Abstract
In this review, we report on the recent developments made using both genetics and functional genomics approaches in the discovery of genes controlling root development in rice. QTL detection in classical biparental mapping populations initially enabled the identification of a very large number of large chromosomal segments carrying root genes. Two segments with large effects have been positionally cloned, allowing the identification of two major genes. One of these genes conferred a tolerance to low phosphate content in soil, while the other conferred a tolerance to drought by controlling root gravitropism, resulting in root system expansion deep in the soil. Findings based on the higher-resolution QTL detection offered by the development of association mapping are discussed. In parallel with genetics approaches, efforts have been made to screen mutant libraries for lines presenting alterations in root development, allowing for the identification of several genes that control different steps of root development, such as crown root and lateral root initiation and emergence, meristem patterning, and the control of root growth. Some of these genes are closely phylogenetically related to Arabidopsis genes involved in the control of lateral root initiation. This close relationship stresses the conservation among plant species of an auxin responsive core gene regulatory network involved in the control of post-embryonic root initiation. In addition, we report on several genetic regulatory pathways that have been described only in rice. The complementarities and the expected convergence of the direct and reverse genetic approaches used to decipher the genetic determinants of root development in rice are discussed in regards to the high diversity characterizing this species and to the adaptations of rice root system architecture to different edaphic environments.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-014-0030-5) contains supplementary material, which is available to authorized users.
Keywords: Rice, Crown root, Lateral root, Branching, QTL mapping, Association mapping, Gene cloning, Direct genetics, Reverse genetics
Introduction
Roots are essential organs for exploring and exploiting soil resources, such as water and mineral nutrients. Different root architecture ideotypes that are adapted to different soil mineral nutrient balances or water statuses have been proposed. It is generally acknowledged that a deeper, thicker and more branched root system with a high root to shoot ratio can enhance the tolerance of rice to water deficits (Fukai and Cooper, [1995]; Gowda et al. [2011]). Roots develop primarily underground and are inaccessible to direct observation. This limitation partly explains why root architecture is not a widespread breeding objective. Because of this lack of past investment, there is room for improvement of rice root systems. This task is particularly important in a context of climate change, which will likely increase the occurrence of unfavorable conditions (Den Herder et al. [2010]). An improved root system can be obtained more easily using indirect selection methods, such as marker-assisted selection. It is therefore necessary to identify the main genetic determinants governing root development. Root development is a complex process that involves constitutive and adaptive mechanisms and regulatory correlations with the shoot part of the plant (Puig et al., [2012]). In this review, we focus on genetic determinants of root development in rice that were identified using complementary direct and reverse genetic approaches, such as QTL detection, mutant analysis and transcriptomics. The involvement of these genes in plant adaptation to stress conditions is discussed when such information is available and relevant.
Review
Main characteristics of the rice root system
The rice root system is composed of a seminal root and postembryonic shoot borne-roots termed crown roots (Rebouillat et al. [2009]; Coudert et al. [2010]). These two types of roots can branch and form long lateral roots or short lateral roots. Crown roots differentiate in the stem from a radial ground meristem that has common characteristics with the root pericycle (Coudert et al. [2013b]; Itoh et al. [2005]). Lateral roots differentiate from the root pericycle and, in part, from the endoderm (Rebouillat et al. [2009]; Orman-Ligeza et al. [2013]). The radial structure of the rice roots comprises the following tissues, from the center to periphery: the stele, including phloem and xylem vessels and the pericycle; the endoderm; the cortex, whose cells can undergo apoptosis to constitute the aerenchyma; the sclerenchyma; the exodermis; and the epidermis (Rebouillat et al. [2009]). This radial structure reflects the capacity of rice roots to grow in aerobic as well as anaerobic conditions. Notably, the aerenchyma enables gas exchange with the shoot when the plant is growing under anaerobic conditions. Rice was domesticated approximately 10 000 years ago (Sweeney and McCouch [2007]) and has since adapted to a large variety of ecosystems (irrigated, rainfed lowland, upland, flood-prone, and mangrove) and production systems (from traditional low-input systems to intensive high-input systems). More than 100 000 rice genotypes are available in the Genebank of the International Rice Research Institute (IRRI), Philippines. The phenotypic evaluation of a small sample of this diversity showed that the tested genotypes exhibited a great deal of root architecture variation (O’Toole and Bland [1987]; Lafitte et al. [2001]). The remaining unexplored extensive variability constitutes a very significant opportunity for the identification of new genes involved in root development.
Detection of QTLs for root development in rice
To identify genes controlling root system architecture, the direct genetic approach has historically preceded the reverse genetic approach. The direct genetic approach was first based on QTL detection in mapping populations. The first study on root architecture in rice was undertaken in 1995 (Champoux et al. [1995]), and a large number of studies followed. These studies were reviewed by Kamoshita et al. ([2008]) for those concerning the CT9993/IR622266 population and [Khowaja et al. (2009)] for those concerning Bala/Azucena. Courtois et al. ([2009]) analyzed the conditions of detection of 675 QTLs in 24 studies conducted on 12 different populations. Simple phenotyping systems, generally based on hydroponic culture conditions, pots or soil columns, characterized this period of work. In the majority of the cases (368 among the 675 QTLs), these phenotyping conditions were favorable in order to estimate the crop’s genetic potential; only a few studies specifically analyzed response QTLs in conditions of water stress or root penetration constraints (147 and 160 QTLs, respectively). The measured traits were those that were the most easily observable, such as maximum root length, root thickness, root number, mass of roots at different depths, or root to shoot ratio. The preferred mapping populations were recombinant inbred lines (RILs) or doubled haploid populations because the fixed nature of these lines enabled replicate measurements. To maximize the phenotypic differences between parents and simplify the identification of polymorphisms, most of the populations (8 populations representing 560 QTLs) were derived from indica x japonica crosses. The mapping population sizes were generally relatively small (between 100 and 150 accessions), enabling the identification of several QTLs with large effects but offering little resolution. Although large QTL x environment interactions were recorded (Kamoshita et al. [2002]; MacMillan et al. [2006]), some chromosomal regions appeared to be detected regularly in various populations and/or environment, as shown by meta QTL analyses (Courtois et al. [2009]; [Khowaja et al. 2009]). For the traits for which the highest number of QTL were detected, QTL clusters were found on chromosomes 1 (30–40 Mb), 2 (25–35 Mb), 3 (0–5 Mb), 4 (30–35 Mb), and 9 (15–20 Mb), as shown in Figure 1. More recently, the progress on high-throughput root phenotyping and 3D image reconstruction has led to a renewed interest in QTL detection in mapping populations, focusing on original traits (e.g., the root network bushiness or its convex hull volume), in addition to more classical traits, such as specific root length or specific root area (Topp et al. [2013]). This last study using the Bala/Azucena population revealed QTLs that had already been detected for simpler traits and identified new QTLs as well.
Figure 1.
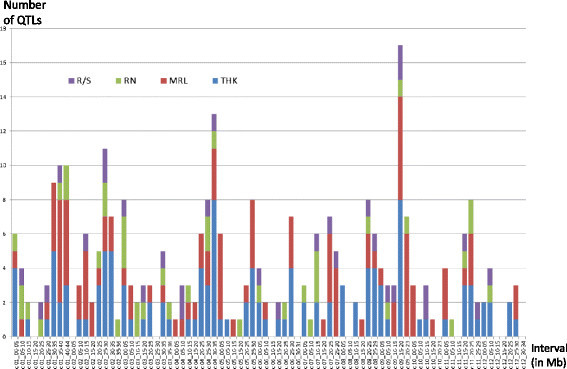
Number of root related QTLs detected in rice per chromosome interval. QTL for root to shoot ratio (R/S, violet), root number (RN, green) maximum root length (MRL, red), and root thickness (THK, blue). The figure was built from data reported in Courtois et al. ([2009]).
The main limitation with QTL detection in these mapping populations is the large size of the confidence interval. Even meta-analyses, on average, did not enable a decrease in the meta-QTL confidence intervals to less than half the original size of the segment (Courtois et al. [2009]). Genome-wide association studies (GWAS) in natural populations showing fast linkage disequilibrium decay have recently appeared as powerful tools to improve the resolution in localizing QTLs compared to mapping populations (Zhu et al. [2008]). GWAS requires a very high marker density, which is permitted by the simultaneous development of next-generation sequencing technology. The first results of GWAS analyzing root traits in rice have recently been published (Clark et al. [2013]; Courtois et al. [2013]). Because of the strong bipolar structure of O. sativa, which induces high risk of false-positive associations, the analyses were conducted on panels composed of accessions belonging to a single varietal group. This type of panel gives access to new varietal group-specific QTLs that cannot be detected by classical mapping in indica x japonica populations. However, even if the resolution on QTL position is clearly improved in GWAS compared with mapping populations, the number of possible candidate genes is still large and requires complementary evidence to determine the functional gene(s).
Positional cloning of genes underlying root QTLs
The results of QTL mapping were pursued through the development of NILs introgressed with QTLs in different backgrounds and QTL fine mapping with the aim of positional cloning (Shen et al. [2001]; Steele et al. [2013]; Suji et al. [2012]; Steele et al. [2006]). The most interesting QTLs for which the importance of the phenotypic effect was confirmed were positionally cloned. The first of these was PHOSPHORUS UPTAKE 1 (PUP1), a QTL contributing to phosphorus (P) uptake in low P content soils. The gene underlying the QTL, later termed PHOSPHORUS-STARVATION TOLERANCE 1 (PSTOL1), was cloned and appeared to encode a receptor-like cytoplasmic kinase (Gamuyao et al. [2012]). The gene does not exist in Nipponbare. When overexpressed, the gene significantly increases the plant P content and enhances grain yield by more than 60% under P-deficient conditions. The gene acts by enhancing early root growth, conferring a globally larger root system to the plant, with enhanced root length and root surface area, contributing to an increased uptake of P, and of other nutrients (N and K). The gene is expressed in the zone of crown root initiation and crown root meristems in stem bases. This localization suggests that the gene is involved in the regulation of crown root formation and elongation.
Another cloned gene related to a root development QTL is DEEPER ROOTING 1 (DRO1), which controls root growth angle and enhances deep rooting (Uga et al. [2013]). DRO1 encodes an unknown protein that is associated with plasmalemma. DRO1 expression is regulated by auxin via AUXIN RESPONSE FACTOR (ARF) transcription factors. DRO1 modulates root gravitropic response, likely via a modulation of epidermal cell elongation that enables roots to orientate their growth relative to the pull of gravity. The expression of DRO1 in the IR64 background increases the angle between roots and the horizontal axis, inducing deeper rooting. Compared to IR64, the near isogenic lines carrying DRO1 showed less deleterious effects, such as leaf wilting or delayed flowering, under moderate to severe drought stresses and better yield under stress, with no yield penalty under no stress conditions.
The cloning of these two QTLs that directly or indirectly affect root architecture exemplifies the difficulties that can occur in QTL cloning and can partly explain why the number of cloned root QTLs is still low. The first difficulty, which is intrinsic to the cloning process whatever the trait, is the lack of precision in positioning QTLs in mapping populations or, to a much lesser extent, in association panels. Therefore, lengthy operations are needed to reduce the size of the intervals to the gene level. A second difficulty in gene cloning is due to the importance of structural differences between varietal groups (Schatz et al. [2014]) and, within varietal groups, between varieties, as is illustrated by the absence of genes such as PSTOL1 in the reference cultivar Nipponbare. A final reason is that many genes do not have an assigned function. For example, the DRO1 product has no similarity to known proteins (Uga et al. [2013]). For these reasons, it is essential to complement these genetic studies that exploit the rice diversity using functional analysis approaches.
The sequencing and annotation of the rice genome (Itoh et al. [2007]; Yu et al. [2002]) as well as the creation of mutant collections (Hirochika et al. [2004]; Krishnan et al. [2009]; Wei et al. [2013]; Lorieux et al. [2012]; Guiderdoni and Gantet [2012]) and their screening for altered root phenotypes allowed the identification of major genes regulating the initiation, emergence or development of rice roots (Rebouillat et al. [2009]; Coudert et al. [2010]; Orman-Ligeza et al. [2013]). The gene networks involved in rice root development discussed below are schematized in Figure 2.
Figure 2.
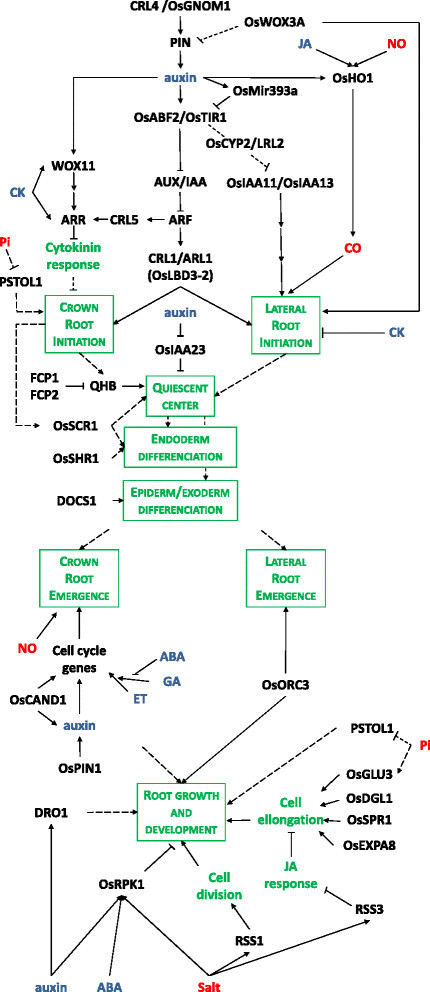
Gene networks controlling root development in rice . Arrows represent the positive regulatory action of one element of the network on another one. A line ending with a trait represents the negative regulatory action of one element of the network on another one. Dotted line represent hypothetical link between two elements. Text color code: genes, black; hormones, blue; signals, red; biological processes, green.
A conserved core gene regulatory network regulates post-embryonic root initiation in response to auxin
The crown root less 4 (crl4) mutant is impaired in OsGNOM1, which encodes a membrane-associated guanine-nucleotide exchange factor for the ADP-ribosylation factor G protein (Kitomi et al. [2008]; Liu et al. [2009]). This latter factor is a homolog of GNOM1, which regulates the intracellular traffic of the PINFORMED1 (PIN1) auxin efflux carrier protein in Arabidopsis. In turn, PIN1 is necessary to concentrate auxin in lateral root funder cells (Kitomi et al. [2008]; Liu et al. [2009]; Steinmann et al. [1999]). Crl4 and Osgnom1 mutants are characterized by an absence of crown roots and a reduced number of lateral roots. In these mutants, the expression of several OsPIN genes is altered. In the crl4 mutant, auxin transport is impaired, causing auxin distribution to be modified in the stem bases (Kitomi et al. [2008]). This result suggests that local auxin transport is involved in the initiation of crown and lateral roots in rice. WUSCHEL-related homeobox 3A (OsWOX3A) is encoded by NARROW LEAF2 (NAL2) and NAL3, a pair of duplicated genes. The nal2/nal3 double mutant shows a complex phenotype, with altered development of different parts of the plant and, notably, a strong reduction of the density of lateral roots. This latter effect is associated with an increase in root hair number and length (Cho et al. [2013]). In nal2/nal3 mutants, the expression of different OsPIN genes is modified. It is hypothesized that this modification results in a preferential allocation of auxin to the epidermis, where root hair formation is promoted. This occurs to the detriment of the pericycle, where lateral roots initiate (Yoo et al. [2013]). OsTIR1 and OsAFB2, the rice orthologs of the Arabidopsis auxin receptors TRANSPORT INHIBITOR RESPONSE 1(TIR1) and AUXIN SIGNALING F-BOX2 (AFB2), are negatively regulated by OsMir393a and OsMir393b (Bian et al. [2012]; Xia et al. [2012]). Plants overexpressing OsMir393 exhibit a pleiotropic altered auxin signaling phenotype, including a reduced number of crown roots. OsMir393a is expressed in the initiation sites of crown and lateral roots, suggesting that OsMir393a is involved in the regulation of post-embryonic root formation via a negative regulation of the rice auxin receptors. Interestingly, the TIR1 and AFB2 auxin receptors are localized to the nucleus, where they can interact with OsIAA1, an AUXIN/INDOLE ACETIC ACID (AUX/IAA) regulatory protein (Bian et al. [2012]; Xia et al. [2012]). AUX/IAA proteins interact with and inhibit the auxin response factors (ARF) that function as transcription factors. In Arabidopsis, when the TIR1 receptor is complexed with auxin, it can interact with AUX/IAA proteins. This binding results in the recruitment of a SKP1-CULLIN-FBOX (SCF) ubiquitin ligase, which ubiquitinates the AUX/IAA protein, targeting it for degradation. These interactions require other chaperone and co-chaperone proteins (Guilfoyle and Hagen [2007]; Vanneste and Friml [2009]; Mockaitis and Estelle [2008]; Lavenus et al. [2013]). In rice, a mutant screen for a loss of auxin sensitivity resulted in the characterization of two mutants, Oscyclophilin2 (Oscyp2) and lateral rootless2 (lrl2), which are characterized by an absence of lateral roots and are caused by mutations in the same gene (Kang et al. [2013]; Zheng et al. [2013]). In addition, Oscyp2 exhibits a pleiotropic auxin-insensitive phenotype that correlates with a reduced degradation of OsIAA11 in response to auxin (Kang et al. [2013]). OsCYP2/LRL2 is a chaperone protein that can interact with the co-chaperone SUPPRESSOR OF G2 ALLELE OF SKP1 (OsSGT1). It has been proposed that this complex is involved in the degradation of OsIAA11 by SCFTIR in response to auxin during the early step of lateral root initiation (Kang et al. [2013]). OsIAA11 or OsIAA13 gain of function mutations, in which amino acids involved in degradation of the protein in response to auxin are modified, exhibit a lateral rootless phenotype (Kitomi et al. [2012]; Zhu et al. [2012]). OsIAA11 and OsIAA13 are required for lateral root initiation. The crown root less 1 (Inukai et al. [2005]) and adventitious less 1 (Liu et al. [2005]) mutants are both impaired in the OsLBD3-2 gene, which encodes a LATERAL ORGAN BOUNDARY DOMAIN (LBD) transcription factor belonging to a specific clade regrouping LBD proteins involved in the control of post-embryonic root initiation in different species (Coudert et al. [2013a]). Crl1 and arl1 mutants are characterized by a total absence of crown root initiation and a reduced number of lateral roots. The expression of CRL1 is induced by auxin. This induction is lost in plants overexpressing a modified OsIAA3 protein that cannot be degraded after auxin treatment, showing that the induction of the expression of CRL1 by auxin is dependent on the degradation of AUX/IAA proteins. In addition, the CRL1 promoter contains an auxin response sequence (ARE) that interacts in vitro with the OsARF16 transcription factor (Inukai et al. [2005]). Taken together, these data show that the initiation of crown and lateral roots in rice involves a core auxin regulated pathway that includes TIR1/AFB2 auxin receptors, AUX/IAA regulatory proteins, as well as specific ARF and LBD transcription factors. This gene regulatory network that controls post-embryonic root development is well conserved in different dicot and monocot plant species (Peret et al. [2009]; Lavenus et al. [2013]; Hochholdinger and Tuberosa [2009]). In rice, some elements of this regulatory network are common for the initiation of crown and lateral roots, whereas others are specific to the initiation of only one root type. For this reason, rice constitutes a promising model to investigate how this auxin-dependent gene regulatory network has evolved to control post-embryonic root development in the context of both stems and roots (Coudert et al. [2013b]).
Post-embryonic root initiation in rice involves genes that mediate other hormone signals
The crown root less5 (crl5) mutant exhibits a reduced number of crown roots due to a reduction in crown root initiation (Kitomi et al. [2011]). CRL5 encodes an APETALA/ETHYLENE RESPONSE FACTOR (AP2/ERF) transcription factor. Its expression is induced by auxin via an ARF transcription factor. Cytokinins negatively regulate the initiation of crown and lateral roots in rice (Rani Debi et al. [2005]; Kitomi et al. [2011]), but plants that constitutively express CRL5 can initiate crown roots normally. CRL5 activates the expression of two type-A RESPONSE REGULATORs (ARRs) that are repressors of cytokinin signaling (To et al. [2004]). Similarly, the WUSCHEL-related Homeobox transcription factor WOX11 controls the expression of different ARRs and regulates both crown root initiation and development (Zhao et al. [2009]). WOX11 expression is induced by auxin and cytokinin. This result shows that CRL5 and WOX11 are involved in the crossway of the antagonistic auxin and cytokinin pathways in the regulation of crown-root initiation.
The HEME OXYGENASE 1 (HOX1) gene controls the formation of lateral roots via the production of carbon monoxide. HOX1 is regulated by auxin and stress related signals, such as jasmonate and nitric oxide (Chen et al. [2012]; Hsu et al. [2012]), which suggests that this pathway can contribute to the modulation of root architecture in response to stress.
Genes involved in root meristem formation
After the initiation step, cells divide and become organized to shape a root meristem. In root meristems, the quiescent center (QC), which is composed of a small number of mitotically inactive cells, is indispensable for the maintenance of the undifferentiated status of the surrounding stem cells from which root tissues are produced (van den Berg et al. [1997]). The maintenance by auxin of the QC in rice is mediated by OsIAA23 (Jun et al. [2011]). The QUIESCENT CENTER HOMEOBOX (QHB) gene is an ortholog to the WUSCHEL-related WOX5, a QC-specific gene in Arabidopsis that contributes to QC and root stem cell maintenance (Kamiya et al. [2003b]; Breuninger et al. [2008]; Ditengou et al. [2008]). QHB is expressed early during root formation in the QC, and its expression is dependent on CRL1/ARL1 during crown root formation (Inukai et al. [2005]; Ni et al. [2014]; Kamiya et al. [2003b]; Liu et al. [2005]). The CLAVATA3 (CLV3)/ENDOSPERM SURROUNDING REGION (ESR)-related (CLE) FON2-LIKE CLE PROTEIN1 (FCP1) and FCP2 proteins inhibit the expression of QHB in the QC (Chu et al. [2013]; Suzaki et al. [2008]). This relationship suggests that a CLAVATA/WOX signaling module regulates the activity of the root apical meristem in rice. Such a module has already been described in Arabidopsis for the regulation of the shoot apical meristem activity. In this context, the transduction of the CLV3 peptide signal is ensured by LEUCINE RICH REPEAT RECEPTOR-LIKE KINASE (LRR RLK) CLV1 (Muller et al. [2006]). In rice lines overexpressing OsRPK1, which encodes an LRR RLK, the expression levels of different OsPIN genes are down regulated, polar auxin transport is perturbed and the length of the seminal root is reduced, as is the number of crown roots and the density of lateral roots (Zou et al. [2014]). Nevertheless, it is undetermined at which step of root development (i.e., initiation, meristem maintenance or other development processes) this gene is involved. Interestingly OsRPK1 is regulated by auxin, abscisic acid and salt stress. Its role in stress-adaptive root morphogenesis should be further studied (Zou et al. [2014]).
In Arabidopsis, SCARECROW (SCR), a transcription factor from the GRAS family, also contributes to the specification of the QC (Sabatini et al. [2003]). In rice, OsSCR1 is expressed in the QC and may also be involved in QC determination (Ni et al. [2014]). The expression of OsSCR1 is down-regulated in stem bases of the arl1 mutant (Liu et al. [2005]). Together with SHORTROOT (SHR, another GRAS transcription factor), SCARECROW controls the division of the endoderm/cortex initial cells and the differentiation of endoderm in Arabidopsis (Di Laurenzio et al. [1996]; Cui et al. [2007]). In rice, this mechanism is likely conserved given that OsSCR1 is also expressed in endoderm/cortex initial cells and in early differentiated endodermal cells (Ni et al., [2014]). In addition, OsSCR1 can interact with OsSHR1, which is expressed in the stele but may likely move to endodermal initial cells (Cui et al. [2007]; Ni et al. [2014]; Kamiya et al. [2003a]). Rice roots are characterized by an exodermis as well as sclerenchyma cell layers just inside the epidermis (Rebouillat et al. [2009]; Coudert et al. [2010]). An aluminum-sensitive mutant, c68, has been identified (Huang et al. [2009]). In this mutant, the differentiation of the epidermis and exodermis is perturbed. The corresponding gene, DEFECTIVE IN OUTERCELL LAYER SPECIFICATION 1 (DOCS1), encodes an LRR RLK protein (Huang et al. [2012]). This result illustrates that a better knowledge of genes involved in root meristem functioning and root radial tissue patterning should help identify new sources of resistance and improve the adaptation of rice to soil containing toxic elements.
Genes involved in post-embryonic root emergence
When formed, the root meristem must grow out through the stem (for crown roots) or the root (for lateral roots) tissues. Crown root emergence is stimulated by submergence. This stimulation is mediated by ethylene, which induces the expression of cell cycle regulatory genes in crown root meristems and promotes, in synergy with gibberellic acid, the death of epidermal cells at the site of root emergence (Lorbiecke and Sauter [1999]; Mergemann and Sauter [2000]; Steffens et al. [2006]). Nitric oxide also promotes crown root emergence, whereas abscisic acid inhibits emergence (Steffens et al. [2006]; Xiong et al. [2009]). A functional auxin polarized transport, largely mediated by OsPIN1, is also required for crown root emergence and development (Xu et al. [2005]). In the Oscand1 mutant, crown root meristems are properly initiated and formed but do not emerge (Wang et al. [2010]). In Arabidopsis, OsCAND1 encodes a CULLIN-ASSOCIATED AND NEDDYLATION-DISSOCIATED 1 (CAND1) SCFTIR1 ubiquitin ligase involved in the degradation of AUX/IAA in response to auxin (Cheng et al. [2004]). In the Oscand1 mutant, auxin distribution is altered in crown root meristems, as is the expression of cell cycle regulatory genes. In origin recognition complex subunit3 (orc3) knock-down plants, the emergence of lateral roots is blocked due to a perturbation of cell cycle activity in newly formed lateral root meristems (Chen et al. [2013]). OsORC3 is also expressed in primary root and crown root primordia, likely playing a more general role in root system development. Root emergence and root initiation are essential processes that determine the number of developed roots of the plant. In this regard, it is also essential to identify the major genetic determinants governing this process to engineer more branched root systems.
Genes involved in root development
Different root development mutants permitted the identification of genes involved in different biological functions. Some of these genes alter root development by affecting root cell wall constitution. OsDGL1 encodes DOLICHYL DIPHOSPHOOLIGOSACCHARIDE-PROTEIN GLYCOSYLTRANSFERASE 48 kDa subunit precursor necessary for the proper structuration of the polysaccharide matrix of root cell walls. Osdgl1 mutant plants exhibit a short root phenotype due to a defect in root cell elongation and division (Qin et al. [2013]). Another gene affecting root cell wall constitution was isolated using short root mutant screening. This gene, OsGLU3, encodes a putative MEMBRANE-BOUND ENDO- 1,4-B-GLUCANASE (Zhang et al. [2012b]). In Osglu3 mutants, the crystalline cellulose content is reduced in the root cell wall and root cell elongation is inhibited. It has been suggested that OsGLU3 can contribute to the modulation of root growth in response to phosphate starvation (Zhang et al. [2012a]). Expansins are also key actors of cell elongation. OsEXPANSIN8 (OsEXPA8) is specifically expressed in root tips (Shin et al. [2005]). Plants overexpressing OsEXPA8 exhibit better global growth. This improvement is notable in their root system, where root elongation and branching are strongly stimulated (Ma et al. [2013]). This phenotype results in a general increase in cell wall elongation capacity. The Oryza sativa short postembryonic roots 1 (Osspr1) mutant also exhibits a short root phenotype that is due to defective root cell elongation and is characterized by a perturbation in iron homeostasis (Jia et al. [2011]). OsSPR1 encodes a mitochondrial protein with an Armadillo-like repeat domain, but its precise molecular function is uncharacterized. In the rice salt sensitive3 (rss3) mutant, the elongation of root cells is strongly inhibited under salt stress conditions, resulting in a short root phenotype (Toda et al. [2013]). RSS3 is a nuclear factor that can interact with key molecular components of the jasmonic acid transduction pathway, a stress hormone known to inhibit root growth. In roots, RSS3 likely contributes to repress the jasmonic acid response in salt stress conditions to maintain root growth. Similarly, the rss1 mutant exhibits strongly reduced root and shoot growth in salt stress conditions (Ogawa et al. [2011]). RSS1 plays a role in maintaining cell division in meristematic zones under salt stress conditions, likely by interfering with the cytokinin response pathway. The targeted overexpression of NO APICAL MERISTEM, ATAF1-2, CUP-SHAPED COTYLEDON (NAC) transcription factors OsNAC5, OsNAC9 and OsNAC10 in roots enhances the water stress tolerance of plants (Jeong et al. [2010]; Redillas et al. [2012]; Jeong et al. [2013]). The overexpression of these factors correlates with an augmentation in root diameter that may facilitate soil penetration by roots and water transport. All of these genes involved in root growth are promising targets in the creation of deeper root systems that can help plants to avoid water deficits and, as illustrated above in different examples, can modulate root growth in response to different edaphic conditions.
Transcriptomic studies will facilitate root gene discovery
The first comprehensive transcriptome study of the rice root system was published in 2012 (Takehisa et al. [2012]). In this study, eight developmental stages along the longitudinal axis of a crown root were investigated as well as different radial tissues: epidermis, exodermis and sclerenchyma; cortex; and endodermis, pericycle and stele. This analysis allowed the identification of genes specifically expressed in root caps, in the initiation or developmental zone of lateral roots, and those involved in water and nutrient transport. Data are available in the RiceXPro data base (Sato et al. [2013]). Data comparing genes expressed in roots of different rice varieties have begun being generated (Chen et al. [2014]). Some authors investigated the genes that were miss-regulated relative to wild type plants in rice root-related mutants such as Osiaa13, c68 or crl1 (Kitomi et al. [2012]; Huang et al. [2012]; Coudert et al. [2011]). In the latter case, the authors performed a second screen based on auxin induction of CRL1 expression. The aim of this screen was to identify CRL1-dependent auxin regulated genes. One of the identified genes, FLATTENED SHOOT MERISTEM (FSM) encodes a component of the CHROMATIN ASSEMBLY FACTOR1 that contributes to crown root formation (Abe et al. [2008]). It is clear that comparison of the increasing number of rice root-specific transcriptome data sets obtained using different experimental approaches will allow the identification of new important genes involved in root development in rice.
Conclusions
In recent years, our knowledge of the genetic control of root development in rice has progressed considerably. Genetic approaches based on the investigation of rice genotypic and phenotypic diversity have allowed the identification of major genes involved in the development of different types of root system architectures, such as DRO1 and PSTOL1, with functions related to drought tolerance or adaptation to soil with low phosphate content, respectively (Gamuyao et al. [2012]; Uga et al. [2013]). The identification of PSTOL1, which was only possible using a traditional variety, stresses that the effort to explore the diversity of O. sativa that has adapted to a large range of ecosystems must continue. Functional genomics approaches have begun to provide a better understanding of the mechanisms involved in root branching, elongation and radial tissue differentiation. Branching is the fundamental process by which the root system can increase its root number. Together with elongation, branching allows the root system to explore and exploit a larger portion of soil. Radial tissue patterning in rice is fundamental for the adaptation of root growth in anoxic conditions and for the control of the diffusion of toxic elements, such as salt or metals, into roots. The genes governing root meristem activity also constitute promising targets for adapting root growth to different soil stress conditions. Only a better knowledge of the mechanisms that regulate the expression of such genes will allow the design of new breeding strategies that target the root system to improve rice resistance to soil related stresses. The transcriptome analysis of root systems in plants grown in different conditions will help to achieve this objective and will provide a large catalogue of root-specific genes that are expressed at different developmental stages and in response to different stresses.
These approaches (QTL detection, gene functional characterization, global expression studies) need to be more closely linked to accelerate the discovery of new genetic determinants, which will result in the generation of varieties with improved root architecture. We hope that this review helps to link the data obtained by these complementary approaches.
The results of genetic and genomic approaches can converge with the identification of alleles of interest at genes with known function and the introgression of these alleles into elite lines. An example of such a strategy is given by the work conducted on SUB1, a gene controlling submergence tolerance, in various Asian countries (Bailey-Serres et al. [2010]). In less than five years, the allele of interest was introgressed into major elite varieties. There is no doubt that the work conducted on root QTLs will lead to the same efforts (Steele et al. [2013]). DRO1 and PSTOL1 have begun to be used in several rice improvement-breeding programs. In addition, a targeted diversity analysis of the genes governing different aspects of root growth and development also needs to be conducted. The identification of the allelic variants of genes that are associated with different root system phenotypes should rapidly provide breeders with the necessary tools to finely modulate root architecture in order to address specific needs. However, the very large GxE interactions expected between roots and soil due to variations in physical, chemical and microorganismal soil composition implies that the observed effect of QTL/gene presence in field grown plants might not be as strong as expected. Therefore, there will be significant opportunities for agronomists and farmers to use their skill to optimize the expression of the root potential of the selected varieties under various conditions.
Acknowledgements
NTPP is supported by a Global Rice International Science Partnership PhD fellowship, MG by a fellowship of IRD and the Ministère des Affaires Etrangères Français. The Vietnamese Ministry of Agriculture and Rural Development supports our research on root development in Vietnam.
Abbreviations
- ABA
Abscissic acid
- AFB2
Auxin signaling F-Box2
- ARF
Auxin response factor
- ARL
Adventitious root less
- ARR
Type-A response regulator
- AUX/IAA
Auxin/Indole-3-Acetic acid
- CAND1
Cullin-Associated and Neddylation-Dissociated 1
- CK
Cytokinin
- CO
Carbon monoxide
- CRL
Crown root less
- CYP2
Oscyclophilin2
- DRO1
Deeper rooting 1
- DGL1
Dolichyl Diphosphooligosaccharide-Protein Glycosyltransfer Glycosyltransferase
- DOCS1
Defective in outercell layer specification 1
- ET
Ethylene
- EXPA8
Expansin8
- FCP
Clavata3 (CLV3)/Endosperm Surrounding Region (ESR)-related (CLE) Fon2-Like CLE PROTEIN
- GA
Gibberelic acid, GLU3, Membrane-Bound Endo- 1,4-B-Glucanase
- GNOM1
Membrane-Associated Guanine-Nucleotide Exchange Factor of the ADP-Ribosylation Factor G Protein (ARF-GEF)
- HO1
Heme oxygenase 1
- JA
Jasmonic acid
- LLR2
Lateral root less 2
- NO
Nitrite oxide
- ORC3
Origin recognition complex Subunit3
- Pi
Inorganic phosphate
- PIN
Pinformed auxin efflux carrier proteins
- RPK1
Receptor protein kinase 1
- PSTOL1
Phosphorus-Starvation tolerance 1
- QHB
Quiescent center homeobox
- RSS
Rice salt sensitive
- SCR
Scarecrow
- SHR
Shortroot
- SPR1
Short postembryonic roots
- TIR1
Transport inhibitor response 1
- WOX
Wushel-related homeobox
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors contributed to collect information and write the manuscript.
Contributor Information
Chung D Mai, Email: mdchungduc@gmail.com.
Nhung TP Phung, Email: nhungphungphuong@gmail.com.
Huong TM To, Email: maihuongbk@yahoo.com.
Mathieu Gonin, Email: gonin.mathieu@live.fr.
Giang T Hoang, Email: nuocngamos@yahoo.com.
Khanh L Nguyen, Email: khanhseu@gmail.com.
Vinh N Do, Email: donangvinh@netnam.vn.
Brigitte Courtois, Email: brigitte.courtois@cirad.fr.
Pascal Gantet, Email: pascal.gantet@univ-montp2.fr.
References
- Abe M, Kuroshita H, Umeda M, Itoh J, Nagato Y. The rice flattened shoot meristem, encoding CAF-1 p150 subunit, is required for meristem maintenance by regulating the cell-cycle period. Dev Biol. 2008;319:384–393. doi: 10.1016/j.ydbio.2008.04.040. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D. Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice. 2010;3:138–147. [Google Scholar]
- Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa) New Phytol. 2012;196:149–161. doi: 10.1111/j.1469-8137.2012.04248.x. [DOI] [PubMed] [Google Scholar]
- Breuninger H, Rikirsch E, Hermann M, Ueda M, Laux T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev Cell. 2008;14:867–876. doi: 10.1016/j.devcel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Champoux M, Wang G, Sarkarung S, Mackill DJ, O’Toole JC, Huang N, McCouch SR. Locating genes associated with root morphology and drought avoidance in rice via linkage to molecular marker. Theor Appl Genet. 1995;90:969–981. doi: 10.1007/BF00222910. [DOI] [PubMed] [Google Scholar]
- Chen JS, Lin SC, Chen CY, Hsieh YT, Pai PH, Chen LK, Lee S. Development of a microarray for two rice subspecies: characterization and validation of gene expression in rice tissues. BMC Res Notes. 2014;7:15. doi: 10.1186/1756-0500-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shi J, Hao X, Liu H, Wu Y, Wu Z, Chen M, Wu P, Mao C. OsORC3 is required for lateral root development in rice. Plant J. 2013;74:339–350. doi: 10.1111/tpj.12126. [DOI] [PubMed] [Google Scholar]
- Chen YH, Chao YY, Hsu YY, Hong CY, Kao CH. Heme oxygenase is involved in nitric oxide- and auxin-induced lateral root formation in rice. Plant Cell Rep. 2012;31:1085–1091. doi: 10.1007/s00299-012-1228-x. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol. 2004;135:1020–1026. doi: 10.1104/pp.104.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Yoo SC, Zhang H, Pandeya D, Koh HJ, Hwang JY, Kim GT, Paek NC. The rice narrow leaf2 and narrow leaf3 loci encode WUSCHEL-related homeobox 3A (OsWOX3A) and function in leaf, spikelet, tiller and lateral root development. New Phytol. 2013;198:1071–1084. doi: 10.1111/nph.12231. [DOI] [PubMed] [Google Scholar]
- Chu H, Liang W, Li J, Hong F, Wu Y, Wang L, Wang J, Wu P, Liu C, Zhang Q, Xu J, Zhang D. A CLE-WOX signalling module regulates root meristem maintenance and vascular tissue development in rice. J Exp Bot. 2013;64:5359–5369. doi: 10.1093/jxb/ert301. [DOI] [PubMed] [Google Scholar]
- Clark RT, Famoso AN, Zhao K, Shaff JE, Craft EJ, Bustamante CD, McCouch SR, Aneshansley DJ, Kochian LV. High-throughput two-dimensional root system phenotyping platform facilitates genetic analysis of root growth and development. Plant Cell Environ. 2013;36:454–466. doi: 10.1111/j.1365-3040.2012.02587.x. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Bes M, Le Thi VA, Pre M, Guiderdoni E, Gantet P. Transcript profiling of crown root less 1 stem base reveals new elements associated with crown root development in rice. BMC Genomics. 2011;12:e387. doi: 10.1186/1471-2164-12-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudert Y, Dievart A, Droc G, Gantet P. ASL/LBD phylogeny suggests that genetic mechanisms of root initiation downstream of auxin are distinct in lycophytes and euphyllophytes. Mol Biol Evol. 2013;30:569–572. doi: 10.1093/molbev/mss250. [DOI] [PubMed] [Google Scholar]
- Coudert Y, Le Thi VA, Gantet P. Rice: a model plant to decipher the hidden origin of adventitious roots. In: Eshel A, Beeckman T, editors. Plant roots: the hidden half. 4. Boca Raton, USA: CCR press, Taylor and Francis group, LLC; 2013b. pp. 157–166. [Google Scholar]
- Coudert Y, Perin C, Courtois B, Khong NG, Gantet P. Genetic control of root development in rice, the model cereal. Trends Plant Sci. 2010;15:219–226. doi: 10.1016/j.tplants.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Courtois B, Ahmadi N, Khowaja F, Price AH, Rami J-F, Frouin J, Hamelin C, Ruiz M. Rice root genetic architecture: meta-analysis from a drought QTL database. Rice. 2009;2:115–128. [Google Scholar]
- Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Goze E, Kilian A, Ahmadi N, Dingkuhn M. Genome-wide association mapping of root traits in a japonica rice panel. PLoS One. 2013;8:e78037. doi: 10.1371/journal.pone.0078037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Den Herder G, Van Isterdael G, Beeckman T, De Smet I. The roots of a new green revolution. Trends Plant Sci. 2010;15:600–607. doi: 10.1016/j.tplants.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, Laux T, Palme K. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:18818–18823. doi: 10.1073/pnas.0807814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai S, Cooper M. Development of drought resistant cultivars using physio-morphological traits in rice. Field Crops Res. 1995;40:67–87. [Google Scholar]
- Gamuyao R, Chin JH, Pariasca-Tanaka J, Pesaresi P, Catausan S, Dalid C, Slamet-Loedin I, Tecson-Mendoza EM, Wissuwa M, Heuer S. The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature. 2012;488:535–539. doi: 10.1038/nature11346. [DOI] [PubMed] [Google Scholar]
- Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R. Root biology and genetic improvement for drought avoidance in rice. Field Crop Res. 2011;122:1–13. [Google Scholar]
- Guiderdoni E, Gantet P. Ac-Ds solutions for rice insertion mutagenesis. Methods Mol Biol. 2012;859:177–187. doi: 10.1007/978-1-61779-603-6_10. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, Sundaresan V, Leung H. Rice mutant resources for gene discovery. Plant Mol Biol. 2004;54:325–334. doi: 10.1023/B:PLAN.0000036368.74758.66. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Tuberosa R. Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol. 2009;12:172–177. doi: 10.1016/j.pbi.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Hsu YY, Chao YY, Kao CH. Methyl jasmonate-induced lateral root formation in rice: The role of heme oxygenase and calcium. Journal of plant physiol. 2012;170:63–69. doi: 10.1016/j.jplph.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Nishimura M, Tajima S, Ma JF. A rice mutant sensitive to Al toxicity is defective in the specification of root outer cell layers. Plant Cell Physiol. 2009;50:976–985. doi: 10.1093/pcp/pcp050. [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Ono K, Ma JF. A leucine-rich repeat receptor-like kinase gene is involved in the specification of outer cell layers in rice roots. Plant J. 2012;69:565–576. doi: 10.1111/j.1365-313X.2011.04824.x. [DOI] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R, Bureau T, Burr F, Costa de Oliveira A, Fuks G, Habara T, Haberer G, Han B, Harada E, Hiraki AT, Hirochika H, Hoen D, Hokari H, Hosokawa S, Hsing YI, Ikawa H, Ikeo K, Imanishi T, Ito Y, Jaiswal P, Kanno M, Kawahara Y, Kawamura T, Kawashima H, Khurana JP, Kikuchi S, Komatsu S, Koyanagi KO, Kubooka H, Lieberherr D, Lin YC, Lonsdale D, Matsumoto T, Matsuya A, McCombie WR, Messing J, Miyao A, Mulder N, Nagamura Y, Nam J, Namiki N, Numa H, Nurimoto S, O’Donovan C, Ohyanagi H, Okido T, Oota S, Osato N, Palmer LE, Quetier F, Raghuvanshi S, Saichi N, Sakai H, Sakai Y, Sakata K, Sakurai T, Sato F, Sato Y, Schoof H, Seki M, Shibata M, Shimizu Y, Shinozaki K, Shinso Y, Singh NK, Smith-White B, Takeda J, Tanino M, Tatusova T, Thongjuea S, Todokoro F, Tsugane M, Tyagi AK, Vanavichit A, Wang A, Wing RA, Yamaguchi K, Yamamoto M, Yamamoto N, Yu Y, Zhang H, Zhao Q, Higo K, Burr B, Gojobori T, Sasaki T. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Redillas MC, Jang G, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J. 2013;11(1):101–114. doi: 10.1111/pbi.12011. [DOI] [PubMed] [Google Scholar]
- Jia L, Wu Z, Hao X, Carrie C, Zheng L, Whelan J, Wu Y, Wang S, Wu P, Mao C. Identification of a novel mitochondrial protein, short postembryonic roots 1 (SPR1), involved in root development and iron homeostasis in Oryza sativa. New Phytol. 2011;189:843–855. doi: 10.1111/j.1469-8137.2010.03513.x. [DOI] [PubMed] [Google Scholar]
- Jun N, Gaohang W, Zhenxing Z, Huanhuan Z, Yunrong W, Ping W. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011;68:433–442. doi: 10.1111/j.1365-313X.2011.04698.x. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Itoh J, Morikami A, Nagato Y, Matsuoka M. The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 2003;36:45–54. doi: 10.1046/j.1365-313x.2003.01856.x. [DOI] [PubMed] [Google Scholar]
- Kamiya N, Nagasaki H, Morikami A, Sato Y, Matsuoka M. Isolation and characterization of a rice WUSCHEL-type homeobox gene that is specifically expressed in the central cells of a quiescent center in the root apical meristem. Plant J. 2003;35:429–441. doi: 10.1046/j.1365-313x.2003.01816.x. [DOI] [PubMed] [Google Scholar]
- Kamoshita A, Chandra Babu R, Boopathi MN, Fukai S. Phenotypic and genotypic analysis of drought-resistance traits for development of rice cultivars adapted to rainfed environments. Field Crop Research. 2008;109:1–23. [Google Scholar]
- Kamoshita A, Zhang J, Siopongco J, Sarkarung S, Nguyen HT, Wade LJ. Effects of phenotyping environment on identification of quantitative trait loci for rice root morphology under anaerobic conditions. Crop Sci. 2002;42:255–265. doi: 10.2135/cropsci2002.2550. [DOI] [PubMed] [Google Scholar]
- Kang B, Zhang Z, Wang L, Zheng L, Mao W, Li M, Wu Y, Wu P, Mo X. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 2013;74:86–97. doi: 10.1111/tpj.12106. [DOI] [PubMed] [Google Scholar]
- Khowaja FS, Norton GJ, Courtois B, Price AH. Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis. BMC Genomics. 2009;10:e276. doi: 10.1186/1471-2164-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitomi Y, Inahashi H, Takehisa H, Sato Y, Inukai Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012;190:116–122. doi: 10.1016/j.plantsci.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011;67:472–484. doi: 10.1111/j.1365-313X.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Ogawa A, Kitano H, Inukai Y. CRL4 regulates crown root formation through auxin transport in rice. Plant Root. 2008;2:19–28. [Google Scholar]
- Khowaja FS, Norton GJ, Courtois B, Price AH. Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis. BMC Genomics. 2009;10(276):1–14. doi: 10.1186/1471-2164-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, Yu SM, Upadhyaya N, Ramachandran S, Zhang Q, Sundaresan V, Hirochika H, Leung H, Pereira A. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafitte H, Champoux M, McLaren G, O’Toole J. Rice root morphological traits are related to isozyme group and adaptation. Field Crop Research. 2001;71:57–70. [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P, Shou H. Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 2009;19:1110–1119. doi: 10.1038/cr.2009.70. [DOI] [PubMed] [Google Scholar]
- Lorbiecke R, Sauter M. Adventitious root growth and cell-cycle induction in deepwater rice. Plant Physiol. 1999;119:21–30. doi: 10.1104/pp.119.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M, Blein M, Lozano J, Bouniol M, Droc G, Dievart A, Perin C, Mieulet D, Lanau N, Bes M, Rouviere C, Gay C, Piffanelli P, Larmande P, Michel C, Barnola I, Biderre-Petit C, Sallaud C, Perez P, Bourgis F, Ghesquiere A, Gantet P, Tohme J, Morel JB, Guiderdoni E. In-depth molecular and phenotypic characterization in a rice insertion line library facilitates gene identification through reverse and forward genetics approaches. Plant Biotechnol J. 2012;10:555–568. doi: 10.1111/j.1467-7652.2012.00689.x. [DOI] [PubMed] [Google Scholar]
- Ma N, Wang Y, Qiu S, Kang Z, Che S, Wang G, Huang J. Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS One. 2013;8:e75997. doi: 10.1371/journal.pone.0075997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan K, Emrich K, Piepho HP, Mullins CE, Price AH. Assessing the importance of genotype x environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. Theor Appl Genet. 2006;113:953–964. doi: 10.1007/s00122-006-0357-4. [DOI] [PubMed] [Google Scholar]
- Mergemann H, Sauter M. Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 2000;124:609–614. doi: 10.1104/pp.124.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. doi: 10.1146/annurev.cellbio.23.090506.123214. [DOI] [PubMed] [Google Scholar]
- Muller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Shen Y, Zhang Y, Wu P (2014) Definition and stabilisation of the quiescent center in rice roots. Plant Biol, in press Ni J, Shen Y, Zhang Y, Wu P (2014) Definition and stabilisation of the quiescent center in rice roots. Plant Biol, in press [DOI] [PubMed]
- O’Toole J, Bland W. Genotypic variation in crop plant root systems. Adv Agron. 1987;41:91–145. [Google Scholar]
- Ogawa D, Abe K, Miyao A, Kojima M, Sakakibara H, Mizutani M, Morita H, Toda Y, Hobo T, Sato Y, Hattori T, Hirochika H, Takeda S. RSS1 regulates the cell cycle and maintains meristematic activity under stress conditions in rice. Nat Commun. 2011;2:278. doi: 10.1038/ncomms1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman-Ligeza B, Parizot B, Gantet P, Beeckman T, Bennett MJ, Draye X. Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci. 2013;18:459–467. doi: 10.1016/j.tplants.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ. Arabidopsis lateral root development: an emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- Puig J, Pauluzzi G, Guiderdoni E, Gantet P. Regulation of shoot and root development trought mutual signaling. Mol Plant. 2012;5:574–583. doi: 10.1093/mp/sss047. [DOI] [PubMed] [Google Scholar]
- Qin C, Li Y, Gan J, Wang W, Zhang H, Liu Y, Wu P. OsDGL1, a homolog of an oligosaccharyltransferase complex subunit, is involved in N-glycosylation and root development in rice. Plant Cell Physiol. 2013;54:129–137. doi: 10.1093/pcp/pcs159. [DOI] [PubMed] [Google Scholar]
- Rani Debi B, Taketa S, Ichii M. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (Oryza sativa) J Plant Physiol. 2005;162:507–515. doi: 10.1016/j.jplph.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Rebouillat A, Dievart A, Verdeil JL, Escoute J, Giese G, Breitler JC, Gantet P, Espeout S, Guiderdoni E, Périn C. Molecular genetic of rice root development. Rice. 2009;2:15–34. [Google Scholar]
- Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J. 2012;10:792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41(Database issue):D1206–D1213. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Maron LG, Stein JC, Hernandez Wentz A, Gurtowski J, Biggers E, Lee H, Kramer M, Antoniou E, Ghiban E, Wright MH, Chia JM, Ware DM, McCouch SR, McCombie WR. New whole genome de novo assemblies of three divergent strains of rice documents novel gene space of aus and indica. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Courtois B, McNally K, Robin S, Li Z. Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor Appl Genet. 2001;103:75–83. [Google Scholar]
- Shin JH, Jeong DH, Park MC, An G. Characterization and transcriptional expression of the alpha-expansin gene family in rice. Mol Cells. 2005;20:210–218. [PubMed] [Google Scholar]
- Steele K, Price A, Shashidhar H, Witcombe J. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet. 2006;112:208–221. doi: 10.1007/s00122-005-0110-4. [DOI] [PubMed] [Google Scholar]
- Steele KA, Price AH, Witcombe JR, Shrestha R, Singh BN, Gibbons JM, Virk DS. QTLs associated with root traits increase yield in upland rice when transferred through marker-assisted selection. Theor Appl Genet. 2013;126:101–108. doi: 10.1007/s00122-012-1963-y. [DOI] [PubMed] [Google Scholar]
- Steffens B, Wang J, Sauter M. Interactions between ethylene, gibberellin and abscisic acid regulate emergence and growth rate of adventitious roots in deepwater rice. Planta. 2006;223:604–612. doi: 10.1007/s00425-005-0111-1. [DOI] [PubMed] [Google Scholar]
- Steinmann T, Geldner N, Grebe M, Mangold S, Jackson CL, Paris S, Galweiler L, Palme K, Jurgens G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286:316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- Suji KK, Prince KSJ, Mankhar PS, Kanagaraj P, Poornima R, Amutha K, Kavitha S, Biji KR, Gomez SM, Chandra Babu R. Evaluation of rice near isogenic lines with root QTLs for plant production and root traits in rainfed target populations of environment. Field Crop Res. 2012;137:89–96. [Google Scholar]
- Suzaki T, Yoshida A, Hirano HY. Functional diversification of CLAVATA3-related CLE proteins in meristem maintenance in rice. Plant Cell. 2008;20:2049–2058. doi: 10.1105/tpc.107.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M, McCouch S. The complex history of the domestication of rice. Ann Bot. 2007;100:951–957. doi: 10.1093/aob/mcm128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, Minami H, Namiki N, Inukai Y, Nakazono M, Nagamura Y. Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J. 2012;69:126–140. doi: 10.1111/j.1365-313X.2011.04777.x. [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:658–671. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda Y, Tanaka M, Ogawa D, Kurata K, Kurotani K, Habu Y, Ando T, Sugimoto K, Mitsuda N, Katoh E, Abe K, Miyao A, Hirochika H, Hattori T, Takeda S. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25:1709–1725. doi: 10.1105/tpc.113.112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, Moore BT, Harer J, Edelsbrunner H, Mitchell-Olds T, Weitz JS, Benfey PN. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci U S A. 2013;110:1695–1704. doi: 10.1073/pnas.1304354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Vanneste S, Friml J. Auxin: a trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Wang XF, He FF, Ma XX, Mao CZ, Hodgman C, Lu CG, Wu P. OsCAND1 is required for crown root emergence in rice. Mol Plant. 2010;4:289–299. doi: 10.1093/mp/ssq068. [DOI] [PubMed] [Google Scholar]
- Wei FJ, Droc G, Guiderdoni E, Hsing YI. International consortium of rice mutagenesis: resources and beyond. Rice: 2013;6:39. doi: 10.1186/1939-8433-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012;7:e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Tao L, Zhu C. Does nitric oxide play a pivotal role downstream of auxin in promoting crown root primordia initiation in monocots? Plant Signal Behav. 2009;4:999–1001. doi: 10.4161/psb.4.10.9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou H, Wu P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005;46:1674–1681. doi: 10.1093/pcp/pci183. [DOI] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Paek NC. Rice WUSCHEL-related homeobox 3A (OsWOX3A) modulates auxin-transport gene expression in lateral root and root hair development. Plant Signal Behav. 2013;8:e25929. doi: 10.4161/psb.25929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, Cao M, Liu J, Sun J, Tang J, Chen Y, Huang X, Lin W, Ye C, Tong W, Cong L, Geng J, Han Y, Li L, Li W, Hu G, Li J, Liu Z, Qi Q, Li T, Wang X, Lu H, Wu T, Zhu M, Ni P, Han H, Dong W, Ren X, Feng X, Cui P, Li X, Wang H, Xu X, Zhai W, Xu Z, Zhang J, He S, Xu J, Zhang K, Zheng X, Dong J, Zeng W, Tao L, Ye J, Tan J, Chen X, He J, Liu D, Tian W, Tian C, Xia H, Bao Q, Li G, Gao H, Cao T, Zhao W, Li P, Chen W, Zhang Y, Hu J, Liu S, Yang J, Zhang G, Xiong Y, Li Z, Mao L, Zhou C, Zhu Z, Chen R, Hao B, Zheng W, Chen S, Guo W, Tao M, Zhu L, Yuan L, Yang H. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu L, Wang F, Deng M, Yi K. Modulating the root elongation by phosphate/nitrogen starvation in an OsGLU3 dependant way in rice. Plant Signal Behav. 2012;7:1144–1145. doi: 10.4161/psb.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JW, Xu L, Wu YR, Chen XA, Liu Y, Zhu SH, Ding WN, Wu P, Yi KK. OsGLU3, a putative membrane-bound endo-1,4-beta-glucanase, is required for root cell elongation and division in rice (Oryza sativa L.) Mol Plant. 2012;5:176–186. doi: 10.1093/mp/ssr084. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009;21:736–748. doi: 10.1105/tpc.108.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Li S, Ren B, Zhang J, Ichii M, Taketa S, Tao Y, Zuo J, Wang H. LATERAL ROOTLESS2, a cyclophilin protein, regulates lateral root initiation and auxin signaling pathway in rice. Mol Plant. 2013;6:1719–1721. doi: 10.1093/mp/sst052. [DOI] [PubMed] [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. The Plant Genome. 2008;1:2–20. [Google Scholar]
- Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, Wu P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol Plant. 2012;5(1):154–161. doi: 10.1093/mp/ssr074. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu X, Wang Q, Chen Y, Liu C, Qiu Y, Zhang W. OsRPK1, a novel leucine-rich repeat receptor-like kinase, negatively regulates polar auxin transport and root development in rice. Biochim Biophys Acta. 2014;ᅟ:ᅟ. doi: 10.1016/j.bbagen.2014.01.003. [DOI] [PubMed] [Google Scholar]


