NFκB is a master regulator of protein quality control. It helps the cells to survive proteotoxicity by modulating autophagy via up-regulation of BAG3 and HspB8 expression, a molecular mechanism relevant to protein conformational diseases.
Abstract
During cell life, proteins often misfold, depending on particular mutations or environmental changes, which may lead to protein aggregates that are toxic for the cell. Such protein aggregates are the root cause of numerous diseases called “protein conformational diseases,” such as myofibrillar myopathy and familial amyotrophic lateral sclerosis. To fight against aggregates, cells are equipped with protein quality control mechanisms. Here we report that NFκB transcription factor is activated by misincorporation of amino acid analogues into proteins, inhibition of proteasomal activity, expression of the R120G mutated form of HspB5 (associated with myofibrillar myopathy), or expression of the G985R and G93A mutated forms of superoxide dismutase 1 (linked to familial amyotrophic lateral sclerosis). This noncanonical stimulation of NFκB triggers the up-regulation of BAG3 and HspB8 expression, two activators of selective autophagy, which relocalize to protein aggregates. Then NFκB-dependent autophagy allows the clearance of protein aggregates. Thus NFκB appears as a central and major regulator of protein aggregate clearance by modulating autophagic activity. In this context, the pharmacological stimulation of this quality control pathway might represent a valuable strategy for therapies against protein conformational diseases.
INTRODUCTION
Proteins fold into specific three-dimensional structures to acquire their functional native states. However, proteins can misfold as a result of inefficient protein biogenesis, mutations, or inefficient translocated secretory precursors (Herczenik and Gebbink, 2008). In addition, protein misfolding can be promoted by environmental stresses such as elevated temperatures, exposure to chemicals or heavy metals, bacterial/viral infection, and production of reactive oxygen species, as well as by physiological changes such as aging (Haigis and Yankner, 2010; Chen et al., 2011). Because of their altered spatial arrangement, misfolded proteins are nonfunctional (loss-of-function effect) and tend to exhibit their hydrophobic domains, driving inappropriate protein association and protein aggregation (Herczenik and Gebbink, 2008). Moreover, because protein aggregates can trap other functional cellular components, they are often toxic for the cell (gain-of-function effect), as evidenced by the growing list of so-called protein conformational diseases, which result from the cellular accumulation of proteotoxic misfolded/aggregated proteins (Hightower, 1991; Holmes et al., 2014). Consequently, cells can detect the presence of abnormal proteins (for a pioneering study, see Edington et al., 1989) and have developed machinery that maintains the health of its proteome by refolding, sequestering, or degrading misfolded proteins. Refolding is achieved by molecular chaperones (Vabulas et al., 2010); for example, spatial sequestering is fulfilled by addressing aggregates to the microtubule-organizing center (MTOC), forming a large, “storage-bin” structure called an aggresome (Garcia-Mata et al., 1999). Degradation is principally accomplished by the ubiquitin proteasome system (UPS), which clears most soluble misfolded proteins (Goldberg, 2003), and lysosomal compartments, such as the autophagy–lysosomal pathway.
Macroautophagy (hereafter referred to as autophagy) is a self-eating process starting with the formation of double-membrane autophagosomes that engulf parts of the cytoplasm and fuse with acidic lysosomes to form autolysosomes (Wirth et al., 2013; Wilson et al., 2014). Then the enclosed material is degraded by acid hydrolases and released for recycling. Autophagy occurs at a basal level in most tissues and allows the degradation of long-lived proteins and cytoplasmic organelles, contributing to the routine turnover of cellular trash (Mehrpour et al., 2010). Under nutrient starvation, autophagy is up-regulated as an energy salvage process (Kuma et al., 2004), but it can also be activated under nonmetabolic stresses such as accumulation of aggregated proteins. Of interest, loss of autophagy function may cause an accumulation of misfolded/aggregated proteins (Rubinsztein, 2006; Williams et al., 2006; Villella et al., 2013). Although autophagy has long been described as a bulk degradation process, it is now largely accepted that autophagy can be selective via the use of autophagic receptors and adaptors (such as p62 or BAG3), linking cargoes to the autophagosomal membrane protein LC3-II (Behrends and Fulda, 2012). We showed that selective autophagy is activated by protein aggregates generated by hyperthermia via the stimulation of NFκB (Nivon et al., 2009, 2012).
NFκB is a key transcription factor of the cellular stress response. This inducible factor is inactivated by its binding to members of the IκB family (Hayden and Ghosh, 2004); hence activation of NFκB requires its dissociation from IκB subunits. This event mostly occurs through the “canonical” pathway consisting of IκB kinase (IKK) phosphorylation, which in turn leads to phosphorylation, ubiquitination, and degradation of IκB by the 26S proteasome (Hayden and Ghosh, 2004). However, some inducers stimulate NFκB through noncanonical pathways. Indeed, ultraviolet (UV) or amino acid analogue treatments induce IκB degradation without prior phosphorylation (Kretz-Remy et al., 1998; Li and Karin, 1998). We also demonstrated that heat stress induces NFκB/IκB dissociation independently of IκB phosphorylation or degradation (Nivon et al., 2009).
Here we sought to determine whether NFκB could act as a central modulator of protein quality control (PQC) upon proteotoxic stress. We tested NFκB involvement in PQC after various protein aggregation stresses such as amino acid analogue treatment, proteasome inhibition by MG132, and overexpression of mutated forms of proteins prone to form aggregates. The amino acid analogues canavanine and azetidine mimic natural arginine and proline, respectively. They can escape detection by the protein synthesis machinery and thus are incorporated into nascent polypeptides, promoting translational errors and irreversible protein misfolding, resulting in protein aggregation (Hightower, 1980; Kretz-Remy et al., 1998; Rodgers and Shiozawa, 2008). MG132 reversibly inhibits the chymotrypsin- and caspase-like activities of the 26S proteasome and thus increases the level of polyubiquitinated proteins and induces protein aggregates and aggresome formation (Shen et al., 2011). Of interest, cardiac dysfunctions, cataract formation, and neurodegenerative diseases (Alzheimer, Parkinson, and Huntington diseases and amyotrophic lateral sclerosis [ALS]) are associated with decreased proteasome activity (Dahlmann, 2007). HspB5 (CRYAB gene) is a member of the small heat shock protein family. It is abundantly expressed in lens, cardiac, and skeletal muscles and, by chaperoning desmin and actin, participates in maintaining cytoskeletal integrity (Simon et al., 2013). The R120G missense mutation of HspB5 was the first to be identified in human patients and causes cataract, cardiomyopathy, and myofibrillar myopathy (Vicart et al., 1998). At the cellular level, this mutation promotes HspB5 aggregation with intermediate filaments (desmin, vimentin), which ultimately leads to aggresome formation (Simon et al., 2007b; Elliott et al., 2013). Cytoplasmic Cu, Zn-superoxide dismutase (SOD1) is an enzyme responsible for scavenging free radicals. More than 140 point mutations found in the SOD1 peptide sequence are associated with 20% of familial ALS cases (Rosen et al., 1993). ALS is a progressive and fatal adult-onset motor neuron disease characterized by premature loss of spinal, cranial, and cortical neurons, muscle weakness, atrophy, and paralysis. At the cellular level, most SOD1 mutations alter its tertiary structure, leading to protein aggregation (Turner et al., 2005; Kim et al., 2014).
In this study, we show that NFκB-depleted cells are unable to induce autophagy and accumulate more aggregates than control cells in response to a wide range of protein aggregation stresses. Our data indicate that, in addition to heat stress, at least four other different protein aggregation stresses can activate NFκB transcription factor through a noncanonical pathway that, in turn, induces the autophagic process. Moreover, we demonstrate that upon those stresses, NFκB activation enhances the expression of BAG3 and HspB8 proteins, which relocalize to a vast majority of protein aggregates and activate selective autophagy and protein aggregate clearance. Thus NFκB can be considered as a central regulator of PQC via regulation of autophagy in response to protein aggregation stresses.
RESULTS
Amino acid analogues, MG132, and the expression of mutated forms of HspB5 or SOD1 induce the formation of protein aggregates
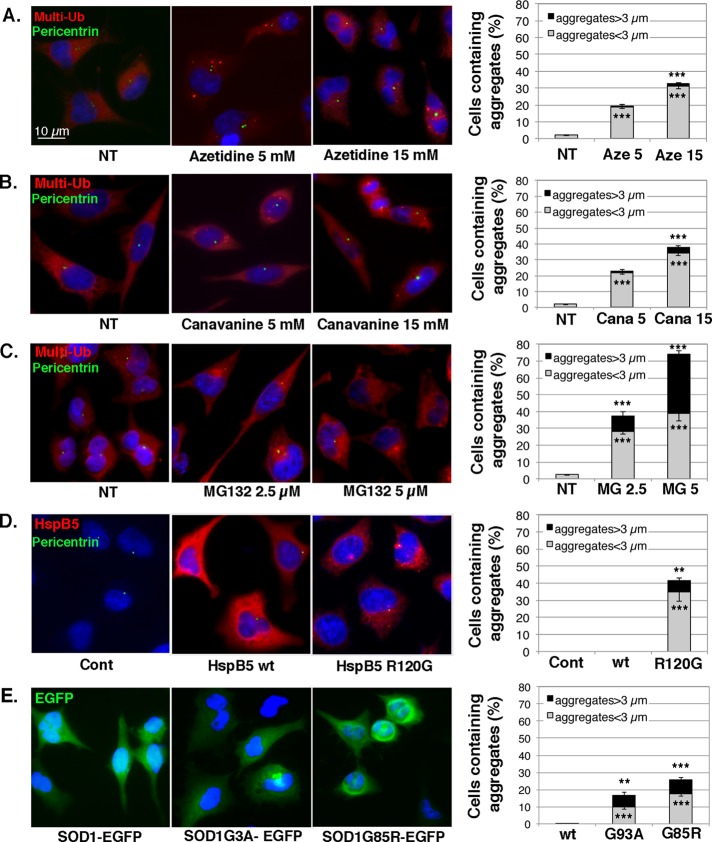
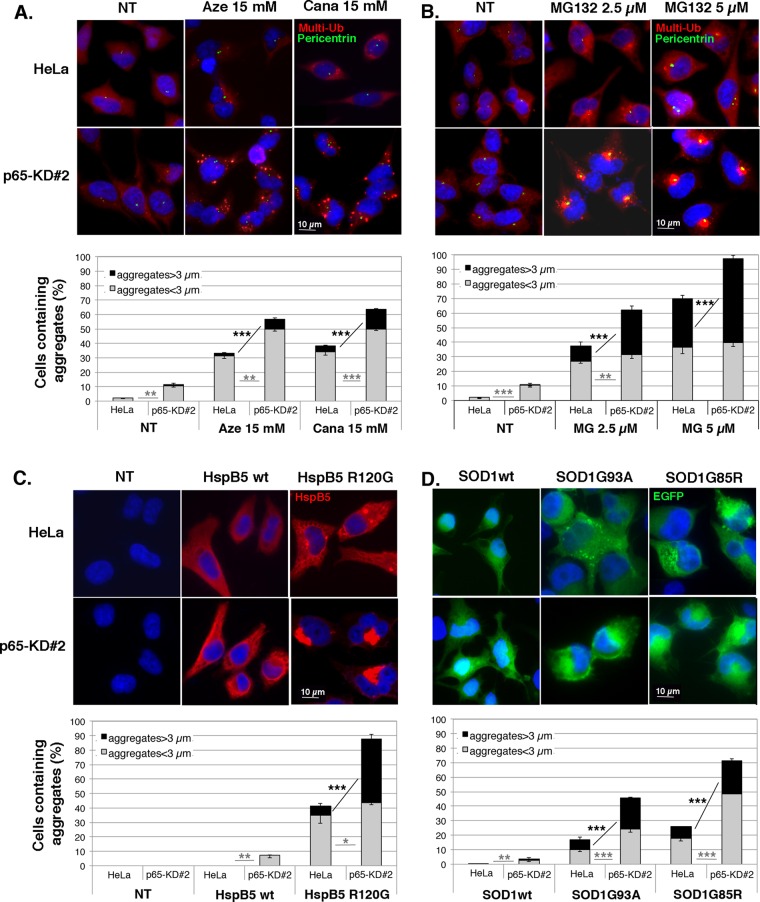
We previously reported that heat stress activates NFκB transcription factor via the induction of protein aggregation (Kretz-Remy et al., 2001). We therefore investigated whether other types of protein aggregation stresses could activate NFκB. We first analyzed, by immunofluorescence (Figure 1) and filter trap assay (Supplemental Figure S1), the protein aggregation status of HeLa cells in response to four different protein aggregation stresses: amino acid analogue and MG132 treatment and expression of the mutated forms of HspB5 and SOD1, which are prone to form aggregates. The distribution of multiubiquitinated proteins was rather diffused in the cytoplasm in nontreated cells (Figure 1, A–C, NT). In contrast, 5 mM canavanine or azetidine treatment induced the formation of small cytosolic granules containing multiubiquitinated proteins in ∼20% of the cells (Figure 1, A and B, graphs). Moreover, 15 mM of the same compounds induced, in a higher percentage of cells (33–38%), aggregates of larger size, colocalized with pericentrin, a component of the MTOC, and sometimes gathered around the nucleus, suggesting aggresome formation (Figure 1, A and B). Filter trap analysis confirmed the accumulation of protein aggregates with greater than fourfold increase in the level of multiubiquitinated protein aggregates after amino acid analogue treatment in comparison to nontreated cells (Supplemental Figure 1A). Similarly, we found that increasing concentration of MG132 induced the formation of multiubiquitin positive inclusions of larger size and increased frequency in the perinuclear region (aggresome-like structures) of treated cells (Figure 1C). Indeed, 38% of 2.5 μM MG132–treated cells contained aggregates, with 10% of these cells containing aggregates >3 μm. In comparison, 74% of 5 μM MG132–treated cells contained aggregates, with 43% of these cells containing aggregates with a size >3 μm. Filter trap assay indicated at least eightfold-increased level of multiubiquitinated protein aggregates after 2.5 or 5 μM MG132 treatment (Supplemental Figure S1B). In overexpression experiments, pcDNA3-HspB5WT–transfected cells (HspB5wt) showed a homogeneous and diffuse localization of HspB5 (Figure 1D), as previously described (Simon et al., 2007a). In contrast, when cells expressed the missense-mutated form of HspB5 (HspB5R120G, found in human disease), the protein was localized in cytoplasmic aggregates in 40% of the cells, with 14% of the aggregates >3 μm. Moreover, some of the aggregates were gathered in the perinuclear region (colocalization with pericentrin). Filter trap data indicated approximately eightfold increase of HspB5 aggregates in those cells in comparison to cells expressing wild-type HspB5 (Supplemental Figure S1C). Similar observations were made with SOD1 protein expressed in HeLa cells. The localization of wild-type SOD1-EGFP was diffuse in the cytoplasm and the nucleus (Figure 1E). In contrast, the expression of the G93A mutated form of SOD1 induced the appearance of cytoplasmic or juxtanuclear enhanced green fluorescent protein (EGFP) inclusions in 17% of the cells. SOD1G85R expression induced similar inclusions with a higher frequency of appearance (27%) but with a similar percentage of cells containing aggregates with a size >3 μm (∼9%). Filter trap analysis indicated that SOD1G85R and SOD1G93A proteins increased the level of protein aggregates respectively by eightfold and fourfold compared with wild-type SOD1 (Supplemental Figure S1D). Hence, by submitting HeLa cells to these diverse stresses, we were able to control protein misfolding and induce various types (quantitatively and qualitatively) of protein aggregates in the cells.
FIGURE 1:
Amino acid analogue or MG132 treatment and overexpression of mutated forms of HspB5 and SOD1 induce the formation of protein aggregates. HeLa cells were untreated (NT) or subjected to 5 and 15 mM azetidine (A) or canavanine (B) treatment or 2.5 and 5 μM MG132 treatment (C) for 6 h and allowed to recover in fresh culture medium for 16 h. HeLa cells were transiently transfected with control (Cont) plasmids (see Materials and Methods) or plasmids expressing wild-type HspB5 or its mutated form, HspB5R120G (D), or wild-type SOD1-EGFP or its mutated forms, SOD1G93A-EGFP and SOD1G85R-EGFP (E), and were analyzed 48 h after transfection. Cells were fixed, permeabilized, and stained with antibodies against multiubiquitin (red staining) and pericentrin (indicator of MTOC localization; green staining; A–C) or HspB5 (red staining; D) and pericentrin (green staining; D). Nuclei were stained with Hoechst (blue), and cells were analyzed with a fluorescence microscope. The percentage of cells containing aggregates larger or smaller than 3 μm is reported in the histograms. n = 4.
Amino acid analogues, MG132 treatment, and expression of HspB5 and SOD1 mutants activate NFκB through a noncanonical pathway
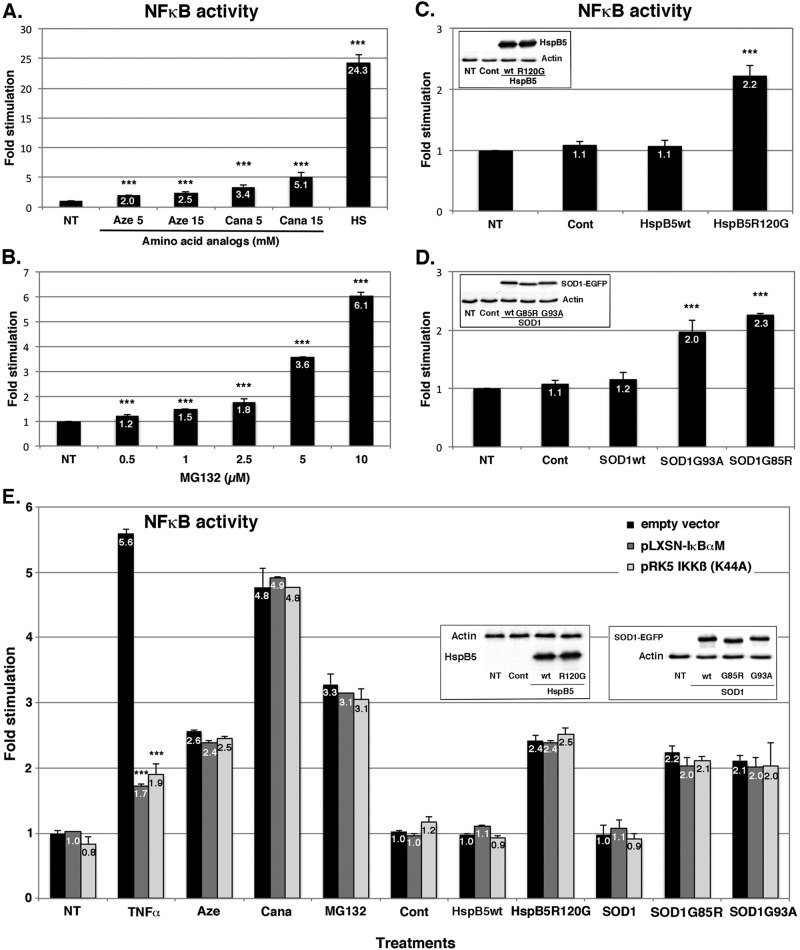
We then tested whether protein aggregation stresses could activate NFκB transcription factor, as we previously reported for heat and amino acid analogue treatments (Kretz-Remy et al., 1998, 2001). NFκB activity was quantified in HeLa cells constitutively expressing pNFκBluc reporter vector (see Materials and Methods) and subjected to increasing concentrations of azetidine, canavanine, and MG132 or transfected with wild-type or mutated HspB5 and SOD1 expression vectors (Figure 2). The κB-dependent luciferase reporter gene revealed that increasing concentrations of azetidine or canavanine gradually stimulated NFκB activity by ∼2.5-fold for azetinine and 5-fold for canavanine treatment (Figure 2A), as we previously showed (Kretz-Remy et al., 1998). Note that this activation was not as intense as that observed after a 43°C heat shock treatment (∼24-fold). Similarly, increasing concentrations of MG132 gradually activated NFκB activity up to 6-fold (Figure 2B). Moreover, overexpression of the mutated form of HspB5 stimulated NFκB activity by ∼2-fold, whereas overexpression of wild-type HspB5 did not. Indeed, NFκB activity in HspB5wt-expressing cells was similar to that in untreated or empty vector–transfected (Cont) cells (Figure 2C). The equivalent expression levels of wild-type and mutated HspB5 were established by Western bot analysis (Figure 2C, inset). Similarly, equivalent expression of G93A and G85R mutated forms of SOD1 increased NFκB activity approximately twofold, whereas the wild-type form did not (Figure 2D and inset). We then performed the same experiment in cells expressing either a dominant-negative mutant of the IKKβ subunit of IKK (pRK5-IKKβ(K44A)) or a dominant-negative mutant of IκBα (pLXSN IκBαM), which both abrogate IκBα phosphorylation and thus the canonical activation pathway of NFκB (Figure 2E). NFκB stimulation by protein aggregation stresses was similar in the presence and absence of those dominant-negative mutants. In contrast, NFκB induction by tumor necrosis factor α (TNFα) treatment, which depends on the canonical pathway, was strongly impaired by the presence of those dominant-negative mutants (Figure 2E). Thus NFκB activation by these protein aggregation stresses appears to be independent of IκBα phosphorylation/degradation. Taken together, our results demonstrate that various protein aggregation stresses are able to activate NFκB via a noncanonical pathway. Moreover, they suggest that NFκB is a central sensor of cellular protein aggregation stress.
FIGURE 2:
Protein aggregation stresses stimulate NFκB activity. pNFκBLuc stable HeLa transformants were untreated (NT) or treated with 5 or 15 mM azetidine (Aze) or canavanine (Cana; A) or with increasing concentration (0.5–10 μM) of MG132 (B) for 6 h. As a positive control for NFκB induction, cells were also subjected to 90-min heat shock treatment (HS) at 43°C (A). (C, D) HeLa cells were transiently transfected or not (NT) with control plasmids (Cont) or plasmids expressing wild-type HspB5 or its mutated form, HspB5R120G (C), or wild- type SOD1-EGFP or its mutated forms, SOD1G93A-EGFP and SOD1G85R-EGFP (D). (E) pNFκBLuc HeLa cells were either transfected with an empty vector or with expression vectors for dominant-negative mutants of IκBα (pLXSN-IκBα) or of IKKβ (pRK5-IKKβ(K44A)). The next day, cells were treated with 2000 U/ml TNFα for 4 h, followed by 16 h of recovery or subjected to the same treatments as described (15 mM Aze or Cana, 5 μM MG132, or expression of wild-type and mutated forms of HspB5 and SOD1). Insets, immunoblot analysis of HspB5 and SOD1-EGFP levels in transfected HeLa cells 48 h after transfection. Luciferase activity was measured 16 h after treatment or 48 h after transfection. Results are presented as fold stimulation (ratio between luciferase activity after treatment vs. luciferase activity in NT counterpart). Results are presented as mean ± SD. n ≥ 3.
Amino acid analogues, MG132 treatment, and expression of mutated forms of HspB5 or SOD1 activate autophagy flux in an NFκB-dependent manner
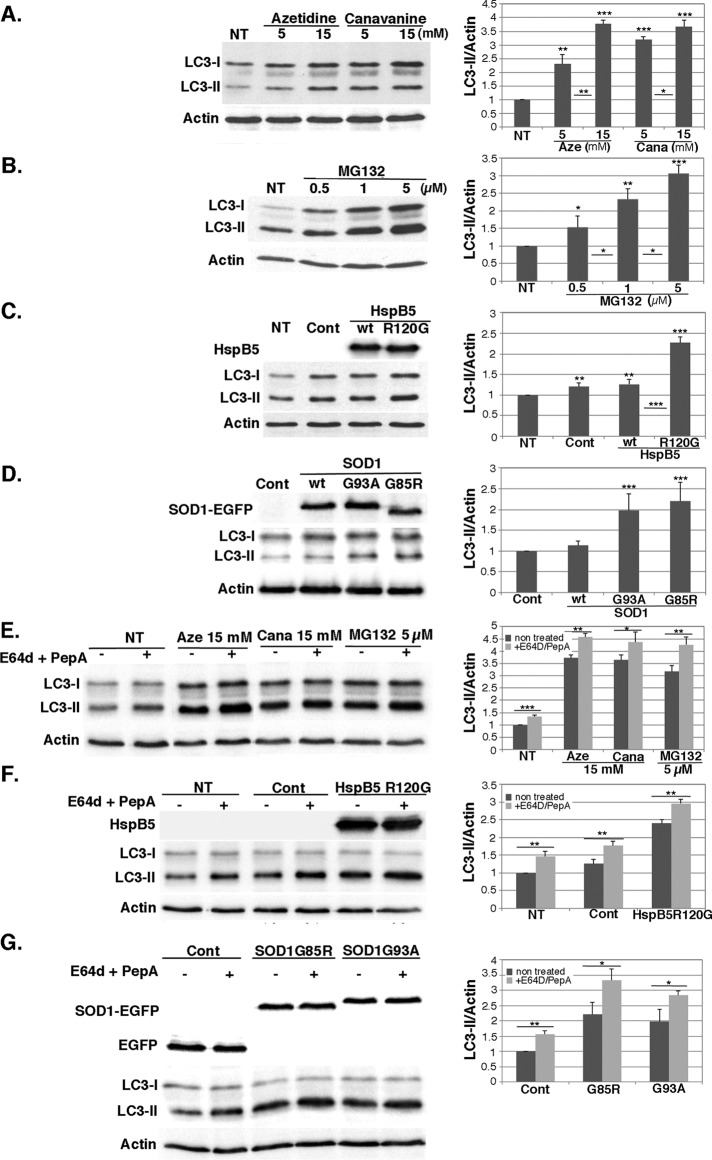
We previously reported that protein aggregation stress induced by heat shock activates NFκB-dependent autophagy. We thus wondered whether the aggregation stresses studied here could induce the same process. To this end, we submitted HeLa cells to increasing concentrations of azetidine, canavanine, or MG132 and quantified the LC3-II level by Western blot as a read-out of the abundance of autophagosomes in cells. We observed that LC3-II level increased in cells treated with increasing concentrations of amino acid analogues (Figure 3A) or MG132 (Figure 3B) compared with nontreated cells. Regarding overexpression of HspB5, transfection by control plasmid or overexpression of wild-type HspB5 did not nearly modify LC3-II levels in comparison to nontransfected cells (1.2 ratio vs. 1 for nontransfected cells). In contrast, overexpression of HspB5R120G doubled the LC3-II level (Figure 3C). The same results were obtained with G93A and G85R mutated forms of SOD1, whose overexpression approximately doubled the LC3-II levels, whereas that of wild-type SOD1 did not (Figure 3D). These results were confirmed by immunofluorescence analysis in cells expressing GFP-LC3 or labeled with LC3 antibody (Supplemental Figure S2). Indeed, whereas we observed diffuse fluorescence in nontreated cells or in cells expressing wild-type HspB5 or SOD1 (Supplemental Figure S2, A–C, NT, Cont and wt), we observed the formation of numerous fluorescent puncta in cells treated by amino acid analogues and by MG132 (Supplemental Figure S2A) and in cells overexpressing HspB5R120G (Supplemental Figure S2B) or SOD1G93A-EGFP and SOD1G85R-EGFP (Supplemental Figure S2C). An increased number of autophagosomes can be the consequence of either increased autophagic flux or inhibition of the maturation/degradation of autophagic vesicles. To distinguish between those two events, we stressed HeLa cells as described earlier in the presence of 5 μM E64d and pepstatin A, two inhibitors of lysosomal cathepsins that abrogate autophagosome maturation into autolysosomes (Figure 3, E–G). On stimulation of protein aggregation, addition of these inhibitors further enhanced the already elevated level of LC3-II (see Western blots and quantifications), indicating that treatment by amino acid analogue or MG132 and overexpression of HspB5R120G, SOD1G85R, or SOD1G93A activated the autophagic flux.
FIGURE 3:
Amino acid analogue or MG132 treatment and overexpression of mutated forms of HspB5 and SOD1 increase autophagic flux. HeLa cells were either nontreated (NT) or subjected to azetidine or canavanine (5 or 15 mM) treatment (A) or MG132 (0.5–5 μM) treatment (B) for 6 h. (C, D) Cells were left untreated (NT) or transiently transfected with control plasmids (Cont) or plasmids expressing wild-type HspB5 and its mutated form, HspB5R120G (C), or wild-type SOD1-EGFP and its mutated forms, SOD1G93A-EGFP and SOD1G85R-EGFP (D). (E–G) HeLa cells were subjected to the same treatments as described in the presence of 5 μM cathepsin inhibitors pepstatin A (PepA) and E64d. The inhibitors were added concomitantly to the treatment or just after transfection and were not removed until protein extraction. Total protein extracts were prepared 16 h after treatment and 24 h after transfection and subjected to SDS–PAGE. Immunoblots were probed with antibodies against LC3-I and -II, HspB5, EGFP, and actin (as a loading control). The histograms show LC3-II/actin ratios, which were calculated from quantifications of LC3-II and actin bands of Western blots by ImageJ (n = 3; ratio was set at 1.0 for NT or Cont-transfected cells). Results are representative of three independent experiments.
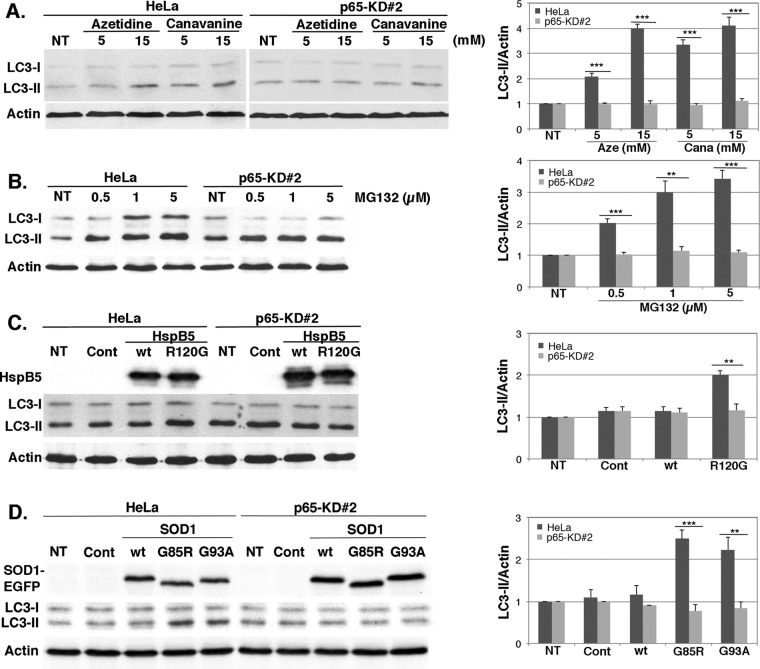
We next determined whether this increased autophagic flux upon protein aggregation stresses was NFκB dependent, by quantifying autophagic activity in control HeLa cells and in HeLa cells depleted of the p65 subunit of NFκB (p65-KD#2). We observed that in nontreated p65-KD#2 cells, the LC3-II level was very slightly increased, as observed in a previous study in which we demonstrated a minor role of NFκB in autophagosome maturation (Nivon et al., 2012). However, those cells were still able to induce autophagy, for example, after TNFα treatment (Nivon et al., 2009). After azetidine, canavanine (Figure 4A), and MG132 treatment (Figure 4B), LC3-II level increased ∼3-fold in HeLa cells but not in p65-depleted cells (p65-KD#2). Similarly, expression of HspB5R120G (Figure 4C) or of SOD1G85R and SOD1G93A (Figure 4D) induced a 2- to 2.5-fold increase of LC3-II levels in control HeLa cells but not in p65-KD#2 cells subjected to the same treatment. Therefore our results indicate that NFκB is a major activator of autophagic flux in response to diverse protein aggregation stresses.
FIGURE 4:
p65 depletion alters autophagy induction by protein aggregation stresses. Control (HeLa) and p65-depleted cell lines (p65-KD#2) were left untreated or subjected to amino acid analogue (azetidine and canavanine; A) or MG132 treatment (B) as described in Figure 3. (C, D) Cell lines were untreated (NT) or transiently transfected with control plasmids (Cont) or plasmid expressing wild-type or mutated forms of HspB5 (C) and SOD1-EGFP (D) as described in Figure 3. Total protein extracts were analyzed in immunoblots probed with LC3, HSPB5, EGFP, and actin antibodies. The histograms show LC3-II/Actin ratios calculated as described in Figure 3. Results are representative of four independent experiments.
NFκB-dependent autophagy is involved in the clearance of protein aggregates generated by treatment with amino acid analogues or MG132 or by expression of HspB5 and SOD1 mutants
Autophagy is a process involved in PQC via degradation of protein aggregates (Sridhar et al., 2012). We previously reported that NFκB-dependent autophagy helps the cell to recover from thermal injury by clearing protein aggregates. We therefore analyzed the protein aggregation status in control and p65-depleted HeLa cells subjected to various protein aggregation stresses (Figure 5). Cell lines were subjected to amino acid analogue treatment, and protein aggregation was monitored by immunofluorescence and analysis of multiubiquitinated protein labeling (Figure 5A). In untreated control and p65-depleted HeLa cells (NT), labeling of multiubiquitinated proteins was faint and diffuse into the cytoplasm, although some rare and small cytoplasmic inclusions could be detected in p65-deficient cells. This is consistent with the fact that NFκB has a minor role in basal protein quality control by modulating autophagosome maturation, as we previously demonstrated (Nivon et al., 2012). After 15 mM azetidine or canavanine treatment, we could distinguish some small inclusions in respectively 31 and 38% of control HeLa cells, with rare (1.8 and 3.8%) aggregates of large size (Figure 5A). In contrast, in p65-depleted cells, those inclusions were more abundant and larger. Indeed 58–62% of the cells contained aggregates, with aggregates >3 μm in 7% of azetidine-treated and 14% of canavanine-treated cells. We made similar observations in cells treated with MG132 (Figure 5B), in which we distinguished inclusions in 38 and 70% of control HeLa cells treated with 2.5 or 5 μM MG132, respectively. This proportion of aggregate-containing cells increased to 70% (2.5 μM) or 98% (5 μM) in NFκB-deficient cells, with also an increased proportion of aggregates of larger sizes. Those large inclusions colocalized with pericentrin, indicating the formation of an aggresome-like structure. After cell transfection with pcDNA3-αBCWT (HspB5wt), we observed a diffuse distribution of HspB5 into the cytoplasm, with the appearance of denser structures in only 7.5% of p65-depleted cells. Of interest, expression of the R120G mutated form of HspB5 induced the formation of small inclusions in only 41% of the control HeLa cells, with only 6.5% of the cells containing large inclusions. In contrast, this mutated protein induced the formation of aggresome-like structures in nearly every p65-deficient cell, with 50% of the cells containing large inclusions (Figure 5C). Finally, the expression of SOD1G93A and SOD1G85R induced mostly the formation of small cytoplasmic inclusions in respectively 15 and 25% of control HeLa cells (Figure 5D). However, inclusions were detected in 45 and 70%, of p65-deficient cells expressing SOD1G93A and SOD1G85R rather than the diffuse and widespread fluorescence observed in control or p65-deficient cells expressing SOD1wt. In addition, the percentage of cells containing large inclusions increased approximately threefold in the p65-KD#2 cell line. Taken together, our observations indicate that NFκB-deficient cells have impaired autophagy induction in response to aggregation stresses, coinciding with altered PQC and clearance of protein aggregates.
FIGURE 5:
p65-deficient cells show increased protein aggregation in response to various protein aggregation stresses. Control (HeLa) and p65-depleted cell lines (p65-KD#2) were either untreated (NT) or treated for 6 h with 15 mM azetidine (Aze) or canavanine (Cana; A) or with 2.5 and 5 μM MG132 (B). (C, D) Cell lines were untreated or transiently transfected with plasmids expressing HspB5wt and HspB5R120G (C) or SOD1wt-EGFP, SOD1G93A-EGFP, and SOD1G85R-EGFP (D). Immunofluorescence analyses were performed 16 h after treatment or 48 h after transfection. Cells were fixed, permeabilized, and stained with multiubiquitin (red staining) and pericentrin (green staining) antibodies (A, B) or with HspB5 antibody (red staining; C). Fluorescent antibodies, SOD1-EGFP proteins, and nuclei stained with Hoechst (blue staining) were visualized under a fluorescence microscope. Percentage of cells containing aggregates larger or smaller than 3 μm is shown in the histograms (mean ± SD); n = 3.
NFκB modulates BAG3 and HspB8 expression in response to various protein aggregation stresses
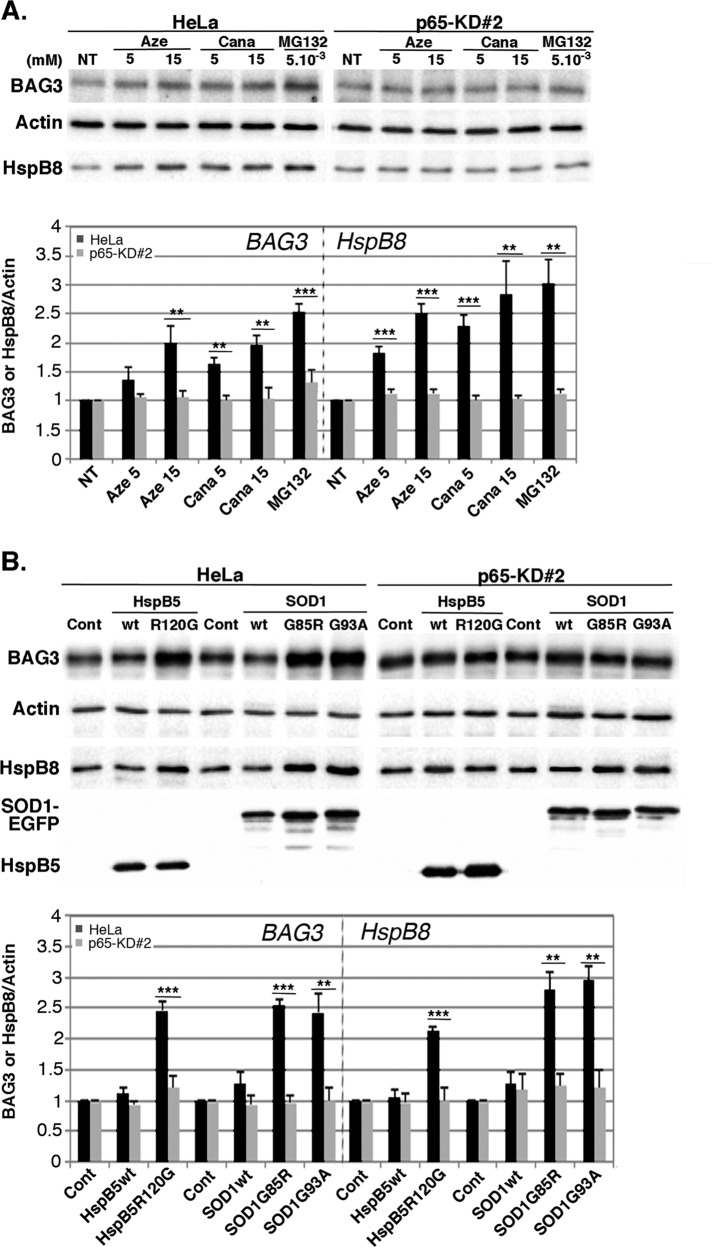
Studies have described the role of the BAG3-HspB8 complex in the selective removal of misfolded proteins by autophagy (Carra et al., 2008a, b; Carra, 2009). Moreover, we demonstrated in a previous work that upon heat shock, NFκB modulates BAG3 and HspB8 levels (Nivon et al., 2012). We thus wondered whether other types of protein aggregation stress could act in the same way. We quantified the level of BAG3 and HspB8 proteins in control and p65-depleted HeLa cells subjected to azetidine, canavanine, and MG132 treatment (Figure 6A). We observed 1.6- to 3-fold increases of BAG3 and HspB8 levels (Figure 6A) in control cells treated with amino acid analogues or proteasome inhibitor. This increase was strongly reduced in p65-depleted cells. Of interest, the expression of HspB5R120G mutant or SOD1G85R and SOD1G93A also increased BAG3 and HspB8 levels (two- to threefold) in control HeLa cells (Figure 6B), whereas expressions of their wild-type counterparts had no effect. In contrast, BAG3 and HspB8 levels were steady in p65-depleted cells expressing HspB5 and SOD1 mutated forms (Figure 6B). Our results thus indicate that protein aggregation stresses activate NFκB, which in turn enhances BAG3 and HspB8 expression, two proteins known to be involved in the stimulation of selective autophagy.
FIGURE 6:
NFκB up-regulates BAG3 and HspB8 expression after various protein aggregation stresses. (A) Control (HeLa) and p65-depleted (p65-KD#2) cell lines were either untreated (NT) or treated with 5 and 15 mM azetidine (Aze) and canavanine (Cana) or 5 μM MG132 for 6 h. At 16 h after treatments, total protein extracts were prepared and analyzed by immunoblots probed with antibodies against BAG3, HspB8, and actin (as a loading control). (B) As in A, but with HeLa and p65-KD#2 cell lines transiently transfected with control plasmids (Cont) or plasmids expressing HspB5wt, HspB5R120G, SOD1wt-EGFP, SOD1G93A-EGFP, or SOD1G85R-EGFP. Analysis was performed 35 h posttransfection. Expression of HspB5 and SOD1-EGFP was checked by probing immunoblots with anti-HspB5 and anti-EGFP antibodies. BAG3 and HspB8 levels were quantified; the densitometric measurements were normalized to the corresponding actin bands, and ratios were calculated between HeLa or p65-KD#2-treated cells vs. nontreated counterparts and are reported in the graphs (n = 3).
HspB8 and BAG3 colocalize with the protein aggregates generated by amino acid analogues, MG132 treatment, and expression of HspB5 and SOD1 mutants
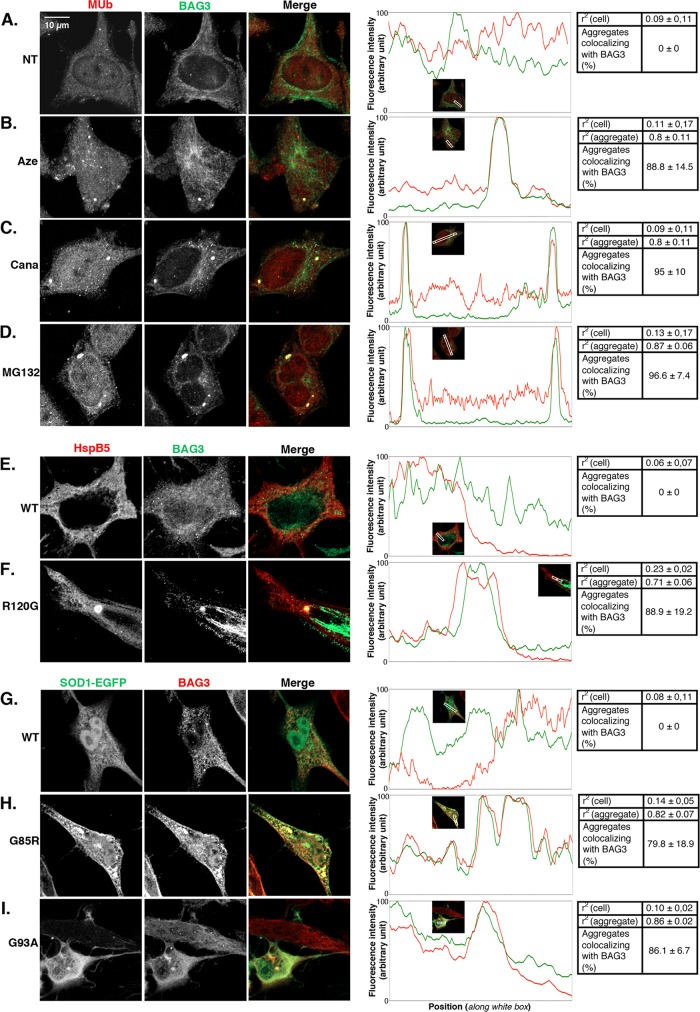
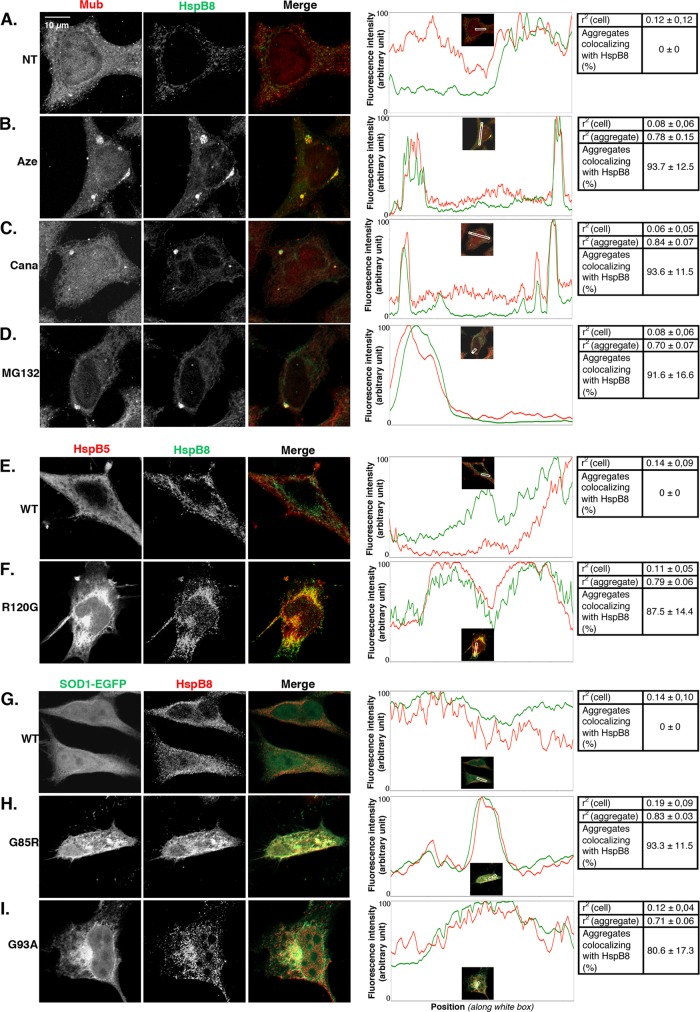
Because BAG3 and HspB8 expression is up-regulated via NFκB upon proteotoxicity, we next asked whether they could localize to protein aggregates. We analyzed BAG3 and HspB8 localization by confocal immunofluorescence in untreated cells and cells submitted to azetidine, canavanine, and MG132 treatment (Figures 7 and 8, A–D). We observed a diffuse cytoplasmic pattern of expression of BAG3 and multiubiquitinated proteins in nontreated cells, with no colocalization (Figure 7A). In contrast, as previously observed (Figure 1), amino acid analogue and MG132 treatment induced the formation of cytosolic granules containing multiubiquitinated proteins. Of interest, these aggregates colocalized with BAG3, as shown by nearly total overlay of BAG3 and multiubiquitin fluorescence peaks corresponding to aggregates (Figure 7, B–D). In addition, Pearson’s r2 in aggregate-containing portions of the cells was ∼0.8–0.9 (Figure 7, B–D). In contrast, r2 was 0.09 or 0.11 in nontreated cells or in portions of cells devoid of aggregates. Moreover, we found that 88–96% of the multiubiquitinated aggregates detected after amino acid analogue or MG132 treatment colocalized with BAG3 (Figure 7, B–D). As for HspB8 (Figure 8), its distribution was rather diffuse in the cytoplasm and less abundant in the nucleus; moreover, it did not colocalize with multiubiquitin in nontreated cells (Figure 8A, r2 = 0.12). However, upon amino acid analogue and MG132 treatment, HspB8 redistributed to multiubiquitinated granules, as evidenced by channel overlays, fluorescence intensity graphs, and r2 = 0.7–0.84. In addition, HspB8 colocalized with >90% of the multiubiquitinated aggregates detected (Figure 8, B–D). pcDNA3-HspB5WT-transfected cells (HspB5wt) showed a homogeneous and diffuse cytoplasmic localization of HspB5 (Figures 7E and 8E); moreover BAG3 (Figure 7E) and HspB8 (Figure 8E) distribution in those cells was similar to the that in nontreated cells (NT, Figures 7A and 8A), with no obvious colocalization of BAG3 or HspB8 with HspB5 (r2 < 0.14; see respective tables). In contrast, the expression of HspB5R120G generated aggregates colocalizing with BAG3 (Figure 7F, r2 = 0.71) and HspB8 (Figure 8F, r2 = 0.79) proteins. In addition, this colocalization was observed in ∼88% of the aggregates detected (Figures 7F and 8F). We made similar observations after expression of SOD1 protein in HeLa cells. BAG3 (Figure 7G) and HspB8 (Figure 8G) localization was not modified by the expression of wild-type SOD1. In contrast, the proteins predominantly relocalized to the inclusions generated by the expression of SOD1G85R-EGFP (Figures 7H and 8H) and SOD1G93A-EGFP (Figures 7I and 8I). Indeed, ∼80–93% of the SOD1G85R and SOD1G93A aggregates detected colocalized with BAG3 or HspB8 proteins (r2 = 0.71–0.86, in comparison to r2 < 0.14 in wtSOD1-expressing cells; see respective tables). Hence our results indicate that BAG3 and HspB8 redistribute toward a vast majority of the protein aggregates generated by diverse proteotoxic stresses.
FIGURE 7:
BAG3 colocalizes with protein aggregates induced by various aggregation stresses. (A–D) HeLa cells were either nontreated (A) or treated with 15 mM azetidine (B) and canavanine (C) or with 5 μM MG132 (D) for 6 h. At 16 h after treatment, cells were fixed and analyzed by confocal microscopy for BAG3 and multiubiquitin localization. (E–I) Cells were transiently transfected with plasmids expressing HspB5wt (E) or HspB5R120G (F) or SOD1 wt-EGFP (G), SOD1G85R-EGFP (H), and SOD1G93A-EGFP (I). At 48 h after transfection, cells were fixed and analyzed by confocal microscopy for BAG3, HspB5, and SOD1-EGFP localization. Single channels and channel overlays (merge) are shown. The intensity of red and green fluorescence measured along portions (white lines in inset pictures) that cross over aggregates is plotted for each condition. Each table indicates the r2 and the percentage of aggregates colocalizing with BAG3, where r2 is the mean of the squares of Pearson’s correlation coefficient r (mean ± SD) in aggregate-containing and random aggregate-devoid portions of the cells; a value close to 0 indicates a lack of colocalization, and a value close to 1 indicates complete colocalization, because all values are positive. The r2 values of the aggregate-containing vs. aggregate-devoid portions are statistically different with p < 0.001. Percentage of colocalization of BAG3 with multiubiquitinated aggregates (mean ± SD) is also indicated in the tables and was determined by geometrical center–based colocalization. Results are representative of three identical experiments.
FIGURE 8:
HspB8 colocalizes with protein aggregates induced by various aggregation stresses. (A–I) Same as in Figure 7, but cells were analyzed by confocal microscopy for multiubiquitin, HspB5, SOD1-EGFP, and HspB8 localization.
Taken together, our results demonstrate that NFκB is a central sensor of protein aggregation and can modulate protein quality control by increasing the expression of BAG3 and HspB8, which redistribute to protein aggregates and favor their selective autophagic clearance.
DISCUSSION
Unchecked protein misfolding and/or aggregation are the root cause of many diverse protein conformational diseases (Herczenik and Gebbink, 2008). Protein aggregation can result in various structural appearances, ranging from unordered amorphous aggregates to highly ordered fibrils called amyloids. Here we studied the involvement of NFκB in the cell response to protein aggregation stress, an event also described as proteotoxicity (Hightower, 1991). To this end, we performed treatments with structural analogues of amino acids that generate randomly abnormal proteins, which are rapidly ubiquitinated, overwhelming chaperones and the proteasome system, thus inducing the formation of cytosolic granules (Dasuri et al., 2011). We indeed observed that amino acid analogue treatment induced the formation of small, multiubiquitinated cytosolic granules but also aggresome-like structures under more drastic treatments (Figure 1). In this respect, it is interesting to note that canavanine treatment seems to induce more frequent and larger aggregates in the cells; this might be related to a better incorporation into proteins of this analogue of arginine, since cells were cultured in medium devoid of arginine but not of proline. Cell treatment by MG132 provoked increased levels of multiubiquitinated proteins and formation of ubiquitin-containing aggresomes, as previously described (Hightower, 1980; Gies et al., 2010). We also induced other kinds of proteotoxicity by expressing mutated proteins that are prone to form aggregates. We observed the formation of perinuclear aggregates after expression of the R120G mutated form of HspB5, which is in line with the description of HspB5R120G aggregation with intermediate filaments leading to aggresome formation containing the amyloid oligomer (Meehan et al., 2007; Simon et al., 2007b). Finally, we expressed the G93A and G85R mutated form of SOD1 in HeLa cells and observed cytoplasmic and perinuclear inclusions. Those aggregates are described to contain various proteins or molecular complexes, such as the copper chaperone for superoxide dismutase (CCS), Hsp70, and proteasomal proteins. Moreover, studies indicated that SOD1 aggregates might be heterogeneous, ranging from amorphous aggregates to amyloid fibrils (Matsumoto et al., 2005; Oztug Durer et al., 2009). Of interest, the aggregates formed in response to the various stresses are of different sizes. This is a quite common observation, and this variability in size and also in structure was illustrated by the following observation: depending on the stress induced (temperature, pH, pressure) but also of the intracellular environment (e.g., protein concentration), a given protein can aggregate into diverse morphological/structural and toxic states (Wang et al., 2010). Moreover, many studies show that if the clearance system, such as UPS or autophagy, is overwhelmed, the aggregated proteins accumulate and usually gather at specific sites, generating larger aggregates such as aggresomes at MTOCs to minimize their toxic effects (Garcia-Mata et al., 1999). In this study, we therefore used various treatments that allow the induction of diverse types of protein aggregation stress—random or selective aggregation of proteins, amorphous aggregates, or amyloid fibrils—and were able to demonstrate that although these aggregates are diverse in origin, structure, and size, they all trigger NFκB activation.
Of interest, MG132 is usually described as an inhibitor of the canonical activation pathway of NFκB via its inhibitory effect on IκB degradation by the proteasome (Zanotto-Filho et al., 2012). Surprisingly, we observed that MG132 treatment could activate NFκB, suggesting the involvement of an alternative activation pathway. We also demonstrated that this stimulation does not occur through the canonical NFκB transduction pathway, since, by the use of dominant-negative mutants, we observed that NFκB activation by MG132 or mutated HspB5 and SOD1 proteins is independent of IKKβ activity or IκBα phosphorylation/degradation (Figure 2). Hence our results suggest that NFκB is a central sensor of protein aggregation stresses, which is activated via a noncanonical pathway. However, we found that the intensity of NFκB activation varies depending on the type of treatment, with maximal induction after heat stress and minimal induction after overexpression of mutated proteins prone to form aggregates. One could hypothesize that heat stress or MG132 treatment affects all (heat stress) or a wide range (MG132) of proteins and induces numerous aggregates, whereas mutated proteins induce specific and less numerous aggregates. Yet this explanation might not reflect reality, since toxicity of aggregates and thus cell response to those aggregates not only depends on their quantity but also correlates with their ability to promote aberrant protein interactions. In addition, different types of aggregates can interact with and/or sequester various types of proteins; these interactions are determined by specific sequence features and/or enrichment of disordered regions of the coaggregating proteins (Olzscha et al., 2011). Nonetheless, even if current knowledge does not allow a direct comparison between aggregate sizes or levels and their capacity to stimulate NFκB activity, our data highlight a new aspect of NFκB function; indeed, NFκB might be considered as a primary regulator of protein aggregation stress response, adding another item to the growing list of stress responses in which NFκB is involved (viral infection, bacterial lipids, DNA damage, oxidative stress, ER stress, chemotherapeutic agents, etc.; Piva et al., 2006; Tam et al., 2012).
We next wondered about the consequences of protein aggregate accumulation and NFκB activation in stressed cells and first focused on autophagy. Indeed, autophagy is widely described to be involved in the clearance of protein aggregates (Martinez-Vicente and Cuervo, 2007; Nivon et al., 2009, 2012). In this study, we demonstrated that amino acid analogue treatment increased autophagic flux (Figure 3 and Supplemental Figure S1). Similarly, we observed that the autophagic flux is activated upon proteasome inhibitor MG132 treatment, as previously observed in cultured astrocytes (Janen et al., 2010). Finally, we found that overexpression of SOD1G85R, SOD1G93A, or HspB5R120G stimulated autophagic flux whereas the wild-type forms of these proteins did not. These results are consistent with a previous analysis with SOD1G93A mutant in NSC34 neuron-like mouse cells (Wei, 2014). As for HspB5R120G, our results are in line with the description of autophagy activation after HspB5R120G expression in cardiomyocytes (Tannous et al., 2008). However, they contrast with reports that autophagy is inhibited (altered autophagosome maturation) in cardiomyocytes and lens from HspB5R120G knock-in mouse models (Maloyan and Robbins, 2010; Wignes et al., 2013). This discrepancy might be explained by the duration of expression of this mutant. Indeed, the previous work examined long-term expression of HspB5R120G, whereas in the present study we performed autophagy measurements 24 h after the beginning of the transient expression of HspB5R120G. In addition, HspB5 is known to chaperone vimentin filaments, which were shown to suppress autophagy dynamics by interacting with Beclin1, a protein crucial for autophagy initiation (Mostowy, 2014). Thus it could be hypothesized that the expression of HspB5R120G, whose chaperone activity is altered, can induce the formation of aggregates with vimentin filaments and provoke the release of Beclin1, which, in turn, could stimulate autophagy. Nonetheless, in the long-term context, the accumulation of HspB5R120G aggregates could overwhelm the autophagic process or the lysosome machinery and alter the autophagy–lysosome fusion process. Next we determined that NFκB-deficient cells subjected to these various protein aggregation stresses exhibited a strong impairment of autophagy stimulation (Figure 4). Thus, even though autophagy was described to be involved in the clearance of protein aggregates generated by MG132 treatment, expression of HspB8R120G in cardiomyocytes, or expression of SOD1G85R and SOD1G93A in a mouse neuroblastoma cell line (Kabuta et al., 2006; Janen et al., 2010; Pattison et al., 2011), to our knowledge, this is the first time that connections have been made between the activation of the NFκB transcription factor by these protein aggregation stresses and the stimulation of autophagy. We thus wondered whether the clearance of protein aggregates was modulated by NFκB-dependent autophagy and analyzed protein aggregation status in NFκB-deficient cells subjected to amino acid analogues, MG132, and expression of HspB5 and SOD1 mutants. We observed a higher accumulation of protein aggregates in these cells than in control cells (Figure 5), leading to the conclusion that NFκB-dependent autophagy is involved in the clearance of various kinds of protein aggregates generated by diverse aggregation stresses.
To determine how NFκB modulated autophagy in response to proteotoxicity, we focused on the BAG3/HspB8 complex. Indeed, the BAG3 cochaperone is a molecular switch that addresses proteins to proteasomal or autophagic degradation; it also increases autophagic activity (Carra, 2009; Gamerdinger et al., 2011; Rapino et al., 2014). Moreover, BAG3 binds to chaperones such as Hsp70 and HspB8, which allow recognition and binding to misfolded proteins (Carra et al., 2008a; Chen et al., 2013). BAG3 is also reported to 1) facilitate the retrograde transport of misfolded proteins to the MTOC, 2) form aggresomes (Gamerdinger et al., 2009), and 3) promote both autophagy induction via its binding to Atg3 (Chen et al., 2013) and selective autophagy through its interaction with p62 (Gamerdinger et al., 2009). Accordingly, the BAG3/HspB8 complex selectively activates autophagic removal of aggregated proteins such as Htt43Q and SOD1G93A (Carra et al., 2008a; Crippa et al., 2013). Of interest, in humans, BAG3 mutations induce severe myofibrillar myopathy associated with cardiomyopathy and neuropathy, whereas HspB8 mutations are associated with neuropathy (Fontaine et al., 2006; Selcen et al., 2009; Konersman et al., 2015). Finally, we previously demonstrated that NFκB activates autophagy in response to heat shock treatments via the up-regulation of BAG3 and HspB8 expression. In this study, we observed that amino acid analogue or MG132 treatment, as well as expression of HspB5R120G, SOD1G85R, and SOD1G93A, induced an increase of BAG3 and HspB8 levels (Figure 6). Hence our results corroborate the increased transcription of bag3 and hspb8 observed after MG132 treatment (Du et al., 2009; Minoia et al., 2014) and the increased levels of BAG3 and HspB8 detected in muscles of SOD1G93A knock-in mouse models (Crippa et al., 2013). Of importance, to our knowledge, our study reports for the first time a modulation of BAG3 and HspB8 levels after protein aggregation stresses such as amino acid analogue treatment or expression of the R120G mutated form of HspB5. Of interest in this context, it was reported that BAG3 interacts with HspB5R120G and suppresses its aggregation and toxicity (Hishiya et al., 2011). Similarly, transfection of HspB8 in cardiomyocytes blocks the amyloid oligomer formed by HspB5R120G (Sanbe et al., 2007). We next investigated whether increased BAG3 and HspB8 levels induced by proteotoxicity were dependent on NFκB activity, similar to what we found after heat shock treatment (Nivon et al., 2012). Impaired stimulation of BAG3 and HspB8 expression was observed after protein aggregation stresses in p65-depleted cells, indicating that NFκB is an important regulator of BAG3 and HspB8 expression in response to amino acid analogue and MG132 treatment or expression of HspB5R120G, SOD1G85R, and SOD1G93A. As for MG132 treatment, these results are consistent with a previous study reporting that the heat shock response, the unfolded protein response, and the NFκB signaling pathway all participate in BAG3 up-regulation upon proteasome inhibition (Minoia et al., 2014); indeed, we observed that the inhibition of BAG3 up-regulation in NFκB-deficient cells is not complete, suggesting the involvement of other regulators. Taken together, our data indicate that NFκB deficiency impairs BAG3/HspB8 up-regulation after various protein aggregation stresses, which is associated with a defective autophagic process and an accumulation of protein aggregates. Of interest, this process seems to be rather restricted to BAG3 and HspB8 partners. Indeed, we could not detect any modulation of the transcript levels of other known players in protein aggregate clearance, such as Alfy, NBR1, or optineurin, in a microarray analysis in control and p65-depleted HeLa cells submitted to heat stress, which is an inducer of the noncanonical NFκB pathway (unpublished data).
Finally, we checked whether up-regulated BAG3 and HspB8 could associate with protein aggregates and observed strong colocalization of BAG3 and HspB8 with the aggregates generated by amino acid analogue and MG132 treatment or expression of HspB5 and SOD1 mutated forms. These observation are in line with the colocalization of BAG3 with multiubiquitinated aggregates described upon MG132 treatment (Minoia et al., 2014), the colocalization of overexpressed BAG3 and HspB5R120G in HEK293 cells (Hishiya et al., 2011), the association of BAG3 and SOD1G85R aggregates (Gamerdinger et al., 2011), and the colocalization of HspB8 and SOD1G93A in mouse models (Crippa et al., 2010), indicating that the NFκB-dependent up-regulation of BAG3 and HspB8 expression might facilitate the recruitment of BAG3/HspB8 to protein aggregates and strengthen the role of BAG3 and HspB8 in the selective removal of aggregates via chaperone-associated selective autophagy.
Taken together, our results further highlight the critical and central role played by NFκB transcription factor in protein quality control. Our data indicate that NFκB is a general sensor of protein aggregation stress that can be activated by random or selective aggregation of proteins, amorphous aggregates, or amyloid fibrils. Its activation induces an on-demand compensatory mechanism, consisting of the up-regulation of the BAG3-HspB8 complex, which in turn associates with protein aggregates and stimulates selective autophagy. This mechanism allows the clearance of toxic protein aggregates and thus helps the cell to recover from proteotoxicity. Hence this molecular pathway may impact therapies for protein conformational diseases, notably cardiac dysfunction, cataract formation, or neurodegenerative diseases associated with proteasome impairment, myofibrillar myopathies associated with HspB5 mutations, or familial ALS linked to SOD1 mutations.
MATERIALS AND METHODS
Cell culture and cell lines
HeLa cells were grown at 37°C and 5% CO2 in DMEM supplemented with 10% decomplemented fetal calf serum (FCS), 1 μg/ml Fungizone, and 50 U/ml penicillin/streptomycin (Nivon et al., 2009). For amino acid analogue treatment, cells were first cultivated for 1 h in DMEM without l-arginine (PAN Biotec, Aidenbach, Germany) supplemented with 10% dialyzed FCS. Then cells were incubated with 5–15 mM of l-canavanine sulfate (abbreviated canavanine) or l-azetidine-2-carboxylic acid (abbreviated azetidine). For E64d and pepstatin A studies, 5 μM inhibitor was added concomitantly with the treatment or just after transfection and not removed until cellular protein extraction. HeLa cells expressing short hairpin RNA (shRNA) directed against the p65 subunit of NF-κB (p65-KD#2) have been described (Nivon et al., 2009). It was shown in previous studies that the p65-KD#2 cell line behaves like other p65-deficient clones in terms of growth, autophagic activity, and response to heat stress; similarly, control HeLa cells used in this study had the same behavior as HeLa cells expressing single-mutated or scrambled-mutated shRNA directed against p65 (Nivon et al., 2009, 2012). Stable HeLa transfectants expressing GFP-LC3 have been described (Bampton et al., 2005).
Reagents and plasmids
E64d, pepstatin A, l-azetidine-2-carboxylic acid, l-canavanine sulfate, Hoechst 33258, and Triton X-100 were from Sigma-Aldrich (Saint Quentin Fallavier, France). MG132 was from Merck (Fontenay sous Bois, France). Mouse monoclonal antibodies used were directed against actin, clone C4 (Millipore, Molsheim, France); p62/SQSTM1, clone 3/p62 lck ligand (Becton Dickinson Biosciences, Le-Pont-de-Claix, France); multiubiquitin, clone FK1 (MBL, Woburn, MA); EGFP, clone 1GFP-2A5 (Euromedex, Souffelweyersheim, France); HspB8, clone 3C5 (Abnova, Walnut, CA); and alphaB crystalline/HspB5, clone 1B6.1-3G4 (Enzo, Villeurbanne, France). Anti-LC3 (L7543; Sigma-Aldrich), BAG3 (Ab47124; Abcam, Paris, France), HspB8 (PAB15424; Abnova), and anti-pericentrin (Ab4448; Abcam) were rabbit polyclonal antibodies. pcDNA3-αBCWT and pcDNA3-αBCR120G are expression vectors for wild-type HspB5 protein or its R120G mutated form (Simon et al., 2007b). pCDNA3.1(+) (Invitrogen, Thermo Fisher, Illkirch, France) was used as the empty plasmid control for those constructs. pEGFP-SOD1WT, pEGFP-SOD1G85R, and pEGFPSOD1G93A are expression vectors for wild-type hSOD1-EGFP and its mutated forms hSOD1G85R-EGFP and hSOD1G93A-EGFP. pEGFP-N1 (Clontech, Paris, France) was used as the empty plasmid control for those constructs (Turner et al., 2005). pLXSN-IκBαM plasmid allows the expression of a dominant-negative mutant of IκBα in which Ser-32, Ser-36, Ser-283, Ser-288, and Ser-293 phosphorylation sites are mutated to alanine (Kretz-Remy et al., 2001). pRK5-IKKß(K44A)-C-Flag plasmid encodes a dominant-negative mutant of IKKβ (Kretz-Remy et al., 2001). pNFκBluc was from Clontech.
Transfection
Cells were seeded the day before transfection at a density of 5 × 105 cells/60-mm dish. They were then transfected with 5 μg of the desired plasmid according to the ExGen 500 (Euromedex) reagent procedure.
Luciferase assay
Stable transfectant HeLa cells expressing pNFκBluc (Clontech) were submitted to various treatments or transfections. Then they were resuspended into phosphate-buffered saline (PBS), and luciferase activity was quantified using the Steady-Glo luciferase assay system according to the manufacturer’s instructions (Promega, Charbonnières-les-Bains, France). The intensity of light produced was quantified with a Victor3 Luminometer (PerkinElmer, Courtaboeuf, France). The related light units produced were reported to 50 μg of total cellular proteins
Gel electrophoresis and immunoblotting
Briefly, 10 μg of total protein extracts was loaded onto SDS–PAGE. After electrophoresis, proteins were transferred onto a Protran BA85 nitrocellulose membrane (Whatman, Perkin Elmer). Blots were hybridized with primary antibodies and horseradish peroxidase–conjugated secondary antibodies (170-65-16 and 170-6515; Bio-Rad, Marnes-la-Coquette, France) and revealed with enhanced chemiluminescence (ECL) blotting detection reagents (Amersham, Velizy-Villacoublay, France) or Clarity Western ECL substrate (Bio-Rad). Western blot imaging was performed with Chemidoc MP (Bio-Rad) based on charge-coupled device detection technology. Image capture and analysis of Western blots were controlled by Image Lab software. Quantification of Western blots was performed by ImageJ software (National Institutes of Health, Bethesda, MD).
Filter trap assay
SDS-insoluble aggregates were analyzed by filter trap analysis as previously described (Nivon et al., 2012). Briefly, cells were scraped in 2% SDS-FTA buffer (FTA: 150 mM NaCl, 50 mM dithiothreitol, 10 mM Tris-HCl, pH 8). Next samples were homogenized by three passages through a 25-gauge needle. The 2.5-μg protein extracts were diluted by a factor of 2–8 (1, 1:2, 1:4, and 1:8) and applied with mild suction into a slot blot apparatus onto a 0.22-μm Protran BA83 nitrocellulose membrane (Schleicher and Schuell, Dutscher, Brumath, France) prewashed with 0.1% SDS-FTA buffer. Then the membrane was washed with 0.1% SDS-FTA buffer and 0.1% Tween-TBS buffer (TBS: 20 mM Tris-HCl, pH 7.6, 137 mM NaCl) and processed for immunoblotting. Equivalent expression levels of wt and mutant constructs for HspB5 and SOD1 were checked on Western blots of total cell lysates in parallel experiments.
Fluorescence microscopy analysis
Nontreated, treated, or transfected cells were grown on glass coverslips, followed by various recovery periods, rinsed in cold PBS, and either fixed for 30 min in 2% paraformaldehyde, pH 7.4 (EGFP-expressing cells), or fixed for 10 min with cold methanol and permeabilized with 0.1% Triton X-100. Cells were stained with primary antibodies (MultiUb, HspB5, HspB8, BAG3, pericentrin, LC3) and fluorescent secondary antibodies (goat anti-mouse Alexa Fluor 568 or 488 nm and goat anti-rabbit Alexa Fluor 488 or 568 nm; Life Technologies, Thermo Fisher). Hoechst 33258 reagent was used to stain nuclei (5 min, 1 ng/ml). For quantification of aggregation, cells were photographed with a Zeiss Axiovert 200M or a Zeiss (Marly-le-Roi, France) Axio Imager Z1 photomicroscope (40× objective). Images were digitized with a camera (CoolSNAP HQ2; Roper Scientific, Lisses, France) and acquired with the Metavue Imaging system. The number of cells containing aggregates smaller or larger than 3 μm was quantified three times on 100 cells. For colocalization experiments, z-stacks were performed with a confocal Zeiss LSM 510 META (63× objective). Colocalization was quantified by measuring on z-stacks Pearson’s r (all r values were positive) and geometrical center–based colocalization with Fiji’s JACoP plug-in on ∼20 cells for each experimental condition.
Statistics
The unpaired Student’s t test was applied to compare results between two sample groups. Differences between groups were considered statistically significant for p < 0.05. *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Material
Acknowledgments
We thank Dominique Guillet for excellent technical assistance, Ludivine Walter for critical reading of the manuscript, Charlotte Scholtès for help with confocal microscopy, and Berangère Pinan-Lucarré and Julien Falk for help with colocalization analysis. This work was supported by the Ligue contre le Cancer, comité du Rhône; the Ligue contre le Cancer, comité de Savoie; the Bonus Qualité Recherche from Université Claude Bernard Lyon 1; the Centre National de la Recherche Scientifique (C.K.); and the Millennium Institute P09-015-F, FONDAP Program 15150012 (C.H.). M.N. was supported by a doctoral fellowship from the French Department of research and the Fondation pour la Recherche Médicale. S.S. was supported by a postdoctoral fellowship from the Association Française contre les Myopathies/Téléthon.
Abbreviations used:
- Alfy
autophagy-linked FYVE protein
- ALS
amyotrophic lateral sclerosis
- CCD
charge-coupled device
- CCS
copper chaperone for superoxyde dismutase
- EGFP
enhanced GFP
- ER
endoplasmic reticulum
- FCS
fetal calf serum
- FTA
filter trap assay
- GFP
green fluorescent protein
- IkB
Inhibitor of kappa B
- IKK
I kB kinase
- MTOC
microtubule-organizing center
- NBR1
neighbor of BRCA1
- NFkB
nuclear factor kappa B
- PBS
phosphate-buffered saline
- PQC
protein quality control
- SOD1
Cu, Zn-superoxide dismutase
- TNF
tumor necrosis factor
- UPS
ubiquitin proteasome system.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-12-0835) on April 13, 2016.
REFERENCES
- Bampton ET, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- Behrends C, Fulda S. Receptor proteins in selective autophagy. Int J Cell Biol. 2012;2012:673290. doi: 10.1155/2012/673290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S. The stress-inducible HspB8-Bag3 complex induces the eIF2alpha kinase pathway: implications for protein quality control and viral factory degradation. Autophagy. 2009;5:428–429. doi: 10.4161/auto.5.3.7894. [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Lambert H, Landry J. HspB8 chaperone activity toward poly(Q)-containing proteins depends on its association with Bag3, a stimulator of macroautophagy. J Biol Chem. 2008a;283:1437–1444. doi: 10.1074/jbc.M706304200. [DOI] [PubMed] [Google Scholar]
- Carra S, Seguin SJ, Landry J. HspB8 and Bag3: a new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy. 2008b;4:237–239. doi: 10.4161/auto.5407. [DOI] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang LN, Cheng L, Tu S, Guo SJ, Le HY, Xiong Q, Mo R, Li CY, Jeong JS, et al. Bcl2-associated athanogene 3 interactome analysis reveals a new role in modulating proteasome activity. Mol Cell Proteomics. 2013;12:2804–2819. doi: 10.1074/mcp.M112.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa V, Boncoraglio A, Galbiati M, Aggarwal T, Rusmini P, Giorgetti E, Cristofani R, Carra S, Pennuto M, Poletti A. Differential autophagy power in the spinal cord and muscle of transgenic ALS mice. Front Cell Neurosci. 2013;7:234. doi: 10.3389/fncel.2013.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E, Marino M, Carra S, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- Dahlmann B. Role of proteasomes in disease. BMC Biochem. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2091-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasuri K, Ebenezer PJ, Uranga RM, Gavilan E, Zhang L, Fernandez-Kim SO, Bruce-Keller AJ, Keller JN. Amino acid analog toxicity in primary rat neuronal and astrocyte cultures: implications for protein misfolding and TDP-43 regulation. J Neurosci Res. 2011;89:1471–1477. doi: 10.1002/jnr.22677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZX, Zhang HY, Meng X, Gao YY, Zou RL, Liu BQ, Guan Y, Wang HQ. Proteasome inhibitor MG132 induces BAG3 expression through activation of heat shock factor 1. J Cell Physiol. 2009;218:631–637. doi: 10.1002/jcp.21634. [DOI] [PubMed] [Google Scholar]
- Edington BV, Whelan SA, Hightower LE. Inhibition of heat shock (stress) protein induction by deuterium oxide and glycerol: additional support for the abnormal protein hypothesis of induction. J Cell Physiol. 1989;139:219–228. doi: 10.1002/jcp.1041390202. [DOI] [PubMed] [Google Scholar]
- Elliott JL, Der Perng M, Prescott AR, Jansen KA, Koenderink GH, Quinlan RA. The specificity of the interaction between alphaB-crystallin and desmin filaments and its impact on filament aggregation and cell viability. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120375. doi: 10.1098/rstb.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine JM, Sun X, Hoppe AD, Simon S, Vicart P, Welsh MJ, Benndorf R. Abnormal small heat shock protein interactions involving neuropathy-associated HSP22 (HSPB8) mutants. FASEB J. 2006;20:2168–2170. doi: 10.1096/fj.06-5911fje. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R, Bebok Z, Sorscher EJ, Sztul ES. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gies E, Wilde I, Winget JM, Brack M, Rotblat B, Novoa CA, Balgi AD, Sorensen PH, Roberge M, Mayor T. Niclosamide prevents the formation of large ubiquitin-containing aggregates caused by proteasome inhibition. PLoS One. 2010;5:e14410. doi: 10.1371/journal.pone.0014410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Herczenik E, Gebbink MF. Molecular and cellular aspects of protein misfolding and disease. FASEB J. 2008;22:2115–2133. doi: 10.1096/fj.07-099671. [DOI] [PubMed] [Google Scholar]
- Hightower LE. Cultured animal cells exposed to amino acid analogues or puromycin rapidly synthesize several polypeptides. J Cell Physiol. 1980;102:407–427. doi: 10.1002/jcp.1041020315. [DOI] [PubMed] [Google Scholar]
- Hightower LE. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell. 1991;66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- Hishiya A, Salman MN, Carra S, Kampinga HH, Takayama S. BAG3 directly interacts with mutated alphaB-crystallin to suppress its aggregation and toxicity. PLoS One. 2011;6:e16828. doi: 10.1371/journal.pone.0016828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WM, Klaips CL, Serio TR. Defining the limits: Protein aggregation and toxicity in vivo. Crit Rev Biochem Mol Biol. 2014;49:294–303. doi: 10.3109/10409238.2014.914151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janen SB, Chaachouay H, Richter-Landsberg C. Autophagy is activated by proteasomal inhibition and involved in aggresome clearance in cultured astrocytes. Glia. 2010;58:1766–1774. doi: 10.1002/glia.21047. [DOI] [PubMed] [Google Scholar]
- Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee H, Lee JH, Kwon DY, Genovesio A, Fenistein D, Ogier A, Brondani V, Grailhe R. Dimerization, oligomerization, and aggregation of human amyotrophic lateral sclerosis copper/zinc superoxide dismutase 1 protein mutant forms in live cells. J Biol Chem. 2014;289:15094–15103. doi: 10.1074/jbc.M113.542613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konersman CG, Bordini BJ, Scharer G, Lawlor MW, Zangwill S, Southern JF, Amos L, Geddes GC, Kliegman R, Collins MP. BAG3 myofibrillar myopathy presenting with cardiomyopathy. Neuromuscul Disord. 2015;25:418–422. doi: 10.1016/j.nmd.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Kretz-Remy C, Bates EE, Arrigo AP. Amino acid analogs activate NF-kappaB through redox-dependent IkappaB-alpha degradation by the proteasome without apparent IkappaB-alpha phosphorylation. Consequence on HIV-1 long terminal repeat activation. J Biol Chem. 1998;273:3180–3191. doi: 10.1074/jbc.273.6.3180. [DOI] [PubMed] [Google Scholar]
- Kretz-Remy C, Munsch B, Arrigo AP. NFkappa B-dependent transcriptional activation during heat shock recovery. Thermolability of the NF-kappaB.Ikappa B complex. J Biol Chem. 2001;276:43723–43733. doi: 10.1074/jbc.M010821200. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Li N, Karin M. Ionizing radiation and short wavelength UV activate NF-kappaB through two distinct mechanisms. Proc Natl Acad Sci USA. 1998;95:13012–13017. doi: 10.1073/pnas.95.22.13012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloyan A, Robbins J. Autophagy in desmin-related cardiomyopathy: thoughts at the halfway point. Autophagy. 2010;6:665–666. doi: 10.1161/CIRCRESAHA.109.212639. [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- Matsumoto G, Stojanovic A, Holmberg CI, Kim S, Morimoto RI. Structural properties and neuronal toxicity of amyotrophic lateral sclerosis-associated Cu/Zn superoxide dismutase 1 aggregates. J Cell Biol. 2005;171:75–85. doi: 10.1083/jcb.200504050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan S, Knowles TP, Baldwin AJ, Smith JF, Squires AM, Clements P, Treweek TM, Ecroyd H, Tartaglia GG, Vendruscolo M, et al. Characterisation of amyloid fibril formation by small heat-shock chaperone proteins human alphaA-, alphaB- and R120G alphaB-crystallins. J Mol Biol. 2007;372:470–484. doi: 10.1016/j.jmb.2007.06.060. [DOI] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview. Am J Physiol Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- Minoia M, Boncoraglio A, Vinet J, Morelli FF, Brunsting JF, Poletti A, Krom S, Reits E, Kampinga HH, Carra S. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: implications for a proteasome-to-autophagy switch. Autophagy. 2014;10:1603–1621. doi: 10.4161/auto.29409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S. Multiple roles of the cytoskeleton in bacterial autophagy. PLoS Pathog. 2014;10:e1004409. doi: 10.1371/journal.ppat.1004409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivon M, Abou-Samra M, Richet E, Guyot B, Arrigo AP, Kretz-Remy C. NF-kappaB regulates protein quality control after heat stress through modulation of the BAG3-HspB8 complex. J Cell Sci. 2012;125:1141–1151. doi: 10.1242/jcs.091041. [DOI] [PubMed] [Google Scholar]
- Nivon M, Richet E, Codogno P, Arrigo AP, Kretz-Remy C. Autophagy activation by NFkappaB is essential for cell survival after heat shock. Autophagy. 2009;5:766–783. doi: 10.4161/auto.8788. [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Oztug Durer ZA, Cohlberg JA, Dinh P, Padua S, Ehrenclou K, Downes S, Tan JK, Nakano Y, Bowman CJ, Hoskins JL, et al. Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One. 2009;4:e5004. doi: 10.1371/journal.pone.0005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison JS, Osinska H, Robbins J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ Res. 2011;109:151–160. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R, Belardo G, Santoro MG. NF-kappaB: a stress-regulated switch for cell survival. Antioxid Redox Signal. 2006;8:478–486. doi: 10.1089/ars.2006.8.478. [DOI] [PubMed] [Google Scholar]
- Rapino F, Jung M, Fulda S. BAG3 induction is required to mitigate proteotoxicity via selective autophagy following inhibition of constitutive protein degradation pathways. Oncogene. 2014;33:1713–1724. doi: 10.1038/onc.2013.110. [DOI] [PubMed] [Google Scholar]
- Rodgers KJ, Shiozawa N. Misincorporation of amino acid analogues into proteins by biosynthesis. Int J Biochem Cell Biol. 2008;40:1452–1466. doi: 10.1016/j.biocel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Yamauchi J, Miyamoto Y, Fujiwara Y, Murabe M, Tanoue A. Interruption of CryAB-amyloid oligomer formation by HSP22. J Biol Chem. 2007;282:555–563. doi: 10.1074/jbc.M605481200. [DOI] [PubMed] [Google Scholar]
- Selcen D, Muntoni F, Burton BK, Pegoraro E, Sewry C, Bite AV, Engel AG. Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann Neurol. 2009;65:83–89. doi: 10.1002/ana.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Coleman J, Chan E, Nicholson TP, Dai L, Sheppard PW, Patton WF. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys. 2011;60:173–185. doi: 10.1007/s12013-010-9138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Dimitrova V, Gibert B, Virot S, Mounier N, Nivon M, Kretz-Remy C, Corset V, Mehlen P, Arrigo AP. Analysis of the dominant effects mediated by wild type or R120G mutant of alphaB-crystallin (HspB5) towards Hsp27 (HspB1) PLoS One. 2013;8:e70545. doi: 10.1371/journal.pone.0070545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Fontaine JM, Martin JL, Sun X, Hoppe AD, Welsh MJ, Benndorf R, Vicart P. Myopathy-associated alphaB-crystallin mutants: abnormal phosphorylation, intracellular location, and interactions with other small heat shock proteins. J Biol Chem. 2007a;282:34276–34287. doi: 10.1074/jbc.M703267200. [DOI] [PubMed] [Google Scholar]
- Simon S, Michiel M, Skouri-Panet F, Lechaire JP, Vicart P, Tardieu A. Residue R120 is essential for the quaternary structure and functional integrity of human alphaB-crystallin. Biochemistry. 2007b;46:9605–9614. doi: 10.1021/bi7003125. [DOI] [PubMed] [Google Scholar]
- Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012;226:255–273. doi: 10.1002/path.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AB, Mercado EL, Hoffmann A, Niwa M. ER stress activates NF-kappaB by integrating functions of basal IKK activity, IRE1 and PERK. PLoS One. 2012;7:e45078. doi: 10.1371/journal.pone.0045078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BJ, Atkin JD, Farg MA, Zang DW, Rembach A, Lopes EC, Patch JD, Hill AF, Cheema SS. Impaired extracellular secretion of mutant superoxide dismutase 1 associates with neurotoxicity in familial amyotrophic lateral sclerosis. J Neurosci. 2005;25:108–117. doi: 10.1523/JNEUROSCI.4253-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicart P, Caron A, Guicheney P, Li Z, Prevost MC, Faure A, Chateau D, Chapon F, Tome F, Dupret JM, et al. A missense mutation in the alphaB-crystallin chaperone gene causes a desmin-related myopathy. Nat Genet. 1998;20:92–95. doi: 10.1038/1765. [DOI] [PubMed] [Google Scholar]
- Villella VR, Esposito S, Bruscia EM, Maiuri MC, Raia V, Kroemer G, Maiuri L. Targeting the intracellular environment in cystic fibrosis: restoring autophagy as a novel strategy to circumvent the CFTR defect. Front Pharmacol. 2013;4:1. doi: 10.3389/fphar.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Schubert D, Sawaya MR, Eisenberg D, Riek R. Multidimensional structure-activity relationship of a protein in its aggregated states. Angew Chem Int Ed Engl. 2010;49:3904–3908. doi: 10.1002/anie.201000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y. Autophagic induction of amyotrophic lateral sclerosis-linked Cu/Zn superoxide dismutase 1 G93A mutant in NSC34 cells. Neural Regen Res. 2014;9:16–24. doi: 10.4103/1673-5374.125325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wignes JA, Goldman JW, Weihl CC, Bartley MG, Andley UP. p62 expression and autophagy in alphaB-crystallin R120G mutant knock-in mouse model of hereditary cataract. Exp Eye Res. 2013;115:263–273. doi: 10.1016/j.exer.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- Wilson MI, Dooley HC, Tooze SA. WIPI2b and Atg16L1: setting the stage for autophagosome formation. Biochem Soc Trans. 2014;42:1327–1334. doi: 10.1042/BST20140177. [DOI] [PubMed] [Google Scholar]
- Wirth M, Joachim J, Tooze SA. Autophagosome formation—the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–309. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Zanotto-Filho A, Braganhol E, Battastini AM, Moreira JC. Proteasome inhibitor MG132 induces selective apoptosis in glioblastoma cells through inhibition of PI3K/Akt and NFkappaB pathways, mitochondrial dysfunction, and activation of p38-JNK1/2 signaling. Invest New Drugs. 2012;30:2252–2262. doi: 10.1007/s10637-012-9804-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.