Abstract
The phytochemical indole-3-carbinol is produced in Cruciferous plants upon tissue rapture and deters herbivores. We recently showed that indole-3-carbinol modulates auxin signaling in root tips. Here we present transcript profiling experiments which further reveal the influence of indole-3-carbinol on auxin signaling in root tips, and also show that I3C affects auxin transporters. Brief treatment with indole-3-carbinol led to a reduction in the amount of PIN1 and to mislocalization of PIN2.
Keywords: Arabidopsis thaliana, auxin, glucosinolate, indole 3 carbinol
Indole-3-carbinol (I3C) is a phytochemical endogenously produced in the Cruciferae plant family. It is formed from the breakdown of indole-3-methylglucosinolate (I3M), which is derived from glucose and tryptophan.1 I3M is the predominant indole glucosinolate, and one of the most prominent glucosinolates detected in roots. The cleavage of I3M to I3C is catalyzed by myrosinase.2,3
In humans, a rich cruciferous vegetables diet has been associated with reduced chances of cancer, and I3C as a therapeutic treatment has potential for both prevention and treatment of a wide verity of cancers, such as leukemia, breast cancer and prostate cancer among others.4-8
In Arabidopsis thaliana I3C is synthesized upon tissue rapture and deters herbivores. The glucosinolates and the myrosinase are normally stored in separate compartments in the plant cells. In response to plant damage or insect attack I3M and the myrosinase are mixed and I3C is synthesized. I3C protects the plant as it is toxic to herbivores, insects and pathogens.9
While the toxic and deterrent effects of glucosinolate breakdown on herbivores and pathogens have been extensively studied, the secondary responses that are induced in the plant by I3C are only now starting to be revealed. Recently we have found that I3C effects plant growth and development by modulating auxin signaling in the root tips.10
In an attempt to reveal a bigger picture of the effect of I3C on auxin signaling in Arabidopsis roots, we carried out microarray experiments that revealed the extensive effect of I3C on the plant at the molecular level in general, and more specifically, on auxin responsive genes. We also used seedlings expressing auxin transporters reporter genes to understand if the effect of I3C is local or wide spread in the root.
The auxin-dependent interaction between SCFTIR1 with the Aux/IAA proteins and subsequent degradation of the Aux/IAA transcriptional repressors regulates the transcription of auxin-induced genes.11-14 As we previously showed that I3C inhibits the interaction of auxin with TIR1,10 we hypothesized that auxin-regulated genes would be misregulated following I3C treatment. We carried out a transcript-profiling experiment on roots tips briefly exposed to I3C. Gene ontology analysis showed that auxin-regulated genes are preferentially misregulated (FDR = 7.00E-05) in roots one hour following I3C treatment. The basal expression of at least 32 genes (12.8%) from a set of 250 auxin regulated genes is misregulated following I3C treatment (Fig. 1).
Figure 1.
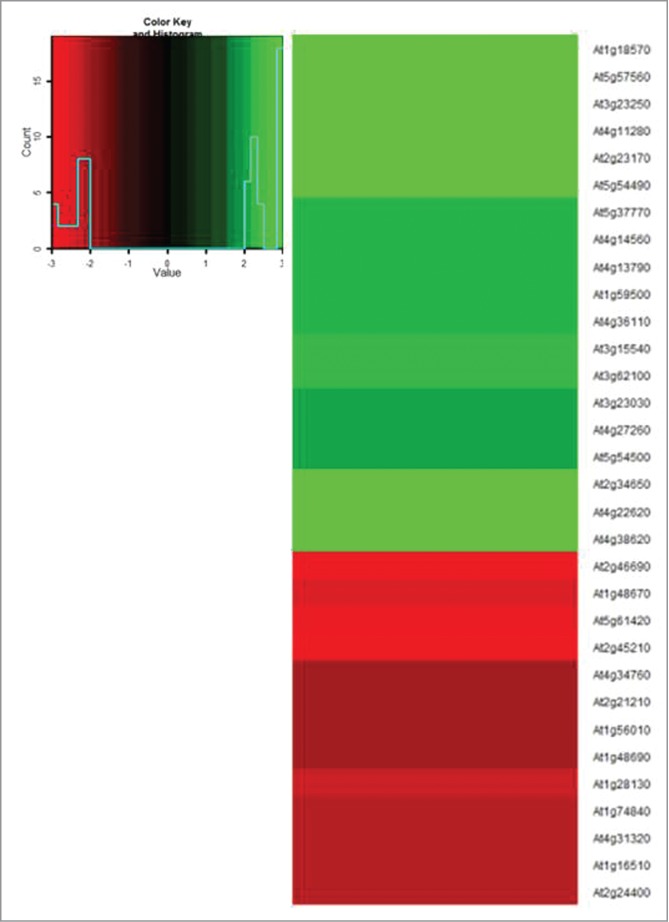
I3C treatment leads to misregulation of auxin responsive genes. Heat map of the expression of known auxin-responsive genes following treatment with 500μM I3C for one hour. RNA was extracted from root tips of 10-day-old seedlings and hybridized to Affymetrix ATH1 chips. A 2 fold cutoff was used. The list of auxin-responsive genes was extracted according to gene ontology in Agrigo [22].
The distribution of auxin in the root is determined mainly by its transporters. The PIN family of active auxin transporters particularly have a major role in regulating auxin distribution,15 and indeed different PIN proteins have a cell-specific polar localization [reviewed in16].
To understand if the effect of I3C on auxin is unique to the root tips or more wide spread, we checked if auxin transport is also affected by I3C treatment. For this purpose we used seedlings expressing PIN1:GFP or PIN2:GFP.17,18 Seedlings were grown on MS medium for 5 to 6 days, treated with 200 µM I3C for 30 minutes, and GFP fluorescence was monitored by confocal microscopy.
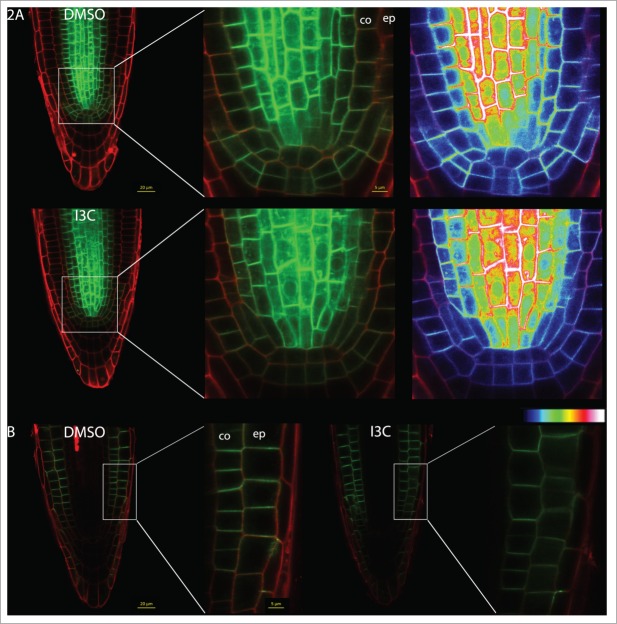
The PIN1 transporter directs auxin to the root tips to create the auxin maxima. Thus under normal conditions PIN1 is localized at the basal side of the cells of the vascular bundle and can also be found in the epidermal and cortical cells that surround the quiescent center19 (Fig. 2A). Following the short I3C treatment, we detected a reduced amount of PIN1 protein in the epidermis and cortex (Fig. 2A). Quantification of the relative integrated density of the GFP fluorescence in these cells showed a significant decrease in the GFP signal following the I3C treatment (50% of the signal of the control plants, P < 0.01; Student's t test).
Figure 2.
I3C treatment affects auxin transporters. (A) Seedlings expressing PIN1:GFP were grown on MS medium for 5 days, treated with 200 μM of I3C or with DMSO for 30 minutes, and imaged using confocal microscopy (Zeiss LSM780, with a 40x water objective). Heat-maps represent GFP density. GFP fluorescence was quantified using ImageJ software. (B) Seedlings expressing PIN2:GFP were grown on MS medium for 6 days, treated with 200 μM of I3C or with DMSO for 30 minutes, and imaged using confocal microscopy (Zeiss LSM780, with a 40x water objective). Cell walls were stained using 0.005mg/ml propidium iodide. co = cortex, ep = epidermis.
The PIN2 transporter is localized at the basal side of the cortical cells and at the apical side of the epidermal cells19 (Fig. 2B). Following the short I3C treatment, we found that even though the amount of the PIN2 protein was not significantly changed following the I3C treatment, its localization was altered by the treatment. I3C treatment caused the PIN2 proteins to be more diffuse in the cells and less oriented to the cell membranes (Fig. 2B).
Based on the Sachs canalization hypothesis,20 it is plausible that the effect of I3C on the PIN proteins is not direct. According to this hypothesis, auxin signaling can affect its own transport by regulating its transporters. Indeed a change in auxin concentration in the root affects the polarization of PIN1 and PIN2.21 Since we have recently shown that I3C modulates auxin signaling,10 this change might cause the mislocalization of PIN2 and the decrease in the amount of PIN1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the following for technical assistance: Dr. Eilon Shani, Iris Tal, and Dr. Daria Bloch. We thank Dr. Sigal Rencus-Lazar for critical reading.
Funding
This research was supported by a grant from the Binational Agricultural Research and Development Fund (BARD, IS-4505-12R) to DAC.
References
- 1.Fahey JW, Zalcmann AT, Talalay P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001. 56:5-51; PMID:11198818; http://dx.doi.org/ 10.1016/S0031-9422(00)00316-2 [DOI] [PubMed] [Google Scholar]
- 2.Agerbirk N, De Vos M, Kim JH, Jander G Indole glucosinolate breakdown and its biological effects. Phytochem Rev 2009; 8:101-20; http://dx.doi.org/ 10.1007/s11101-008-9098-0 [DOI] [Google Scholar]
- 3.Halkier BA, Gershenzon J. Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 2006; 57:303-33; PMID:16669764; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- 4.Meng Q, Goldberg ID, Rosen EM, Fan S. Inhibitory effects of Indole-3-carbinol on invasion and migration in human breast cancer cells. Breast Cancer Res Treat 2000; 63:147-52; PMID:11097090; http://dx.doi.org/ 10.1023/A:1006495824158 [DOI] [PubMed] [Google Scholar]
- 5.Meng Q, Qi M, Chen DZ, Yuan R, Goldberg ID, Rosen EM, Auborn K, Fan S. Suppression of breast cancer invasion and migration by indole-3-carbinol: associated with up-regulation of BRCA1 and E-cadherin/catenin complexes. J Mol Med 2000; 78:155-65; PMID:10868478; http://dx.doi.org/ 10.1007/s001090000088 [DOI] [PubMed] [Google Scholar]
- 6.Firestone GL, Bjeldanes LF. Indole-3-carbinol and 3-3′-diindolylmethane antiproliferative signaling pathways control cell-cycle gene transcription in human breast cancer cells by regulating promoter-Sp1 transcription factor interactions. J Nutr 2003; 133(7Suppl):2448S-55S; PMID:12840223 [DOI] [PubMed] [Google Scholar]
- 7.Sarkar FH, Li Y. Indole-3-carbinol and prostate cancer. J Nutr 2004; 134(12 Suppl):3493S-8S; PMID:15570059 [DOI] [PubMed] [Google Scholar]
- 8.Fan S, Meng Q, Auborn K, Carter T, Rosen EM. BRCA1 and BRCA2 as molecular targets for phytochemicals indole-3-carbinol and genistein in breast and prostate cancer cells. Br J Cancer 2006; 94:407-26; PMID:6434996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Jander G. Myzus persicae (green peach aphid) feeding on Arabidopsis induces the formation of a deterrent indole glucosinolate. Plant J 2007; 49:1008-19; PMID:17257166; http://dx.doi.org/ 10.1111/j.1365-313X.2006.03019.x [DOI] [PubMed] [Google Scholar]
- 10.Katz E, Nisani S, Yadav BS, Woldemariam MG, Shai B, Obolski U, Ehrlich M, Shani E, Jander G, Chamovitz DA. The glucosinolate breakdown product indole-3-carbinol acts as an auxin antagonist in roots of Arabidopsis thaliana. Plant J 2015; 82:547-55; PMID:25758811; http://dx.doi.org/ 10.1111/tpj.12824 [DOI] [PubMed] [Google Scholar]
- 11.Gray WM., Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 2001; 414:271-6; PMID:11713520; http://dx.doi.org/ 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- 12.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 2005; 435:441-5; PMID:15917797; http://dx.doi.org/ 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- 13.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005; 435:446-51; PMID:15917798; http://dx.doi.org/ 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- 14.Villalobos C, Caballero E, Sanz-Blasco S, Núñez L. Study of neurotoxic intracellular calcium signalling triggered by amyloids. Methods Mol Biol 2012; 849:289-302; PMID:22528098; http://dx.doi.org/ 10.1007/978-1-61779-551-0_20 [DOI] [PubMed] [Google Scholar]
- 15.Grieneisen VA, Xu J, Marée AF, Hogeweg P, Scheres B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007; 449:1008-13; PMID:17960234; http://dx.doi.org/ 10.1038/nature06215 [DOI] [PubMed] [Google Scholar]
- 16.Friml J. Auxin transport - shaping the plant. Curr Opin Plant Biol 2003; 6:7-12; PMID:12495745; http://dx.doi.org/ 10.1016/S1369526602000031 [DOI] [PubMed] [Google Scholar]
- 17.Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003; 115:591-602; PMID:14651850; http://dx.doi.org/ 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- 18.Xu JBS. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 2005; 17:525-36; PMID:15659621; http://dx.doi.org/ 10.1105/tpc.104.028449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005; 433:39-44; PMID:15635403; http://dx.doi.org/ 10.1038/nature03184 [DOI] [PubMed] [Google Scholar]
- 20.Sachs T. The control of patterned differentiation of vascular tissues. Adv Bot Res 1981; 9:151-262; http://dx.doi.org/ 10.1016/S0065-2296(08)60351-1 [DOI] [Google Scholar]
- 21.Sauer M, Balla J, Luschnig C, Wisniewska J, Reinöhl V, Friml J, Benková E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 2006; 20:2902-11; PMID:17043314; http://dx.doi.org/ 10.1101/gad.390806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou D, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucl Acids Res 2010; 38:W64-70; PMID:20435677; http://dx.doi.org/ 10.1093/nar/gkq310 [DOI] [PMC free article] [PubMed] [Google Scholar]