Abstract
Argonautes (AGOs) are the effector proteins of the RNA-induced silencing (RISC) complex, formed during the phenomena of small-RNA mediated post-transcriptional gene silencing. AGOs are a large family of proteins; their number varies from a few (4 in Chlamydomonas reinhardtii) to many (18 in Oryza sativa) in plants. Genetics-guided analysis have demonstrated the roles of some of the AGOs during growth and development of plants. Biochemical studies have further revealed differences in functional specificities among AGOs. How the AGO family expanded in different plant species during the course of evolution is starting to emerge. We hypothesized that 4 classes of AGOs evolved after divergence of unicellular green algae when an ancestral AGO underwent duplication events. Evolution of multicellularity may have coincided with the diversification of AGOs. A comparative sequence and structure analysis of the plant AGOs, including those from the mosses and the unicellular algae, show not only conformational differences between those from lower and higher plants, but also functional divergence of important sites.
Keywords: Argonaute, evolution, plants, structure
Small-RNA- (smRNA) mediated gene-silencing pathways, often also referred to as RNA interference (RNAi), are fundamental to the gene regulatory systems.1,2 During the process of recognition of mRNA by smRNAs, which guide cellular and biochemical processes such as development, differentiation, protein synthesis, mRNA stability, and genomic integrity, the Argonaute family of proteins act as effectors of the RNA-induced silencing complex (RISC).1,3-5 AGOs may display a diversity in pattern of change in their gene expressions when plants are subjected to environmental stresses.6-8 Plants display a diverse pool of smRNAs that are reprogrammed to help plants adapt to changes in their environment.9 Specificity in recruitment of smRNAs warrants functional divergence (due to evolutionary constraints) among AGO effectors, which in turn may be possible as a result of changes in protein structure. Elucidation of structures of human and yeast AGOs have indeed started to explain the molecular basis of the recognition of smRNA substrates by AGOs and their actions in eukaryotes.10-13
AGOs of multicellular plants have evolved into 4 phylogenetic classes through 5 successive duplication events in around 3.5 million years, after the divergence of unicellular forms but before the divergence of Bryophytes. The unicellular forms of AGOs might have independently evolved.14 In comparison to other classes, the seed recognition region and the nucleotide specificity loop in the class I AGOs (AGO1 and AGO10) tend to evolve at relatively slow rates. Further, likelihood ratio test suggest that selection constraints have been altered at many sites on AGOs among different classes; maximum being those between classes I and IV (class IV comprising of AGOs 4, 6, 8 and 914).
Evolutionary diversity in residues in the substrate-recognition and catalytic domains (e.g. RFY, DDH) of the 4 classes of plant AGOs has been clearly evident.14 Many such sites show a posterior probability of ≥0 .9. Mapping onto AtAGO4 (class IV representative) for example, the nonpolar G881, adjacent to the L880 that corresponds to A813 of HsAGO2 and critical for the MID-PIWI interface,15 is replaced with polar residue, R, in all the other AGOs (Fig. 1). Similarly, the diverged residue E682 in AtAGO4, corresponding to the D619 of HsAGO2 and important for Trp-pocket formation in PIWI domain,16 is largely replaced with residues D or N in AtAGO1, AtAGO2, AtAGO5, PptAGOlike1 and CrnAGO2 (Fig. 1). The N616, M637 and V640 in AtAGO4, which are in the 5′ binding region of MID domain, or G840 of PIWI domain, are also replaced with functionally divergent residues in other AGOs (Fig. 1).
Figure 1.
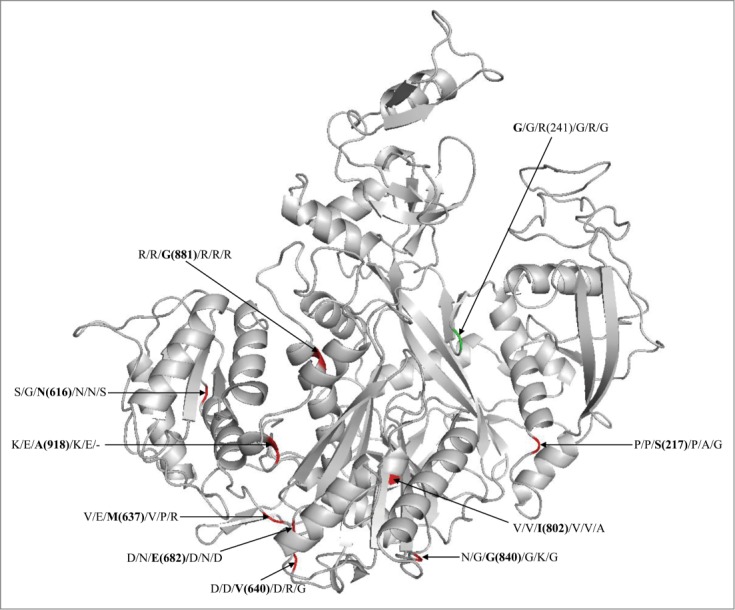
Modeled structure of AtAGO4 showing the functionally diverged sites. The labeled residues separated with ‘/’ at each site are the residues from AtAGO1, AtAGO2, AtAGO4, AtAGO5, PptAGOLike1 and CrnAGO2, respectively. The value in parenthesis is the coordinate of residue in AtAGO4. The sites with red color indicates that it has diveged in AtAGO4 than the other AGOs, while with green color indicate divergence in AtAGO1 than the other AGOs.
These changes in functional residues in the 4 classes of AGOs may affect structural conformations during interaction with diverse pool of smRNAs in the cell. In order to test this hypothesis, we modeled the changes in structures of representative AGOs for the 4 classes in plants (AtAGO1: class I, AtAGO2: class III; AtAGO4: class IV; AtAGO5: class II14) with 2 different RNA substrates that bind eukaryotic AGOs, 4F3T:R and 4W5O:B.10,17 We compared these to the respective substrate- bound AGOs of human (HsAGO2), Physcomitrella patens (PptAGOlike1) and the unicellular alga, Chlamydomonas reinharditii (CrnAGO2). Interestingly, the overall structure of plant AGOs may be largely conserved (Table 1). However, binding sites of plant AGOs, when bound to 2 RNA substrates, displayed diversity (Table 1, Fig. 2) with respect to surface area, volume, charge distribution, and affinities to substrate (interaction energy, hydrogen bond energy and potential energy). For instance, when compared to HsAGO2, AtAGO2 showed least RMSD (1.79) when bound to 4F3T:R. But putative binding of 4W5O:B induced conformational changes (Fig. 2) such that the RMSD was 3.20 (Table 1). On the other hand, RMSDs of HsAGO2 and AtAGO5 did not change with change in the RNA substrate (2.41–2.42; Table 1). Among the plant AGOs, AtAGO4 had the largest substrate-binding pocket (area and volume; Table 1). AtAGO4 indeed displayed the lowest interaction energy among all the plant AGOs for both the substrates, whereas a difference of 2-fold in interaction energy was noticed when AtAGO1 was independently docked with 2 substrates (Table 1). AtAGO5 recruited maximum number of positively charged residues to interact with an RNA substrate, followed by AtAGO1 and AtAGO4 (Table 1). AGOs may indeed have diverse affinities for substrate RNAs.
Table 1.
Analysis of structural diversity of plant AGOs (AtAGO1 (class I), AtAGO5 (class II), AtAGO2 (class III), AtAGO4 (class IV)) in comparison to the HsAGO2, PptAGOlike1 (Physcomitrella patens), and CrnAGO2 (unicellular alga, Chlamydomonas reinhardittii), bound to 2 different RNA substrates., 4F3T:R and 4W5O:B. (nd: could not be determined).
|
A. RMSD (upper diagonal) and TM scores (lower diagonal) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| 4F3T:R | HsAGO2 | AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO1 | CrnAGO2 | ||
| HsAGO2 | 2.12 | 1.79 | 2.12 | 2.42 | 2.04 | 3.84 | |||
| AtAGO1 | 0.95 | 2.59 | 2.50 | 2.85 | 2.46 | 4.23 | |||
| AtAGO2 | 0.96 | 0.90 | 2.57 | 2.97 | 2.34 | 3.92 | |||
| AtAGO4 | 0.93 | 0.90 | 0.90 | 3.21 | 2.81 | 3.82 | |||
| AtAGO5 | 0.94 | 0.90 | 0.88 | 0.86 | 3.24 | 4.34 | |||
| PptAGO1 | 0.94 | 0.90 | 0.90 | 0.90 | 0.89 | 3.83 | |||
| CrnAGO2 | 0.82 | 0.78 | 0.80 | 0.78 | 0.76 | 0.78 | |||
| 4W5O:B | HsAGO2 | AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO1 | CrnAGO2 | ||
| HsAGO2 | 3.21 | 3.20 | 3.42 | 2.41 | 3.44 | 3.82 | |||
| AtAGO1 | 0.91 | 2.47 | 2.55 | 2.87 | 2.33 | 4.28 | |||
| AtAGO2 | 0.90 | 0.90 | 2.63 | 2.93 | 2.39 | 3.86 | |||
| AtAGO4 | 0.88 | 0.90 | 0.90 | 3.18 | 2.90 | 3.84 | |||
| AtAGO5 | 0.94 | 0.91 | 0.88 | 0.86 | 3.28 | 4.30 | |||
| PptAGO1 | 0.88 | 0.90 | 0.90 | 0.90 | 0.89 | 3.90 | |||
| CrnAGO2 | 0.78 | 0.78 | 0.80 | 0.78 | 0.76 | 0.76 | |||
|
B. Area (Å2) of the largest binding pocket | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | 6088 | 7594 | 8165 | 3571 | 4675 | 8341 |
| 4W5O:B | 7898 | 6279 | 8918 | 3533 | 10366 | 4366 |
|
C. Volume (Å3) of the largest binding pocket | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | 15433 | 18034 | 20765 | 7477 | 10351 | 14642 |
| 4W5O:B | 18409 | 13177 | 20849 | 7214 | 22082 | 7235 |
|
D. Number of positively charged residues in MID-PIWI lobe | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | 186 | 145 | 161 | 204 | 182 | 220 |
| 4W5O:B | 178 | 151 | 171 | 195 | 199 | 227 |
|
E. Total interaction energy (Kcal/mol) | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | −6.11 | −6.6 | −11.48 | −6.87 | −4.86 | −9.7 |
| 4W5O:B | −12.37 | −8.72 | −12.41 | nd | −13.14 | −13.35 |
|
F. Total hydrogen bond energy (Kcal/mol) | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | −3.31 | −3.8 | −8.68 | −4.07 | −2.06 | −6.9 |
| 4W5O:B | −9.57 | −5.92 | −9.61 | nd | −10.34 | −10.55 |
|
G. Potential energy (electrostatic coulomb) of the AGO-smRNA complex | ||||||
|---|---|---|---|---|---|---|
| AtAGO1 | AtAGO2 | AtAGO4 | AtAGO5 | PptAGO | CrnAGO2 | |
| 4F3T:R | −2563.5 | −2180.9 | −2578.8 | −2152.2 | −2251.1 | −2554 |
| 4W5O:B | −3374.5 | −2707.7 | −2665.5 | −2778.9 | −2480.2 | −2630.4 |
Figure 2.
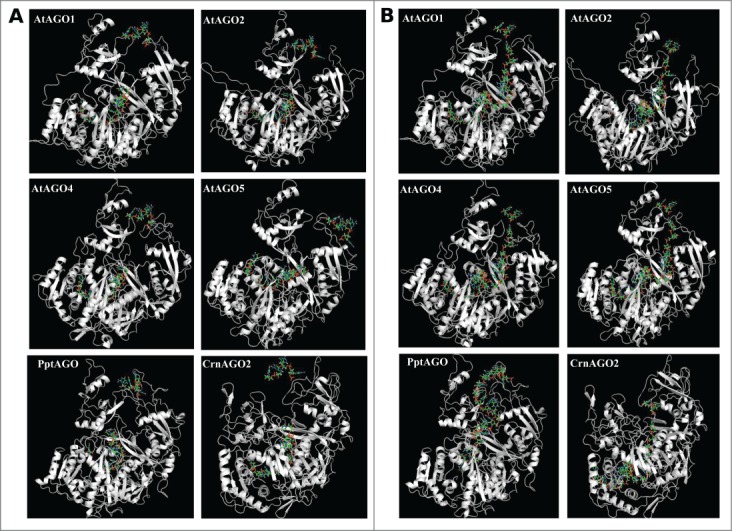
Modeling of interaction of the 4 representative AtAGOs, PptAGOlike1 and CrnAGO2 with the 2 RNA substrates, 4F3T:R (A) and 4W5O:B (B), respectively.
These indicate that different classes of AGOs may adapt variable structural conformations, particularly within the smRNA binding and catalytic domains. Thus, changes in ‘sturcture-function’ relationships due to evolutionary diversification among the 4 classes of plant AGO are bound to have implications on recruitment of smRNAs in a pathway.
Funding
This work is supported by MPG-India partner group program of the Max Planck Society and the Indo-German Center for Science and Technology/ Department of Science and Technology (India), and the WHEAT Competitive Grants Initiative, CIMMYT and the CGIAR (A4031.09.10). Funding support from IISER-Kolkata in the form of ‘startup’ to SPP is also acknowledged.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 2013; 64:137-59; PMID:23330790; http://dx.doi.org/ 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- 2.Baulcombe D. RNA silencing in plants. Nature 2004; 431:356-63; PMID:15372043; http://dx.doi.org/ 10.1038/nature02874 [DOI] [PubMed] [Google Scholar]
- 3.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008; 9:22-32; PMID:18073770; http://dx.doi.org/ 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- 4.Kuhn CD, Joshua-Tor L. Eukaryotic Argonautes come into focus. Trends Biochem Sci 2013; 38:263-71; PMID:23541793; http://dx.doi.org/ 10.1016/j.tibs.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 5.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat Genet 2006; 38 Suppl:S31-6; PMID:16736022; http://dx.doi.org/ 10.1038/ng1791 [DOI] [PubMed] [Google Scholar]
- 6.Kapoor M, Arora R, Lama T, Nijhawan A, Khurana JP, Tyagi AK, Kapoor S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 2008; 9:451; PMID:18826656; http://dx.doi.org/ 10.1186/1471-2164-9-451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shao F, Lu S. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genomics 2013; 14:512; PMID:23889895; http://dx.doi.org/ 10.1186/1471-2164-14-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Zhong J, Ouyang YD, Yao J. The integrative expression and co-expression analysis of the AGO gene family in rice. Gene 2013; 528:221-35; PMID:23872535; http://dx.doi.org/ 10.1016/j.gene.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 9.Pandey SP, Shahi P, Gase K, Baldwin IT. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc Natl Acad Sci U S A 2008; 105:4559-64; PMID:18339806; http://dx.doi.org/ 10.1073/pnas.0711363105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell 2012; 150:100-10; PMID:22682761; http://dx.doi.org/ 10.1016/j.cell.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J 2012; 31:3588-95; PMID:22850669; http://dx.doi.org/ 10.1038/emboj.2012.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature 2012; 486:368-74; PMID:22722195; http://dx.doi.org/ 10.1038/nature11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science 2012; 336:1037-40; PMID:22539551; http://dx.doi.org/ 10.1126/science.1221551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh RK, Gase K, Baldwin IT, Pandey SP. Molecular evolution and diversification of the Argonaute family of proteins in plants. BMC Plant Biol 2015; 15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith MR, Willmann MR, Wu G, Berardini TZ, Moller B, Weijers D, Poethig RS. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc Natl Acad Sci U S A 2009; 106:5424-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji L, Liu X, Yan J, Wang W, Yumul RE, Kim YJ, Dinh TT, Liu J, Cui X, Zheng B, et al.. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet 2011; 7:e1001358; PMID:21483759; http://dx.doi.org/ 10.1371/journal.pgen.1001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science 2014; 346:608-13; PMID:25359968; http://dx.doi.org/ 10.1126/science.1258040 [DOI] [PMC free article] [PubMed] [Google Scholar]


