/pnas.1214848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiao H, Shen Z, Huang SC, Schmitz RJ, Urich MA, Briggs SP, Ecker JR. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 2012; 338:390-3; PMID:22936567; http://dx.doi.org/ 10.1126/science.1225974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wen X, Zhang C, Ji Y, Zhao Q, He W, An F, Jiang L, Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res 2012; 22:1613-6; PMID:23070300; http://dx.doi.org/ 10.1038/cr.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker† JR. Activation of the ethylene gas response pathway in arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 1997; 89:1133-44; PMID:9215635; http://dx.doi.org/ 10.1016/S0092-8674(00)80300-1 [DOI] [PubMed] [Google Scholar]
- 6.Solano R, Stepanova A, Chao Q, Ecker JR, Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 1998; 12:3703-14; PMID:9851977; http://dx.doi.org/ 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell Online 2006; 18:3235-51; PMID:17114350; http://dx.doi.org/ 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shakeel SN, Wang X, Binder BM, Schaller GE. Mechanisms of signal transduction by ethylene: overlapping and non-overlapping signalling roles in a receptor family. Aob Plants 2013; 5:plt010-plt010; PMID:23543258; http://dx.doi.org/ 10.1093/aobpla/plt010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo S-D, Cho Y-H, Tena G, Xiong Y, Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 2008; 451:789-95; PMID:18273012; http://dx.doi.org/ 10.1038/nature06543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adie B, Chico JM, Rubio-Somoza I, Solano R. Modulation of plant defenses by ethylene. J Plant Growth Regul 2007; 26:160-77; http://dx.doi.org/ 10.1007/s00344-007-0012-6 [DOI] [Google Scholar]
- 11.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot 2009; 60:3311-36; PMID:19567479; http://dx.doi.org/ 10.1093/jxb/erp204 [DOI] [PubMed] [Google Scholar]
- 12.Lynch J, Brown KM. Ethylene and plant responses to nutritional stress. Physiol Plant 1997; 100:613-9; http://dx.doi.org/ 10.1111/j.1399-3054.1997.tb03067.x [DOI] [Google Scholar]
- 13.Benlloch-González M, Romera J, Cristescu S, Harren F, Fournier JM, Benlloch M. K+ starvation inhibits water-stress-induced stomatal closure via ethylene synthesis in sunflower plants. J Exp Bot 2010; 61:1139-45; PMID:20054030; http://dx.doi.org/ 10.1093/jxb/erp379 [DOI] [PubMed] [Google Scholar]
- 14.Hermans C, Vuylsteke M, Coppens F, Cristescu SM, Harren FJM, Inzé D, Verbruggen N. Systems analysis of the responses to long-term magnesium deficiency and restoration in Arabidopsis thaliana. New Phytol 2010; 187:132-44; PMID:20412444; http://dx.doi.org/ 10.1111/j.1469-8137.2010.03257.x [DOI] [PubMed] [Google Scholar]
- 15.Vestreng V, Myhre G, Fagerli H, Reis S, Tarrasón L. Twenty-five years of continuous sulphur dioxide emission reduction in Europe. Atmos Chem Phys 2007; 7:3663-81; http://dx.doi.org/ 10.5194/acp-7-3663-2007 [DOI] [Google Scholar]
- 16.Schnug E, Ernst WHO, Kraztz S, Knolle F, Haneklaus S. Aspects of ecotoxicology of sulphur in the Harz region—a guided excursion, Landbauforsch. Voelkenrode 2004; 54:129-43 [Google Scholar]
- 17.Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 2013; 451:145-54; PMID:23535167; http://dx.doi.org/ 10.1042/BJ20121744 [DOI] [PubMed] [Google Scholar]
- 18.Moniuszko G, Skoneczny M, Zientara-Rytter K, Wawrzyńska A, Głów D, Cristescu SM, Harren FJ, Sirko A. Tobacco LSU-like protein couples sulphur-deficiency response with ethylene signalling pathway. J Exp Bot 2013; 5173-82; PMID:24085579; http://dx.doi.org/ 10.1093/jxb/ert309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuchi S, Cesco S, Varanini Z, Pinton R, Astolfi S. Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta 2009; 230:85-94; PMID:19350269; http://dx.doi.org/ 10.1007/s00425-009-0919-1 [DOI] [PubMed] [Google Scholar]
- 20.Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K, Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 2003; 33:651-63; PMID:12609039; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01658.x [DOI] [PubMed] [Google Scholar]
- 21.Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 2003; 33:633-50; PMID:12609038; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01657.x [DOI] [PubMed] [Google Scholar]
- 22.Meldau DG, Meldau S, Hoang LH, Underberg S, Wünsche H, Baldwin IT, Dimethyl disulfide produced by the naturally associated bacterium bacillus sp B55 promotes nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell Online 2013; 25:2731-47; PMID:23903320; http://dx.doi.org/ 10.1105/tpc.113.114744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vannini C, Domingo G, Onelli E, Prinsi B, Marsoni M, Espen L, Bracale M. Morphological and proteomic responses of eruca sativa exposed to silver nanoparticles or silver nitrate. Plos One 2013; 8:e68752; PMID:23874747; http://dx.doi.org/ 10.1371/journal.pone.0068752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008; 2008:http://dx.doi.org/ 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coskun D, Britto DT, Jean Y-K, Schulze LM, Becker A, Kronzucker HJ, Silver ions disrupt K+ homeostasis and cellular integrity in intact barley (Hordeum vulgare L.) roots. J Exp Bot 2012; 63:151-62; PMID:21948852; http://dx.doi.org/ 10.1093/jxb/err267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strader LC, Beisner ER, Bartel B. Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell Online 2009; 21:3585-90; PMID:19903871; http://dx.doi.org/ 10.1105/tpc.108.065185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nocito FF, Lancilli C, Crema B, Fourcroy P, Davidian J-C, Sacchi GA. Heavy metal stress and sulfate uptake in maize roots. Plant Physiol 2006; 141:1138-48; PMID:16698905; http://dx.doi.org/ 10.1104/pp.105.076240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel BK, Binder BM. Ethylene receptor 1 (ETR1) is sufficient and has the predominant role in mediating inhibition of ethylene responses by silver in Arabidopsis thaliana. J Biol Chem 2012; 287:26094-103; PMID:22692214; http://dx.doi.org/ 10.1074/jbc.M112.383034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 1999; 284:2148-52; PMID:10381874; http://dx.doi.org/ 10.1126/science.284.5423.2148 [DOI] [PubMed] [Google Scholar]
- 30.Scharein B, Groth G, Phosphorylation alters the interaction of the Arabidopsis phosphotransfer protein AHP1 with its sensor kinase ETR1. Plos One 2011; 6:e24173; PMID:21912672; http://dx.doi.org/ 10.1371/journal.pone.0024173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hass C, Lohrmann J, Albrecht V, Sweere U, Hummel F, Yoo SD, Hwang I, Zhu T, Schäfer E, Kudla J, et al.. The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. Embo J 2004; 23:3290-302; PMID:15282545; http://dx.doi.org/ 10.1038/sj.emboj.7600337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason MG, Mathews DE, Argyros DA, Maxwell BB, Kieber JJ, Alonso JM, Ecker JR, Schaller GE. Multiple type-B response regulators mediate cytokinin signal transduction in arabidopsis. Plant Cell Online 2005; 17:3007-18; PMID:16227453; http://dx.doi.org/ 10.1105/tpc.105.035451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novikova GV, Moshkov IE, Smith AR, Hall MA. The effect of ethylene on MAPKinase-like activity in Arabidopsis thaliana. Febs Lett 2000; 474:29-32; PMID:10828445; http://dx.doi.org/ 10.1016/S0014-5793(00)01565-9 [DOI] [PubMed] [Google Scholar]
- 34.Ouaked F, Rozhon W, Lecourieux D, Hirt H. A MAPK pathway mediates ethylene signaling in plants. Embo J 2003; 22:1282-8; PMID:12628921; http://dx.doi.org/ 10.1093/emboj/cdg131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall AE, Bleecker AB. Analysis of combinatorial loss-of-function mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell Online 2003; 15:2032-41; PMID:12953109; http://dx.doi.org/ 10.1105/tpc.013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen PB, Cancel JD. Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J 2003; 34:709-18; PMID:12787251; http://dx.doi.org/ 10.1046/j.1365-313X.2003.01762.x [DOI] [PubMed] [Google Scholar]
- 37.Joo S, Liu Y, Lueth A, Zhang S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J 2008; 54:129-40; PMID:18182027; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03404.x [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell Online 2004; 16:3386-99; PMID:15539472; http://dx.doi.org/ 10.1105/tpc.104.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Li Y, Wang Y, Liu H, Lei L, Yang H, Liu G, Ren D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 2008; 283:26996-7006; PMID:18693252; http://dx.doi.org/ 10.1074/jbc.M801392200 [DOI] [PubMed] [Google Scholar]
- 40.Zhao Q, Guo H-W. Paradigms and paradox in the ethylene signaling pathway and interaction network. Mol Plant 2011; 4:626-34; PMID:21690206; http://dx.doi.org/ 10.1093/mp/ssr042 [DOI] [PubMed] [Google Scholar]
- 41.Hahn A, Harter K. Mitogen-activated protein kinase cascades and ethylene: signaling, biosynthesis, or both?. Plant Physiol 2009; 149:1207-10; PMID:19109412; http://dx.doi.org/ 10.1104/pp.108.132241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Pasini R, Dan H, Joshi N, Zhao Y, Leustek T, Zheng ZL. Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J 2014; 77:185-97; PMID:24308460; http://dx.doi.org/ 10.1111/tpj.12376 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


