ABSTRACT
Cytokinins control key aspects of plant growth, including shoot and root meristem development and the timing of senescence of leaves and stems. Cytokinin perception triggers a 2-component signaling mechanism that ultimately leads to phosphorylation-dependent activation of a class of transcriptional regulators called type-B ARRs (RRBs). We have recently shown that the stability of the RRB family member ARR1 is increased in response to elevated cytokinin concentrations. In contrast, cytokinin decreases the stability of the closely related RRB member ARR2. The molecular mechanism governing the differential stability regulation of these 2 closely related RRBs remains unknown.
KEYWORDS: ARR2, ARR1, cytokinin signaling, protein stability, proteasome, type-B ARRs, type-A ARRs
In Arabidopsis, cytokinin signaling is initiated by the cytokinin-induced auto-phosphorylation of histidine kinase receptors (AHKs).1,2 The phosphatidyl group is then transferred via histidine phosphotransfer proteins (AHPs) to response regulators (RRs) that are activated upon phosphorylation. RRs form 2 functionally antagonistic classes: the response promoting type-B RRs (RRBs) and the response inhibiting type-A RRs (RRAs). RRBs are transcriptional activators that regulate the expression of primary cytokinin-response genes, including RRAs. RRAs in turn function as negative feedback regulators of cytokinin signaling through an unknown mechanism.1,2
We have recently shown that the cellular pool of the Arabidopsis RRB member ARR1 is comprised of 2 subpools: a subpool with stable ARR1 that cannot be depleted even after prolonged treatments with protein synthesis inhibitors and an unstable ARR1 subpool that can be detected only after treatments with a proteasome inhibitor.3 In addition, we found that the unstable ARR1 subpool is increased in response to cytokinin treatment, implying that the cytokinin signal inhibits the proteasome-dependent degradation of ARR1. Interestingly, RRA loss of function also caused an increase in ARR1 level, suggesting that the RRAs inhibit cytokinin responses by promoting ARR1 degradation.
Contrary to our results,3 earlier reports showed that ARR1 is an unstable protein, and that its proteasome-dependent degradation is not regulated by cytokinins.4-6 However, these studies analyzed a chimeric ARR1 version in which the protein was fused to a peptide tag for immunodetection, while our study was based on detection of endogenous ARR1. This suggests that ARR1 fusion proteins do not accurately reflect ARR1 stability control. If tagging of ARR1 altered its stability, then the proteasome targeting of other RRBs may also be impacted by the presence of peptide tags. To address this question, we analyzed the stability of ARR2, a less abundant member of the ARR1/ARR2 RRB subfamily.7 In mesophyll protoplasts, a transiently expressed ARR2-hemagglutinin (ARR2-HA) tagged protein was shown to be unstable and its degradation was accelerated by cytokinin treatments.12 To determine the stability of unmodified ARR2, we generated ARR2-specific antibodies against a synthetic peptide NGRSSRKRKEEEVDC using a previously described methodology.3 Contrary to ARR1, which was detectable with specific antisera,3 we were unable to visualize the endogenous ARR2 protein by immunoblotting analyses. Therefore, we generated transgenic plants expressing the full-length ARR2 cDNA from the strong constitutive CaMV 35S promoter (Fig. 1). Prior to protein stability assays, we tested whether the overexpressed ARR2 is indeed functional. It has been shown previously that high-level expression of RRBs alters development and induces cytokinin hypersensitivity.8-10 Indeed, the strong overexpressing lines were dwarfed, had shorter roots (Fig. 1A), increased anthocyanin content (4.00 ± 0.01, and 5.64 ± 0.01 fold increase [n = 3] compared to Col-0 for lines #2 and #3, respectively) and shorter hypocotyls when grown in darkness (72 ± 3% and 76 ± 2% [n = 25] of Col-0 hypocotyl length for lines #2 and #3, respectively). These phenotypes are characteristic of plants treated with cytokinins.8-10 Additionally, both lines were hypersensitive to cytokinin in a shoot induction response assay (Fig. 1B).
Figure 1.
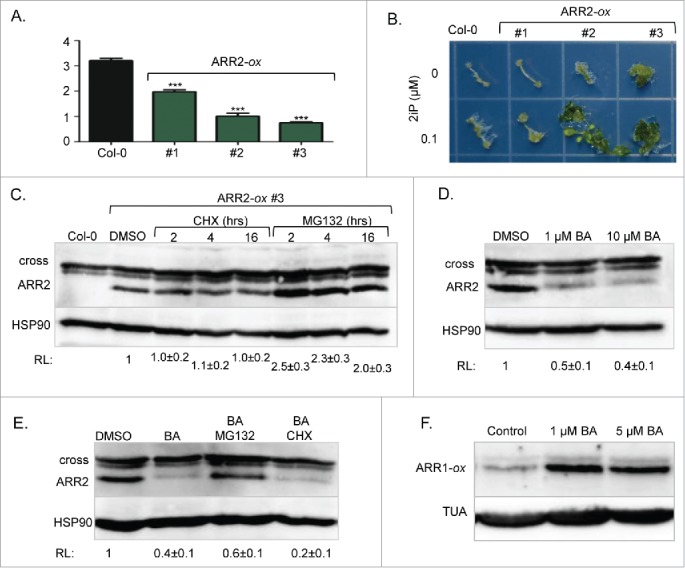
Stability control of ARR2. (A) Effect of ARR2 overexpression (ARR2-ox) on the root length. Seeds were sown and grown on vertically positioned plates with half-strength Murashige and Skoog growth (MS/2) media. After seven days of growth, seedlings were photographed and root length was measured using ImageJ. Data are presented and mean ± SD (n≥12 ). ***, p<0.001 (B) Shoot induction assay. Plants grown on MS/2 media for 7 d under dark conditions were exposed to light for 4 d to strengthen etiolated hypocotyls. Hypocotyls were then excised and placed on MS/2 media containing 1 µM NAA with or without 0.1 µM 2iP. Explants incubated on shoot induction media for 40 d were transferred to a new plate for photography. 2iP, N6-(2-isopentenyl)adenine. (C) Seven-day-old ARR2-ox #3 seedlings were treated for 2, 4 or 16 hours with 200 µM of the protein synthesis inhibitor cycloheximide (CHX) or 100 µM of the proteasome inhibitor MG132 as described.3 Immunoblotting analyses were done as described.3 Proteins were resolved on a 6% acrylamide SDS/PAGE gel. After probing with anti-ARR2 antibodies, membranes were re-probed with anti-HSP90 sera to demonstrate equal loading. Mean ± SD (n≥3 ) of the relative signal intensity (RL) is shown below the blot. (D) Seven-day-old ARR2-ox #3 seedlings were treated for 4 hours with 1 µM or 10 µM of the cytokinin BA. Immunoblotting was done as described in C. (E) Seven-day-old ARR2-ox #3 seedlings were treated for 4 hours with 1 µM of BA in the absence or presence of 100 µM MG132 or 200 µM CHX. (F) Seven-day-old transgenic plants overexpressing ARR1 (ARR1-ox) were treated for 4 hours with 1 µM or 5 µM BA. Proteins were resolved on 7.5% acrylamide SDS/PAGE gels. After probing with anti-ARR1 antibodies, membranes were re-probed with anti-α tubulin (TUA) antibodies.
Analyses of the ARR2 overexpression lines revealed that ARR2 abundance is indeed cytokinin-regulated in a manner opposite to ARR1. Like ARR1,3 the ARR2 steady-state level was not affected by prolonged treatment with the translation inhibitor cyclohexamide (Fig. 1C), suggesting that this ARR2 version is not actively targeted for proteolysis, in contrast to what was reported earlier.12 ARR2 abundance was increased upon treatment with the proteasome inhibitor MG132 suggesting that, like ARR1,3 a fraction of ARR2 is highly unstable and immediately degraded after synthesis (Fig. 1C). However, contrary to the cytokinin-stabilized ARR1, we observed a decrease in ARR2 abundance in response to cytokinin treatment (Fig. 1D). This decrease required 26S proteasome activity as it was blocked by MG132 (Fig. 1E). Thus, contrary to ARR1, ARR2 is destabilized by increased cytokinin action. This result was in agreement with the earlier study that used ARR2-HA, suggesting that this fusion protein retained its cytokinin stability control.12 However, the ARR1-HA version analyzed in the same study was shown to be cytokinin insensitive.12 One possible explanation for this discrepancy is that ARR1-HA was expressed from a strong constitutive promoter and that this caused a deregulation of its cytokinin stability control. However, similar to its effect on endogenous ARR1, cytokinin treatment also increased the abundance of overexpressed and unmodified ARR1, indicating that increased expression of an RRB does not alter its cytokinin-regulated stability (Fig. 1F).
In conclusion, cytokinin-regulated proteolytic control of ARR1 and ARR2 differs; while cytokinin signaling led to accumulation of ARR1, it induced degradation of ARR2. This is unusual considering that in other hormone signaling pathways the perception of the signal led to either degradation or stabilization of functionally related pathway components.11-13 In all described cases, a single E3 ubiquitin ligase or a family of specific E3s is responsible for signal-regulated protein degradation.11-13 A previous study suggested that a family of F-box proteins targets ARR1 and ARR2 for proteasomal degradation.5 However, this earlier report, which used chimeric RRB versions, did not reveal any effect of cytokinin on the stability of these 2 RRB members. This, in combination with our data, would suggest that there may be different RRB-targeting E3s whose actions are regulated by cytokinin. Indeed, cytokinin treatments lead to de novo synthesis of a battery of E3s, and their functions in the regulation of the cytokinin signaling pathway is still poorly understood.14-16 Collectively, these results suggest that our current understanding of the molecular mechanisms that control RRB proteolysis is still incomplete.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from NIFA (1009329), NSF (0919991) and the Kentucky Tobacco Research and Development Center (Lexington, KY, USA). T. Shull was supported by an Undergraduate Summer Research and Creativity Fellowship from the University of Kentucky.
References
- 1.Kieber JJ, Schaller GE. Cytokinins. Arabidopsis Book 2014; 12:e0168; PMID:24465173; http://dx.doi.org/ 10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hwang I, Sheen J, Muller B. Cytokinin signaling networks. Annu Rev Plant Biol 2012; 63:353-80; PMID:22554243; http://dx.doi.org/ 10.1146/annurev-arplant-042811-105503 [DOI] [PubMed] [Google Scholar]
- 3.Kurepa J, Li Y, Smalle JA. Cytokinin signaling stabilizes the response activator ARR1. Plant J 2014; 78:157-68; PMID:24617630; http://dx.doi.org/ 10.1111/tpj.12458` [DOI] [PubMed] [Google Scholar]
- 4.Hill K, Mathews DE, Kim HJ, Street IH, Wildes SL, Chiang YH, Mason MG, Alonso JM, Ecker JR, Kieber JJ, Schaller GE. Functional characterization of type-B response regulators in the Arabidopsis cytokinin response. Plant Physiol 2013; 162:212-24; PMID:23482873; http://dx.doi.org/ 10.1104/pp.112.208736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HJ, Chiang YH, Kieber JJ, Schaller GE. SCFKMD controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc Natl Acad Sci U S A 2013; 110:10028-33; PMID:23720308; http://dx.doi.org/ 10.1073/pnas.1300403110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser MT, Persson S. Cracking the elusive alignment hypothesis: the microtubule-cellulose synthase nexus unraveled. Trends Plant Sci 2012; 17:666-74; PMID:22784824; http://dx.doi.org/ 10.1016/j.tplants.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 2004; 135:927-37; PMID:15173562; http://dx.doi.org/ 10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:89-118; PMID:11337393; http://dx.doi.org/ 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- 9.Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001; 294:1519-21; PMID:11691951; http://dx.doi.org/ 10.1126/science.1065201 [DOI] [PubMed] [Google Scholar]
- 10.van der Graaff EE, Hooykaas PJJ, Auer CA. Altered development of Arabidopsis thaliana carrying the Agrobacterium tumefaciens ipt gene is partially due to ethylene effects. Plant Growth Regulation 2001; 34:305-15; http://dx.doi.org/ 10.1023/A:1013351502643 [DOI] [Google Scholar]
- 11.Irigoyen ML, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, et al.. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014; 26:712-28; PMID:24563205; http://dx.doi.org/ 10.1105/tpc.113.122234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 2001; 414:271-6; PMID:11713520; http://dx.doi.org/ 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Wang B, Jiang L, Liu X, Li X, Lu Z, Meng X, Wang Y, Smith SM, Li J. Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-Like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 2015; 27:3128-42; PMID:26546446; http://dx.doi.org/ 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 2013; 162:272-94; PMID:23524861; http://dx.doi.org/ 10.1104/pp.113.217026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner ED, Feinberg P, Runko S, Coruzzi GM. A mutation in theproteosomal regulatory particle AAA-ATPase-3 in Arabidopsis impairs the light-specific hypocotyl elongation response elicited by a glutamate receptor agonist, BMAA. Plant Mol Biol 2009; 70:523-33; PMID:19412571; http://dx.doi.org/ 10.1007/s11103-009-9489-7 [DOI] [PubMed] [Google Scholar]
- 16.Rashotte AM, Carson SD, To JP, Kieber JJ. Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 2003; 132:1998-2011; PMID:12913156; http://dx.doi.org/ 10.1104/pp.103.021436 [DOI] [PMC free article] [PubMed] [Google Scholar]


