Abstract
Purpose
To investigate how potentially functional genetic variants are coinherited on each of four common complement factor H (CFH) and CFH-related gene haplotypes and to measure expression of these genes in eye and liver tissues.
Methods
We sequenced the CFH region in four individuals (one homozygote for each of four common CFH region haplotypes) to identify all genetic variants. We studied associations between the haplotypes and AMD phenotypes in 2157 cases and 1150 controls. We examined RNA-seq profiles in macular and peripheral retina and retinal pigment epithelium/choroid/sclera (RCS) from eight eye donors and three liver samples.
Results
The haplotypic coinheritance of potentially functional variants (including missense variants, novel splice sites, and the CFHR3–CFHR1 deletion) was described for the four common haplotypes. Expression of the short and long CFH transcripts differed markedly between the retina and liver. We found no expression of any of the five CFH-related genes in the retina or RCS, in contrast to the liver, which is the main source of the circulating proteins.
Conclusions
We identified all genetic variants on common CFH region haplotypes and described their coinheritance. Understanding their functional effects will be key to developing and stratifying AMD therapies. The small scale of our expression study prevented us from investigating the relationships between CFH region haplotypes and their expression, and it will take time and collaboration to develop epidemiologic-scale studies. However, the striking difference between systemic and ocular expression of complement regulators shown in this study suggests important implications for the development of intraocular and systemic treatments.
Keywords: complement factor H, genetic variation, gene expression, genetic association, haplotype
Ten years have passed since the association between variation in the human complement factor H (CFH) gene and risk of age-related macular degeneration (AMD) was discovered,1–3 yet this knowledge has not yet been translated into preventive measures for the aging population or treatments for patients with AMD. New therapeutic agents that modify the function of the pathway are in development and in trials, and stratified medicine approaches to the use of existing treatments may soon require ophthalmologists to choose treatments according to individual patients' inherited genetic variants.
Dysfunction of the complement system (a central part of the innate immune system) is key to the etiology of AMD.4 The complement system comprises a series of proteins, C1 to C9, which are sequentially activated or inhibited by regulatory molecules, including complement factor H (FH), factor D (FD), and factor I (FI). The central component of the process, C3, forms enzymes that accelerate its own activation in a positive feedback loop. When in homeostatic balance, the system facilitates destruction of microbial pathogens, apoptosis, and the removal of cell debris without damage to healthy self-tissues.
Pharmaceuticals currently in clinical trials that inhibit the complement system include lampalizumab (used to inhibit FD, currently in phase III trials for the treatment of “dry” AMD or geographic atrophy in people with specific genotypes), compstatin (an inhibitor of C3, in phase II trials for treatment of AMD), and bikaciomab (an inhibitor of FB that is in phase II trials for treatment of AMD).5 Numerous studies of the impact of patients' genetics on their response to intravitreal anti-VEGF for AMD have been carried out, yet they have primarily focused on the effect of only one polymorphism, Y402H, even though there are four distinct haplotypes of CFH with different functional variants and risk profiles. Even while potential clinical applications are being investigated there are many unanswered questions about how the complement system behaves in people with different inherited genetic variants and in the different tissues of the eye.
The major inhibitor of the system, FH, is produced in the liver and secreted into the serum, where it is highly abundant.6 Rare CFH mutations are associated with early-onset AMD.7 Relatively little is known about how variants influence pathogenesis.8,9 Factor H protein is composed of 20 short consensus repeats (SCRs), each approximately 60 amino acids in length, which share homology at specific residues.10,11 Full-length FH is encoded by 22 exons, and a shorter form, FHL-1, stops after splicing of an alternative 10th exon. Five homologous CFH-related genes (CFHR1 to CFHR5 encoding FHR-1 to FHR-5) exist in tandem on chromosome 1q in a region of strong linkage disequilibrium (Supplementary Fig. S1). Four common haplotypes of the CFH region are inherited in white Europeans, and each confers a different risk of AMD (and other diseases).9,12,13 A common deletion of entire CFH-related genes (CFHR3-CFHR1) has been found between sites of segmental duplication,14 and two distinct isoforms of FHR-1, one acidic and one basic, have been reported.15 Homologous sequences cause significant technical challenges for investigators trying to genotype variants and structural rearrangements.
The research questions that we sought to answer were which of the sequence variations in the CFH region might be functional and therefore may be of interest as prognostic biomarkers, predictive (of therapeutic response) biomarkers, or therapeutic targets for inhibition or synthetic imitation; and in which AMD-relevant human tissues the CFH and CFHR1-5 genes and their splicing variants are expressed (eye tissues for local production and liver for systemic production). In order to answer these questions, we undertook massively parallel genomic sequencing of the CFH region in individuals who were homozygous for each of four common haplotypes. We investigated the risk of AMD-related phenotypes from a genome-wide association study of AMD to illustrate the risk associated with each haplotype. Lastly, we studied RNA expression of CFH and CFHR gene transcripts in central and peripheral retina and retinal pigment epithelium/choroid/sclera (RCS) and in the liver. Our results will help inform the development of a personalized medicine approach to complement system therapeutics.
Materials and Methods
Study Populations
Institutional Review Board (IRB)/Ethics Committee approval was obtained, and informed consent was obtained from all participants in the original studies. The research adhered to the tenets of the Declaration of Helsinki. DNA for the sequencing experiment was from the anonymized DNA, RNA, and Serum Bank in the Centre for Public Health at Queen's University Belfast. Patients had neovascular AMD confirmed by clinical examination, grading of digital fundal color photographs, and fluorescein angiography. No details about the clinical course or treatment were available. Research ethics approval for this bank allows analysis of DNA samples for this purpose (Office for Research Ethics Committees Northern Ireland approval reference 11/NI/0139). The in silico data analyzed for this report were from IRB-approved studies. The characteristics of participants included in these studies are summarized in Supplementary Table S1.
Investigation of CFH and CFH-Related Gene Sequence
To identify functional variants in the CFH gene region, we selected genomic DNA samples from individuals of white European ancestry who were homozygous for each of the four common CFH haplotypes, as described previously.12,13 Full details of the preparatory, sequencing, and bioinformatics methods have been provided (Supplementary Methods). Briefly, we quantified and fragmented genomic DNA, size selected, and indexed it for Illumina sequencing using the Truseq system.16 We used Nimblegen SeqCap EZ capture to target the entire CFH and CFHR genomic region. Sequencing was performed on a HiSequation 2000 (Illumina, San Diego, CA, USA) with 100-bp paired-end reads. We aligned genomic reads to hg19 human reference sequence with the Burroughs-Wheeler Aligner (BWA) 0.5.9 aln algorithm17 and used SAMtools 0.1.1418 for sorting, indexing, and removal of duplicate reads. We employed Genome Analysis Toolkit (GATK) to recalibrate and realign and to call polymorphisms.19 We predicted the effect of coding polymorphisms with PolyPhen-2 (Polymorphism Phenotyping v2).20
Investigation of the Effect of CFH Region Haplotypes on AMD Phenotypes
To investigate the effect of polymorphisms on the differential effect on risk of neovascular AMD compared to drusen only (as a proxy for progression), we conducted a genome-wide case–case study and candidate gene studies using the Michigan, Mayo, AREDS, Pennsylvania (MMAP; dbGAP accession phs000182.v1.p1) AMD study, consisting of 2157 cases of neovascular AMD, geographic atrophy, or drusen and 1150 healthy controls of white European ancestry (Supplementary Table S1).21 The candidate gene studies aimed to investigate the roles of the four CFH haplotypes and a representative polymorphism from each of the other known major AMD loci (CFB, C3, and HTRA1)1–3,22–25 on the differential risk of neovascular AMD compared to drusen. The risks of drusen and neovascular AMD compared to disease-free controls are shown to illustrate the effects associated with each haplotype.
Full details of quality control and analyses are shown in the Supplementary Methods. Briefly, genotyping was conducted on Illumina Human370 microarray chips. Final analyses included 867 individuals with neovascular AMD, 519 with drusen, and 1115 healthy controls. A Q-Q plot is shown in Supplementary Figure S2. We used additive model univariate binary logistic regression in PLINK v1.07.26,27 Statistical significance was accepted at two-sided P < 0.05 after Bonferroni correction for multiple testing in the genome-wide association studies. We did not apply correction to the candidate polymorphisms because their selection was based on prior hypotheses due to their known association with the AMD phenotype. We phased haplotypes of CFH in PLINK and used SNP Annotation and Proxy Search (Broad Institute)28 to identify proxy single nucleotide polymorphisms (SNPs).
Investigation of FH, FHL-1, and FHR-1 to FHR-5 Expression in the Retinal Pigment Epithelium, Choroid, and Sclera and in the Retina
To investigate the relative expression of FH, FHL-1, and FHR-1 to FHR-5 in eye tissues and to investigate the expression of alternatively sliced variants, we aligned RNA-seq reads from our (ML, CAC, DS) previous studies29 to the human reference hg19 genome using GSNAP.30,31 Eight participants of white European ancestry from the United States were included. Full details are described in Supplementary Methods. We used SAMtools (version 0.1.19)18 to sort reads; Picard (version 1.121)32 to mark duplicate reads; and SAMtools18 depth to obtain read depths. Relative read depths were calculated for FH and FHL-1 relative to FH and FHL-1 combined. An estimate of the proportion of FH relative to both FH and FHL-1 combined for all samples was calculated by random effects meta-analysis using metaprop in meta (v4.0-3) in R 3.2.1.33 This method was chosen as it allowed calculation of an appropriately weighted average proportion (taking into account the total number of reads from each individual) with an overall estimate of uncertainty (confidence interval [CI]) for each tissue.
Investigation of FH, FHL-1, and FHR-1 to FHR-5 Expression in Liver
To investigate the relative expression of FH, FHL-1, and FHR-1 to FHR-5 in liver, we accessed liver RNA-seq reads from three individuals (no details of ancestry were available) from the EBI Illumina body map34 and EBI Expression Atlas35 and aligned them to the human reference hg19 chromosome 1 using STAR aligner (version 2.4.0f1).36 We sorted the alignments with SAMtools and viewed sequence in IGV.18,37
Results
Sequencing of CFH and CFH-Related Genes From Genomic DNA
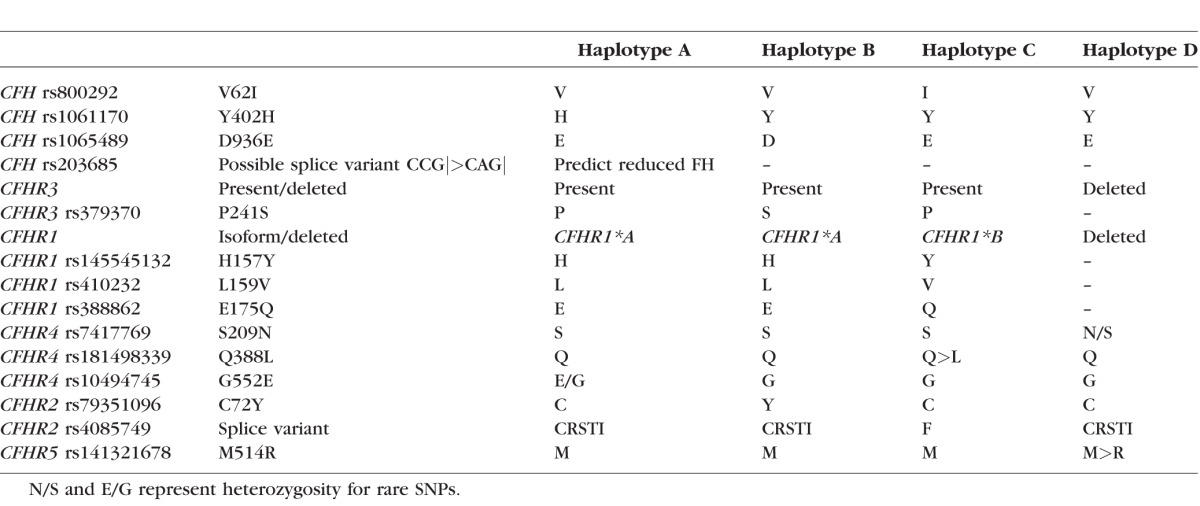
We identified nine missense coding SNPs (cSNPs) within CFH and the CFH-related genes, each of which had a minor allele that was homozygous and restricted to one of the four common haplotypes (Table 1). In addition, four heterozygous missense cSNPs (interpreted as rare SNPs against specific haplotype backgrounds; Table 1) and numerous synonymous cSNPs were also identified (Supplementary Table S2). Haplotype A carried a SNP that was predicted to create a novel splice site that would result in nonsense-mediated decay (rs203685, A) of the transcript (creating CAG|). An ancestral gene conversion event has caused the replacement of part of the CFHR1 gene with part of CFH on haplotype C only, resulting in the FHR-1 157Y, 159V, and 175Q polymorphisms in SCR 3. This means that on haplotype C, SCR 3 of FHR-1 is identical to SCR 18 of FH. These variants correlate with the previously reported15 FHR-1 acidic and basic isoforms (CFHR1*A and CFHR1*B): Our haplotypes A and B produced the acidic isoform, and haplotype C produced the basic isoform. Haplotype D carried the deletion of CFHR3 and CFHR1 that we identified previously, and therefore neither isoform of FHR-1 is produced.14 Haplotype C also carried CFHR2 rs4085749 T, which created a premature splice donor site GCAGG>GTAGG, resulting in replacement of residues 140 to 144 by a single phenylalanine residue, removing one of the key cysteine residues responsible for the SCR structure.
Table 1.
Putatively Functional Polymorphisms Identified on Common CFH Region Haplotypes in Sequenced Homozygotes (n = 1 Homozygous Participant per Haplotype)
Investigation of Risk of AMD Phenotypes
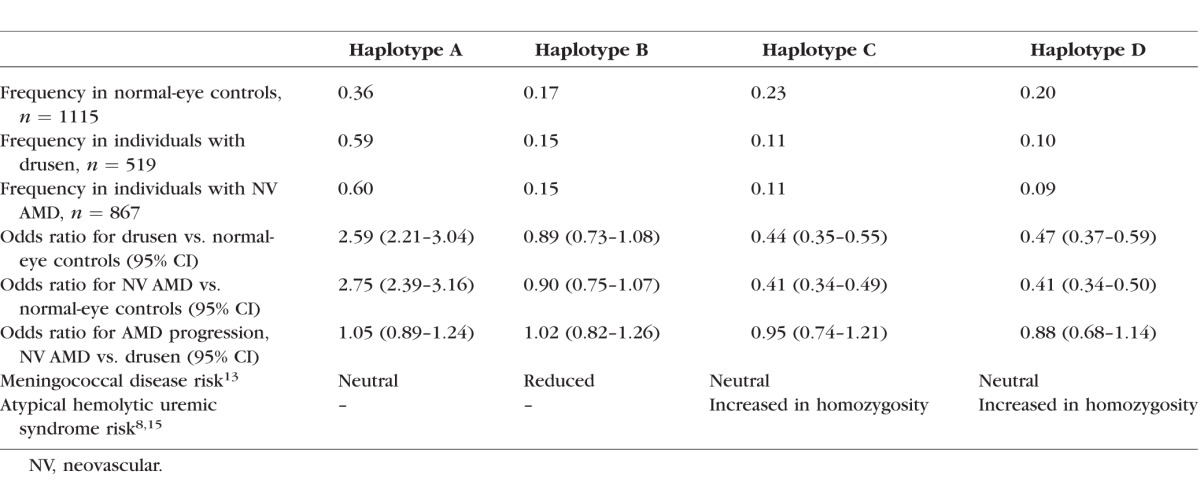
The risks of drusen and neovascular AMD associated with each CFH haplotype were calculated to illustrate the effects of each CFH haplotype (Table 2). We compared genome-wide variation in individuals with neovascular AMD to those with drusen and found that two SNPs were significantly associated with differential risk (a proxy for a “progression” phenotype) after Bonferroni correction for multiple testing: rs932275 and rs2248799, both in HTRA1 (Supplementary Table S3). We identified several SNPs with a suggestive P value < 5×10−5, many of which were in biologically plausible genes (Supplementary Table S3). We also compared individuals with neovascular AMD to those with drusen for previously reported major AMD loci (Supplementary Table S4), but the only association identified from among these candidates was the same one that was identified at the HTRA1 locus in the genome-wide analysis. The complement system gene variants were not significantly different between people with drusen and people with neovascular AMD in this cohort.
Table 2.
CFH Region Haplotype Frequencies and Associated Risk of Neovascular AMD and Drusen (Additive Model Univariate Logistic Regression With Each Haplotype Compared to All Others); the Reported Risk of Invasive Meningococcal Disease and Atypical Hemolytic Uremic Syndrome Shown to Provide Context
Retinal Expression of CFH and CFH-Related Genes
We found no expression of any CFH-related genes (CFHR1-5) in any of the eye tissues studied. Expression of both long and short CFH transcripts was 30- to 50-fold higher in the RCS than in peripheral or macular retina (Table 3). Within the retina, the full-length FH transcripts were considerably more abundant than FHL-1 transcripts in the macula compared to the periphery (Table 3). The RCS showed increased ratio of FH to FHL-1 transcripts in the periphery compared to the macula in seven of eight eyes (Supplementary Table S5).
Table 3.
The Mean Combined Expression of FH and FHL-1 Measured by RNA-seq Read Depth in Four Eye Tissues From Eight Individuals and the Percentage of the Total That Is FH

Expression of CFH and CFH-Related Genes in Liver and Confirmation of the Effect of rs4085749 T on CFHR2 mRNA
Relative to total CFH transcription in the liver, CFHR1 was expressed approximately 2-fold more strongly, CFHR2 at a similar level, and CFHR3 approximately 2-fold less strongly (Supplementary Table S6). CFHR4 and CFHR5 showed lower expression, between 5% and 17% of CFH levels. Relative expression of CFH long (FH) and short (FHL-1) transcripts varied widely, with reduction of FH transcripts in proportion to the number of copies haplotype C carried (Supplementary Table S7).
CFHR2 reads from the haplotype AC heterozygote showed that the early splice donor site created by the T allele of rs4085749 carried on haplotype C was used for 66% of exon-spanning reads, while the original site 12 bases downstream was used for the remaining 34% of reads from this haplotype. Reads from the homozygous haplotype CC sample showed 68% reduction of read depth across the site, and 60% of exon-spanning reads used the novel splice site (Supplementary Fig. S3).
The liver RNA-seq reads also revealed several novel exons that were occasionally found in other transcripts. Of note was an exon within intron 4 of CFHR3 (196,757,875–196,758,074) that was included in about half of all CFHR3 transcripts, despite carrying an in-frame stop codon.
Discussion
The central role of factor H in innate immunity and the effects of its genetic variation on risk of AMD, atypical hemolytic uremic syndrome (aHUS),8 systemic lupus erythematosus,38 rheumatoid arthritis,39 and invasive meningococcal infection9 make it intriguing, and it has yielded many surprises. Our study is the first to investigate the extended CFH region in individuals who are homozygous for the common European haplotypes. The results of this study should provide a clear foundation for future studies of FH function and aid the development of personalized approaches based on patients' own complement system genetics.
Haplotype A confers greatest risk of AMD and carries the 402H risk variant.12 Codon 402 is found in both FH and FHL-1, so both products of the CFH gene are affected.40 This haplotype also carries a possible new splice site (rs203685), which could reduce expression of CFH through nonsense-mediated decay. Haplotype A produces the acidic isoform of FHR-1.
Haplotype B has no effect on AMD risk,12 but confers reduced risk of meningococcal disease, which may be due to altered binding of meningococci to FH or FH-related proteins.9 We showed that the minor alleles of virtually all variants within the CFHR3 region were found on haplotype B. CFH 936D and CFHR3 241S, two variants in the 3′untranslated region (UTR) of CFHR3 (rs402372 and rs390837), and CFHR2 72Y are carried on haplotype B. This haplotypye also produces the acidic isoform of FHR-1.
Haploytype C protects against AMD.12 CFHR1 157Y, 159V, and 175Q, which have arisen by conversion from CFH, code for the basic isoform of FHR-1 on this haplotype. This may result in altered competition between FHR-1 and FH for binding sites, or alteration of the regulatory roles of FHR-1. On haplotype C, CFH 62I encodes FH with increased binding activity for C3b, and increased cofactor activity for complement factor I–mediated cleavage of fluid phase and cell-bound C3b.41 The preferential use of an alternative splice donor site created by CFHR2 rs4085749 on haplotype C may be functionally important.42 This site is predicted to result in omission of one of the four cysteine residues that form the SCR structure and would have a major effect on the protein structure. Haplotype D is missing the entire CFHR3 and CFHR1 genes.14 There are no cSNPs in CFH on this haplotype. Homozygotes for haplotypes C and D are at elevated risk of aHUS.8,15,43,44 Absence of FHR-1 is thought to be critical, resulting in lack of immune tolerance to an epitope of FHR-1 that mimics a neoepitope of FH that may be formed during interaction with bacteria and development of antibodies that cross-react against FH.15,43
Although CFH was expressed in the retina and in the RCS of normal aged eyes, we found no evidence of transcription of any of the CFH-related genes (CFHR1-5) in these tissues. This finding contrasts with reports by Bennis et al.45 and Booij et al.46,47 of a microarray experiment that measured transcriptome expression of six human eyes, which reported CFHR1 transcription at approximately one-fourth the level of CFH (and lower levels of CFHR4, CFHR5, and CFHR2, but not CFHR3). They found no CFHR1 expression in mouse eye tissues, which is consistent with an earlier report by Luo et al.48 in which RT-PCR indicated no expression of CFHR1 in mouse retina or RPE/choroidal tissue. The conflicting results indicate either that our method did not detect CFHR mRNA where it was present, or that the Bennis et al.45 and Booij et al.46,47 experiments falsely detected CFHR mRNA. If CFHR genes are expressed in these tissues, the most likely explanation for their apparent absence in our experiment would be differences in the tissue processing between the two studies. Alternatively, the Bennis et al.45 and Booij et al.46,47 custom microarrays may have suffered cross-reactivity from FH and FHL-1 transcripts (Supplementary Fig. S1), leading to the apparent presence of CFHR genes. Absent expression would not necessarily indicate that CFHR genes have no role in these tissues, as it is likely that they could diffuse across Bruch's membrane, as FHL-1 does (though FH does not).49 Further studies are needed to assess CFHR expression (if any) in AMD eyes and the effect of inflammation. CFH transcripts for FH and FHL-1 showed relatively low expression in the macular and peripheral retina and much greater expression in the RCS, suggesting reduced protection in the retina and greater protection in at least one of the RCS tissues. Our results suggested a greater proportion of FH compared to FHL-1 in the macular retina compared to the peripheral retina. This may be due to regulation by microRNAs. Both transcripts appear to be heavily regulated by microRNAs; however, target sites for miR146a and miR155 are adjacent on the 3′ UTR of CFH and absent from the 3′ UTR of the short (FHL-1) transcript. These are among the microRNAs with greatest influence on immune signalling pathways and immune homeostasis.50
In contrast to the eye, we found that all CFH-related transcripts were present in liver RNA. Both CFH transcripts, CFHR1, CFHR2, and CFHR3 were much more highly expressed than CFHR4 or CFHR5. Our analysis of liver RNA-seq data suggested that haplotype C may be associated with either considerable reduction in FH or increase in FHL-1 mRNA transcript levels compared to the other haplotypes. Unexpectedly, there seems to be poor correlation between levels of transcripts in the liver and the reported concentrations of their translated products in the blood plasma. FHL-1 is reported at only 2% to 8% of FH levels in plasma, despite having transcript levels in the liver greater than that of CFH mRNA.6 It may be important to understand the stability of FH, FHL-1, and their degradation products. Measurement of FH and related proteins is difficult because of the potential for related proteins to cross-react with antibodies used in immunoassays. Reported plasma concentrations of FH also include FHL-1, which reacts with antibodies specific for the N-terminal SCRs common to both proteins, and there is also considerable scope for cross-reactivity with FH-related proteins. Scholl et al.51 reported increased concentrations of most complement proteins and activation products in AMD compared to normal controls, compatible with systemic complement activation and low-grade inflammation Complement activation products were highest in those carrying CFH risk haplotypes for AMD.51 In a study that used an immunoassay with antibodies highly specific for either FH 402H or 402Y, Hakobyan et al.52 showed that plasma FH levels of normal adults were not significantly different in those with FH 402YY, 402YH, and 402HH genotypes. The largest study of genetic influences on plasma FH and AMD was reported by Ansari et al. in 2013.53 They showed a relationship between CFH genotypes and concentrations of FH and of FHR-1, though they reported plasma FHR-1 concentrations in people with no copies of the CFHR1 gene to be >50% of those with two copies of the gene, suggesting that their assay cross-reacted with other proteins.53 It is possible, though difficult, to create antibodies against specific variants, and gene conversion adds another layer of complexity to the measurement and assessment of function in this region.
We conducted a secondary analysis of a genome-wide association study of AMD21 to investigate whether variation was associated with progression from drusen to neovascular AMD. Only SNPs in HTRA1 (near ARMS2) showed an association when comparing individuals with neovascular AMD to those with drusen. Variants in CFH, C3, and CFB increased risk of drusen and AMD equally, but added no extra risk effect to differentiate those with drusen and those with neovascular AMD, suggesting the possibility that complement dysfunction is necessary but not sufficient for progression to neovascular AMD. Interestingly, earlier case–control studies reported similar findings for CFH and ARMS2/HTRA1,54,55 whereas a longitudinal study showed a role for CFH on progression from early to late disease over a period of 6 years.56 It is possible that in the case–control studies, age, sex, smoking, and/or other factors might confound or interact with genetic risk factors in a manner not seen in the cohort study.
The most important limitation of our study is that the number of donated eyes is small, and availability of a greater number of eye samples would improve our ability to draw firm conclusions about the levels of FH and FHL-1 expression in people with different CFH haplotypes. Collaborative studies may be required in the future to bring together enough expression data to fully explore the effects of haplotypes on the levels of expression in different eye tissues.
We have comprehensively characterized the variants that may be the underlying causes of the AMD risk conferred by the four common haplotypes. Taking into account the real-world biological complexity of the region may allow some clearer thought about the mechanisms of disease and protection, as well as the potential for future development of personalized treatments based on these mechanisms.
Supplementary Material
Acknowledgments
We thank the participants in the National Eye Institute Study of Age-Related Macular Degeneration (NEI-AMD) and the NEI-AMD Research Group for their valuable contribution to this research.
The AMD dataset used for the analyses described in this paper was obtained from the NEI-AMD database, through dbGaP accession no. phs000182.v1.p1. Funding support for NEI-AMD was provided by the National Eye Institute.
Supported by Guide Dogs for the Blind Association Grant OR2010-03c (AEH), National Eye Institute Grant R01EY023164, and the Arnold and Mabel Beckman Initiative for Macular Research (CAC and DS).
Disclosure: A.E. Hughes, None; S. Bridgett, None; W. Meng, None; M. Li, None; C.A. Curcio, Genentech (C), Novartis (C), Merck (C), Janssen Cell Therapy (C), IOVS (S); D. Stambolian, None; D.T. Bradley, None
References
- 1. Klein RJ,, Zeiss C,, Chew EY,, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005; 308: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haines JL,, Hauser MA,, Schmidt S,, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005; 308: 419–421. [DOI] [PubMed] [Google Scholar]
- 3. Edwards AO,, Ritter R,, III, Abel KJ,, Manning A,, Panhuysen C,, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308: 421–424. [DOI] [PubMed] [Google Scholar]
- 4. Zipfel PF. Complement and immune defense: from innate immunity to human diseases. Immunol Lett. 2009; 126: 1–7. [DOI] [PubMed] [Google Scholar]
- 5. Melis JPM,, Strumane K,, Ruuls SR,, Beurskens FJ,, Schuurman J,, Parren PW. Complement in therapy and disease: regulating the complement system with antibody-based therapeutics. Mol Immunol. 2015; 67: 117–130. [DOI] [PubMed] [Google Scholar]
- 6. Friese MA,, Hellwage J,, Jokiranta TS,, et al. Different regulation of factor H and FHL-1/reconectin by inflammatory mediators and expression of the two proteins in rheumatoid arthritis (RA). Clin Exp Immunol. 2000; 121: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hughes AE,, Meng W,, Bridgett S,, Bradley DT. Rare CFH mutations and early-onset age-related macular degeneration. Acta Ophthalmol. 2016; 94: e247–e248. [DOI] [PubMed] [Google Scholar]
- 8. Zipfel PF,, Edey M,, Heinen S,, et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007; 3: e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davila S,, Wright VJ,, Khor CC,, et al. Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet. 2010; 42: 772–776. [DOI] [PubMed] [Google Scholar]
- 10. Zipfel PF,, Skerka C. Complement regulators and inhibitory proteins. Nat Rev Immunol. 2009; 9: 729–740. [DOI] [PubMed] [Google Scholar]
- 11. Jozsi M,, Zipfel PF. Factor H family proteins and human diseases. Trends Immunol. 2008; 29: 380–387. [DOI] [PubMed] [Google Scholar]
- 12. Hughes AE,, Orr N,, Patterson C,, et al. Neovascular age-related macular degeneration risk based on CFH, LOC387715/HTRA1, and smoking. PLoS Med. 2007; 4: e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bradley DT,, Bourke TW,, Fairley DJ,, et al. Susceptibility to invasive meningococcal disease: polymorphism of complement system genes and Neisseria meningitidis factor H binding protein. PLoS One. 2015; 10: e0120757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes AE,, Orr N,, Esfandiary H,, Diaz-Torres M,, Goodship T,, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006; 38: 1173–1177. [DOI] [PubMed] [Google Scholar]
- 15. Abarrategui-Garrido C,, Martinez-Barricarte R,, Lopez-Trascasa M,, de Cordoba SR,, Sanchez-Corral P. Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood. 2009; 114: 4261–4271. [DOI] [PubMed] [Google Scholar]
- 16. TruSeq® DNA sample preparation guide (rev. C). 2012. Available at: http://support.illumina.com/content/dam/illumina-support/documents/myillumina/f5f619d3-2c4c-489b-80a3-e0414baa4e89/truseq_dna_sampleprep_guide_15026486_c.pdf. Accessed July 10 2013.
- 17. Li H,, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H,, Handsaker B,, Wysoker A,, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKenna A,, Hanna M,, Banks E,, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adzhubei IA,, Schmidt S,, Peshkin L,, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen W,, Stambolian D,, Edwards AO,, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010; 107: 7401–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gold B,, Merriam JE,, Zernant J,, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006; 38: 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yates JR,, Sepp T,, Matharu BK,, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007; 357: 553–561. [DOI] [PubMed] [Google Scholar]
- 24. Yang Z,, Camp NJ,, Sun H,, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006; 314: 992–993. [DOI] [PubMed] [Google Scholar]
- 25. Dewan A,, Liu M,, Hartman S,, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006; 314: 989–992. [DOI] [PubMed] [Google Scholar]
- 26. Purcell S,, Neale B,, Todd-Brown K,, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purcell S. PLINK. 2009; v1.07. Available at: http://pngu.mgh.harvard.edu/purcell/plink/. Accessed July 31, 2014.
- 28. Johnson AD,, Handsaker RE,, Pulit SL,, Nizzari MM,, O'Donnell CJ,, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008; 24: 2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li M,, Jia C,, Kazmierkiewicz KL,, et al. Comprehensive analysis of gene expression in human retina and supporting tissues. Hum Mol Genet. 2014; 23: 4001–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu TD,, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010; 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeLuca DS,, Levin JZ,, Sivachenko A,, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012; 28: 1530–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Picard tools. Available at: http://broadinstitute.github.io/picard/. Accessed May 8, 2014.
- 33. Metaprop. Available at: http://www.inside-r.org/packages/cran/meta/docs/metaprop. Accessed December 5, 2015.
- 34. E-MTAB-513 - RNA-Seq of human individual tissues and mixture of 16 tissues (Illumina Body Map). Array Express; 2011. Available at: https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513/ Accessed February 27, 2015. [Google Scholar]
- 35. E-MTAB-2836 - RNA-seq of coding RNA from tissue samples of 122 human individuals representing 32 different tissues. Array Express; 2014. Available at: https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2836/ Accessed February 27, 2015. [Google Scholar]
- 36. Dobin A,, Davis CA,, Schlesinger F,, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robinson JT,, Thorvaldsdottir H,, Winckler W,, et al. Integrative genomics viewer. Nat Biotechnol. 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao J,, Wu H,, Khosravi M,, et al. Association of genetic variants in complement factor H and factor H-related genes with systemic lupus erythematosus susceptibility. PLoS Genet. 2011; 7: e1002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Foltyn Zadura A,, Zipfel PF,, Bokarewa MI, et al. Factor H autoantibodies and deletion of complement factor H-related protein-1 in rheumatic diseases in comparison to atypical hemolytic uremic syndrome. Arthritis Res Ther. 2012; 14: R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skerka C,, Lauer N,, Weinberger AA,, et al. Defective complement control of factor H (Y402H) and FHL-1 in age-related macular degeneration. Mol Immunol. 2007; 44: 3398–3406. [DOI] [PubMed] [Google Scholar]
- 41. Tortajada A,, Montes T,, Martinez-Barricarte R,, Morgan BP,, Harris CL,, de Cordoba SR. The disease-protective complement factor H allotypic variant Ile62 shows increased binding affinity for C3b and enhanced cofactor activity. Hum Mol Genet. 2009; 18: 3452–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eberhardt HU,, Buhlmann D,, Hortschansky P,, et al. Human factor H-related protein 2 (CFHR2) regulates complement activation. PLoS One. 2013; 8: e78617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jozsi M,, Strobel S,, Dahse HM,, et al. Anti factor H autoantibodies block C-terminal recognition function of factor H in hemolytic uremic syndrome. Blood. 2007; 110: 1516–1518. [DOI] [PubMed] [Google Scholar]
- 44. Jozsi M,, Licht C,, Strobel S,, et al. Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood. 2008; 111: 1512–1514. [DOI] [PubMed] [Google Scholar]
- 45. Bennis A,, Gorgels TG,, Jacoline B,, et al. Comparison of mouse and human retinal pigment epithelium gene expression profiles: potential implications for age-related macular degeneration. PLoS One. 2015; 10: e0141597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Booij JC,, van Soest S,, Swagemakers SM,, et al. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009; 10: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Booij JC,, Jacoline B,, Swagemakers SM,, et al. A new strategy to identify and annotate human RPE-specific gene expression. PLoS One. 2010; 5: e9341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Luo C,, Chen M,, Xu H. Complement gene expression and regulation in mouse retina and retinal pigment epithelium/choroid. Mol Vis. 2011; 17: 1588–1597. [PMC free article] [PubMed] [Google Scholar]
- 49. Clark SJ,, Schmidt CQ,, White AM,, Hakobyan S,, Morgan BP,, Bishop PN. Identification of factor H-like protein 1 as the predominant complement regulator in bruch's membrane: implications for age-related macular degeneration. J Immunol. 2014; 193: 4962–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo X,, Ranade K,, Talker R,, Jallal B,, Shen N,, Yao Y. MicroRNA-mediated regulation of innate immune response in rheumatic diseases. Arthritis Res Ther. 2013; 15: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scholl HP,, Charbel Issa P,, Walier M,, et al. Systemic complement activation in age-related macular degeneration. PLoS One. 2008; 3: e2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hakobyan S,, Tortajada A,, Harris CL,, de Córdoba SR,, Morgan BP. Variant-specific quantification of factor H in plasma identifies null alleles associated with atypical hemolytic uremic syndrome. Kidney Int. 2010; 78: 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ansari M,, McKeigue PM,, Skerka C,, et al. Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum Mol Genet. 2013; 22: 4857–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Magnusson KP,, Duan S,, Sigurdsson H,, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006; 3: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Farwick A,, Dasch B,, Weber BHF,, Pauleikhoff D,, Stoll M,, Hense H. Variations in five genes and the severity of age-related macular degeneration: results from the Muenster aging and retina study. Eye (Lond). 2009; 23: 2238–2244. [DOI] [PubMed] [Google Scholar]
- 56. Seddon JM,, Francis PJ,, George S,, Schultz DW,, Rosner B,, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007; 297: 1793–1800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


