The formin INF2 is required for stable Glu microtubule formation and inhibition of microtubule dynamics in NIH3T3 cells downstream of mDia1 and LPA. Evidence also shows that the formation of an mDia1/INF2 complex is necessary for microtubule stabilization stimulated by LPA and is regulated by IQGAP1.
Abstract
Multiple formins regulate microtubule (MT) arrays, but whether they function individually or in a common pathway is unknown. Lysophosphatidic acid (LPA) stimulates the formation of stabilized detyrosinated MTs (Glu MTs) in NIH3T3 fibroblasts through RhoA and the formin mDia1. Here we show that another formin, INF2, is necessary for mDia1-mediated induction of Glu MTs and regulation of MT dynamics and that mDia1 can be bypassed by activating INF2. INF2 localized to MTs after LPA treatment in an mDia1-dependent manner, suggesting that mDia1 regulates INF2. Mutants of either formin that disrupt their interaction failed to rescue MT stability in cells depleted of the respective formin, and the mDia1-interacting protein IQGAP1 regulated INF2’s localization to MTs and the induction of Glu MTs by either formin. The N-terminus of IQGAP1 associated with the C-terminus of INF2 directly, suggesting the possibility of a tripartite complex stimulated by LPA. Supporting this, the interaction of mDia1 and INF2 was induced by LPA and dependent on IQGAP1. Our data highlight a unique mechanism of formin action in which mDia1 and INF2 function in series to stabilize MTs and point to IQGAP1 as a scaffold that facilitates the activation of one formin by another.
INTRODUCTION
Formins are multidomain proteins that regulate the dynamics and organization of both actin filaments and microtubules (MTs) in a variety of cellular functions requiring coordinated action of the cytoskeleton (Bartolini and Gundersen, 2010; Chesarone et al., 2010; Breitsprecher and Goode, 2013). Formins nucleate and elongate unbranched actin filaments through the activities of two domains: formin homology 1 (FH1), which binds to actin-bound profilin, and formin homology 2 (FH2), which is involved in actin polymerization. The FH2 domain of formins dimerizes, and this is essential for formin association with the barbed end of growing actin filaments (Moseley et al., 2004; Zigmond, 2004; Otomo et al., 2005). Of the 15 vertebrate formins, the largest subgroup is the diaphanous-related formins (DRFs), characterized by an N-terminal GTPase-binding domain (GBD), an adjacent diaphanous inhibitory domain (DID), and a C-terminal diaphanous autoregulatory domain (DAD). In many cases, the binding of a Rho-GTPase to the GBD regulates DRF activation by releasing an intramolecular interaction between the DID and the DAD domains that keeps the molecule inactive (Alberts, 2001; Amano et al., 2003; Otomo et al., 2005, 2010). However, both activation and deactivation steps are often incomplete or kinetically slow when tested with purified components, suggesting that full activation may require convergence of multiple inputs (Li and Higgs, 2003; Maiti et al., 2012).
Formins also act as positive regulators of MT organization and stability (Bartolini and Gundersen, 2010; Chesarone et al., 2010). In many cases, activation of formins generates a subset of selectively stabilized MTs that accumulates posttranslational modifications of tubulin, such as detyrosinated (or Glu) and acetylated tubulin (Palazzo et al., 2001a, 2004; Gundersen et al., 2004; Goulimari et al., 2005; Andres-Delgado et al., 2012; Thurston et al., 2012). These modified MTs act as specialized tracks for kinesin-dependent transport of cargoes such as vimentin intermediate filaments and endocytic vesicles (Kreitzer et al., 1999; Lin et al., 2002; Reed et al., 2006; Konishi and Setou, 2009; Wickstrom et al., 2010). In some cases, the formin activity toward MTs is independent of the actin polymerization activity (Bartolini et al., 2008; Cheng et al., 2011; Andres-Delgado et al., 2012; Daou et al., 2014; Roth-Johnson et al., 2014).
The FH2 domain of DRFs binds MTs and is typically implicated in the MT activity, although other domains also interact with MTs (Bartolini and Gundersen, 2010; Gaillard et al., 2011). The formin FH2 activity toward MTs seems to be regulated by an autoinhibitory mechanism similar to that toward actin. Active Rho GTP stimulates mDia1’s activity to stabilize MTs (Palazzo et al., 2001a, 2004; Goulimari et al., 2005, 2008), and MT stability can be induced by mDia1 and other formin constructs lacking the DID domain or by expressing their DAD domains (Palazzo et al., 2001a, 2004; Andres-Delgado et al., 2012; Thurston et al., 2012; Daou et al., 2014).
The relatively large number of vertebrate formins raises the question of whether individual formin family members regulate discrete aspects of the actin and MT cytoskeletons. Indeed, it was proposed that individual formin family members might regulate the distinct arrays of actin filaments found in cells (Chhabra and Higgs, 2007), and there is now abundant evidence for this (Chesarone et al., 2010). However, recent studies also reveal that multiple formins collaborate in the regulation of individual actin arrays. For example, FMNL1 and mDia1 have been implicated in the signaling pathway regulating actin assembly during polar body extrusion (Zhang et al., 2015), and both mDia1 and mDia2 RNAi inhibited nuclear actin assembly in NIH3T3 fibroblasts (Baarlink et al., 2013). INF2 was shown to regulate lamellipodial actin dynamics by opposing Rho/mDia-mediated actin polymerization (Sun et al., 2013).
With respect to MTs, which generally form only a single array in cells, there is perhaps an even stronger case for formin collaboration. Most of the formin family members regulate MTs, and there are cases in which multiple formins are required to generate a subset of stable MTs (Bartolini and Gundersen, 2010). For example, in breast carcinoma cells, mDia1, mDia2, and mDia3 function nonredundantly for cortical MT capture (Daou et al., 2014). In T-cells, FMNL1, mDia1, and the closely related formin INF2 are all necessary for the generation of stable detyrosinated microtubules (Glu MTs), which are essential for centrosome reorientation to the immunological synapse (Andres-Delgado et al., 2012). Nonetheless, whether the formins in these cases exert their effects on MTs sequentially or are required independently is unknown.
Two formins that regulate MT stability, mDia1 and INF2, are known to interact through their DAD and DID domains, respectively (Sun et al., 2011, 2013). The interaction of INF2 with mDia1 has been suggested to negatively regulate mDia1’s action on actin polymerization, but whether it is necessary for regulating MTs is unknown (Sun et al., 2011, 2013). In this study, we addressed whether mDia1 and INF2 function individually or together on MTs using the well-established NIH3T3 fibroblast system, in which Rho induces stable MTs through the formin mDia1 (Cook et al., 1998; Palazzo et al., 2001a). We find that mDia1 functions upstream of INF2, stimulating its localization to MTs. The interaction between mDia1’s DAD and INF2’s DID is required for stable MT formation. Both formins interact with the adaptor protein IQGAP1, and IQGAP1 is required for stable MT formation, INF2 localization to MTs, and promotion of mDia1 and INF2 interaction in vivo. These results suggest a novel mechanism of formin activation in which mDia1 activates INF2’s MT-stabilizing activity through the scaffolding function of IQGAP1.
RESULTS
INF2 is required for lysophosphatidic acid stimulation of Glu MTs
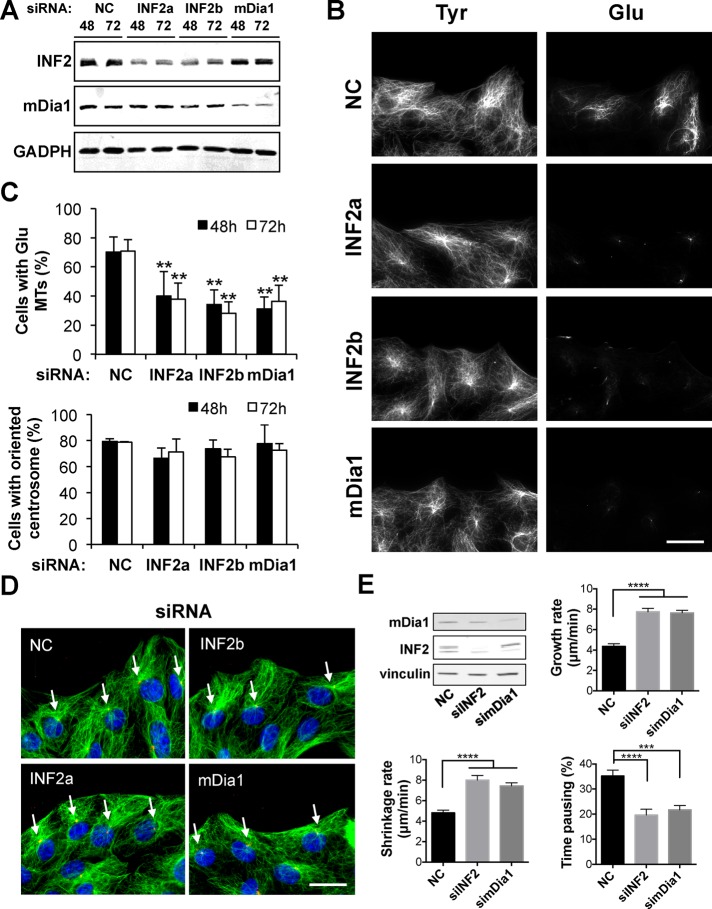
We tested the role of INF2, implicated in the stabilization of MTs in vitro (Gaillard et al., 2011) and T-cells (Andres-Delgado et al., 2012), in the formation of Glu MTs in the well-characterized NIH3T3 fibroblast system. Reducing INF2 expression with two different small interfering RNA (siRNA) oligonucleotides significantly inhibited the percentage of cells with Glu MTs without noticeably affecting dynamic tyrosinated (Tyr) MTs (Figure 1, A–C). The reduction in Glu MTs was comparable to that observed in mDia1 knockdown, and knocking down both formins did not further reduce Glu MTs (Supplemental Figure S1, A and B). Unlike in T-cells, knocking down mDia1, INF2, or both did not affect centrosome reorientation in cells at the edge of wounded monolayers (Figure 1, C and D, and Supplemental Figure S1B). Of note, knocking down either formin also resulted in a significant increase in MT dynamicity by increasing MT growth and shrinkage rates and decreasing pausing (Figure 1E, Supplemental Table S1, and Supplemental Movies S1–S3).
FIGURE 1:
INF2 regulates Glu MTs and MT dynamics but not centrosome orientation in NIH3T3 fibroblasts. (A) Immunoblot analysis of INF2, mDia1, and GAPDH (loading control) in NIH3T3 fibroblasts treated with noncoding control (NC), mDia1, or two INF2 siRNAs for the indicated time (in hours). (B) Glu and Tyr tubulin immunostaining of cells treated as in A and fixed 48 h after siRNA transfection. (C) Quantification of cells with Glu MTs (top) or oriented centrosome (bottom) treated as in A. Data are mean ± SD from four independent experiments (>200 cells/experiment). **p < 0.001 calculated by chi-square test. (D) Tyr tubulin, pericentrin, and 4’,6-diamidino-2-phenylindole (DAPI) staining of cells treated as in A and fixed 48 h after siRNA transfection. Arrows indicate the location of the centrosome in cells at the wound edge. (E) MT dynamics in mDia1- and INF2-knockdown NIH3T3 fibroblasts. Level of knockdown of mDia1 or INF2 in cells treated with the indicated siRNAs at the time movies were recorded. Growth and shrinkage rates and percentage of pausing of MTs in tagRFP-C4 tubulin stably expressing NIH3T3 fibroblasts transfected for 72 h with noncoding control siRNA (NC) or siRNA to either mDia1 (simDia1) or INF2 (siINF2). Data are mean ± SEM. ****p < 0.0001, ***p < 0.001 by two-tailed Student’s t test. No asterisk, p > 0.05. Bars, 20 μm.
Active forms of INF2 induce stable Glu MTs
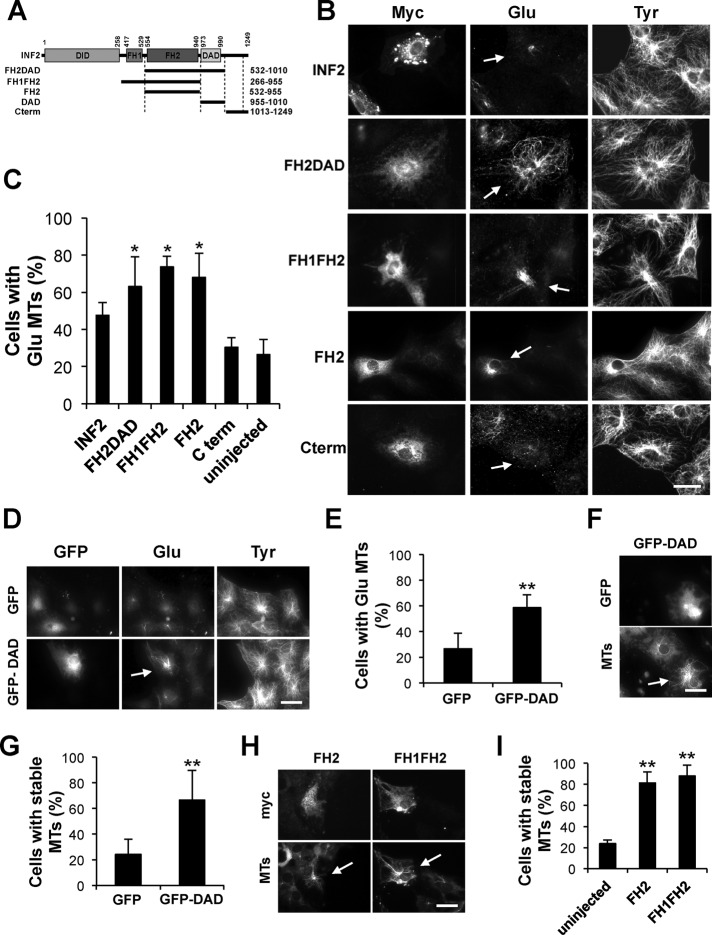
Next we tested the ability of INF2 to form de novo stable Glu MTs in serum-starved cells that have few Glu MTs (Cook et al., 1998; Palazzo et al., 2001a). Overexpression of full-length INF2 only weakly induced Glu MTs, suggesting that INF2’s MT stabilizing activity is negatively regulated by intramolecular inhibition. Indeed, fragments of INF2 containing the FH2 domain but lacking the N-terminal regulatory domain (FH2DAD, FH1FH2, and FH2) all induced stable Glu MTs above background levels (Figure 2, A–C), indicating that the FH2 alone is capable of generating Glu MTs. The FH2DAD was the only fragment that induced Glu MTs extending to the cell periphery, suggesting that peripheral MT stabilization may require the DAD domain, perhaps to induce endogenous INF2. Consistent with this idea, expression of INF2-DAD stimulated Glu MT formation (Figure 2, D and E). The Glu MTs induced by INF2-DAD, FH1FH2, and FH2 alone were all resistant to nocodazole-induced depolymerization, confirming their stability (Figure 2, F–I). INF2 may also be regulated negatively by membrane association, as expression of FH2-DAD constructs containing a C-terminal consensus motif for prenylation did not strongly induce Glu MTs (Supplemental Figure S2).
FIGURE 2:
INF2 induces stable Glu MTs through its FH2 domain. (A) Schematic of the INF2 fragments used in this study. (B) Myc (INF2), Glu, and Tyr tubulin staining of starved cells expressing indicated myc-tagged INF2 fragments. Arrows, expressing cells. (C) Quantification of cells treated as in B that exhibited Glu MTs. (D) GFP, Glu, and Tyr tubulin staining of starved cells expressing GFP or GFP-INF2-DAD. Arrow indicates Glu MTs in cell expressing GFP-INF2-DAD. (E) Quantification of cells treated as in D that exhibited Glu MTs. (F) DM1A and GFP staining of starved cells expressing GFP-INF2-DAD and treated with 2 μM nocodazole (30 min). Arrow, expressing cell. (G) Quantification of cells treated as in F that exhibited DM1A-labeled MTs. (H) DM1A (MTs) and myc staining of starved cells expressing myc-INF2-FH2 or myc-INF2-FH1FH2 and treated with nocodazole as in F. Arrows, expressing cell. (I) Quantification of cells treated as in H that exhibited DM1A-labeled MTs. In C, E, G, and H, data are mean ± SD from three independent experiments (>50 cells/experiment). *p < 0.05, **p < 0.001 by chi-square test. Bars, 20 μm.
Mutation of three leucine residues (Leu976, Leu977, Leu986) to alanine in the DAD of INF2 abrogates its in vitro actin depolymerization but not polymerization activity (Chhabra and Higgs, 2006). Conversely, mutation of Lys792 in the FH2 domain of INF2 to alanine diminishes actin polymerization activity without affecting actin severing and depolymerization (Ramabhadran et al., 2012). We introduced the same mutations alone or in combination in green fluorescent protein (GFP)–INF2 and found that the triple Leu976,977,986, Lys792, or the combined mutants rescued stable Glu MT formation in INF2-depleted cells as well as wild-type INF2 (Supplemental Figure S3A). To confirm this, we expressed an INF2-FH2 construct containing the Ile643/Lys792 mutant shown to severely affect actin polymerization in vitro. Both the Lys792 and the Ile643/Lys792 double mutant retained their ability to induce Glu MTs in serum-starved cells (Supplemental Figure S3B). Thus, similarly to closely related mDia (Bartolini et al., 2008; Cheng et al., 2011; Andres-Delgado et al., 2012; Daou et al., 2014) and as observed in T-cells (Andres-Delgado et al., 2012), INF2 activities toward actin polymerization and MT stabilization can be segregated.
INF2 localizes to Glu MTs in a lysophosphatidic acid–dependent manner
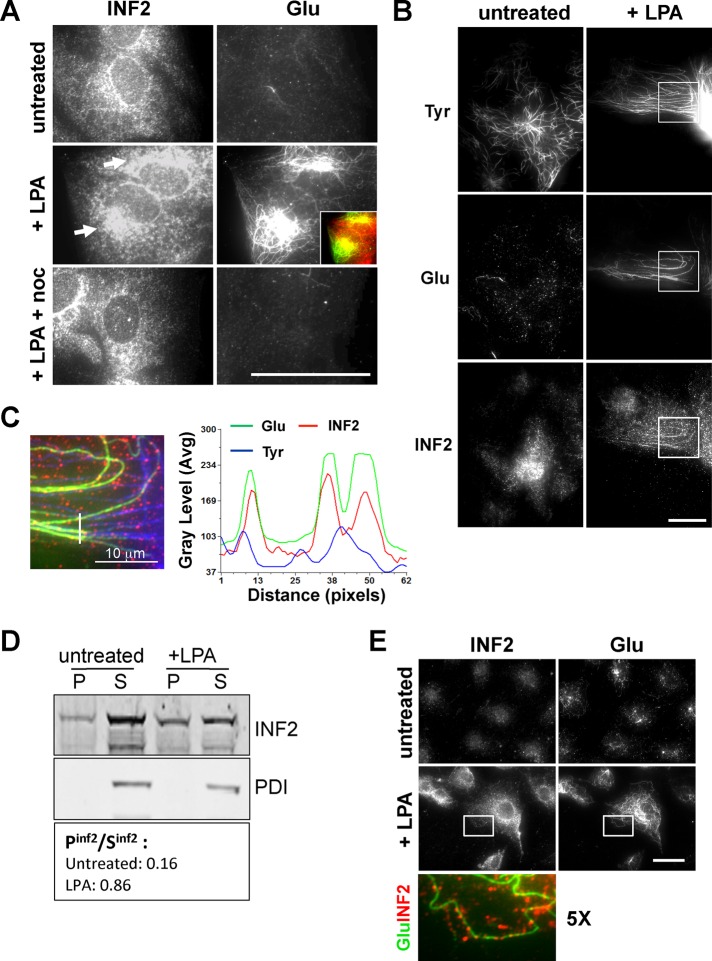
INF2 fragments containing the FH2 domain bind to MTs directly and stabilize them against depolymerization in vitro (Supplemental Figure S4; Gaillard et al., 2011), indicating that MT stabilization in cells might be related to direct MT binding. To test this, we analyzed the localization of endogenous INF2 in starved cells before and after stimulation with lysophosphatidic acid (LPA) to induce stable Glu MTs. Epifluorescence microscopy showed that INF2 accumulated in a pericentriolar region coincident with Glu MTs only in cells stimulated with LPA (Figure 3A). Of importance, this localization was strongly reduced in cells treated with high doses of nocodazole to induce complete depolymerization of dynamic and stable MTs, suggesting that INF2 associated with MTs. To critically test this possibility, we used total internal reflection fluorescence (TIRF) microscopy, which illuminates a narrow (∼100–200 nm) section of the ventral surface of the cell and was previously used to localize Glu MTs at the ventral surface (Wen et al., 2004). By TIRF microscopy, we found that INF2 appeared as linear streaks after LPA stimulation that colocalized preferentially with Glu MTs (Figure 3, B and C, and Supplemental Figure S5). LPA-dependent association of INF2 with MTs was confirmed by enrichment of the INF2 signal on Triton X-100–resistant Glu MTs by Western blot analysis and immunofluorescence of the insoluble cytoskeletal fraction (Figure 3, D and E). Together these results show that INF2 is required for LPA stimulation of stable Glu MTs in NIH3T3 fibroblasts, its activity toward MTs depends on its FH2 domain (but not its actin polymerization activity), and it localizes to Glu MTs in an LPA-dependent manner.
FIGURE 3:
LPA stimulates INF2 localization to Glu MTs. (A) Epifluorescence microscopy of endogenous INF2 and Glu tubulin immunostaining of starved NIH3T3 fibroblasts treated with LPA or LPA and nocodazole (10 μM, 2 h). Arrows indicate accumulated INF2 staining near Glu MTs. Inset, partial colocalization of INF2 (red) with Glu MTs (green). (B) TIRF microscopy of LPA-stimulated NIH3T3 fibroblasts immunostained for INF2, Glu, and Tyr tubulin. The boxed region is shown at higher magnification in C. (C) Merged image of the boxed region in B and linescan showing quantification of INF2, Glu, and Tyr tubulin fluorescence along the white line depicted in the merged image. (D) Western blot analysis of Triton X-100–soluble (S) and –insoluble (P) fractions from NIH3T3 fibroblasts before and after LPA addition. PDI is a marker for the soluble fraction. The ratio of INF2 in the fractions is shown. (E) INF2 and Glu tubulin immunostaining of LPA-stimulated NIH3T3 fibroblasts after brief extraction with Triton X-100 before fixation. Boxes show the region used for the high-magnification merge shown below. Bars, 20 μm.
INF2 functions downstream of mDia1
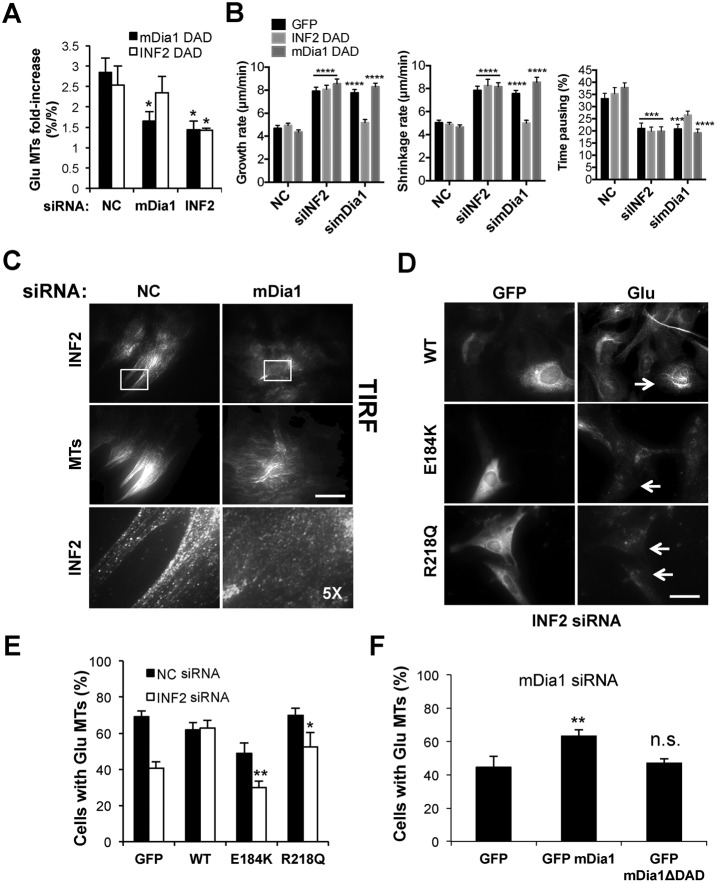
The involvement of two formins in the stabilization of Glu MTs downstream of LPA raised two possibilities: 1) the two formins function in series, with both of them regulating the same subset of MTs and one of them functioning upstream of the other, or 2) the two formins function in parallel, with both of them inducing a subset of stable Glu MTs. In the former case, one of the two formins should depend on the activation of the other for its promotion of MT stability. We tested this possibility by specifically activating each formin by expressing its DAD domain in cells that had been previously depleted of either mDia1 or INF2 by siRNA oligonucleotides. DAD domains activate endogenous DRFs by competing with the intramolecular inhibitory interaction between the DAD and DID domains of the endogenous formin (Alberts, 2001). As expected, expression of the DAD domains of both formins induced Glu MTs in starved cells but not in cells depleted of their parent molecules (Figure 4A). Of interest, whereas INF2-DAD induced Glu MTs in mDia1-depleted cells, mDia1-DAD failed to induce Glu MTs in INF2-depleted cells. These results suggest that INF2 activity is downstream of mDia1 and that activation of INF2 is sufficient to induce Glu MTs in the absence of mDia1.
FIGURE 4:
mDia1 induction of stable MTs requires INF2. (A) Quantification of Glu MTs in serum-starved NIH3T3 fibroblasts treated with NC, mDia1, or INF2 siRNAs before activation of each formin by expression of mDia1-DAD or INF2-DAD. (B) MT dynamics in NIH3T3 fibroblasts stably expressing RFP-tubulin and treated with NC, mDia1, or INF2 siRNAs before expressing mDia1-DAD or INF2-DAD. Data are mean ± SEM. Statistics were performed by two-way ANOVA with Tukey’s multiple comparison test comparing different groups to NC GFP. ***p < 0.001, ****p < 0.0001. (C) INF2 and MT (Tyr tubulin) immunostaining of NIH3T3 fibroblasts depleted of mDia1 imaged by TIRF microscopy. Bottom, higher magnifications of the boxed regions shown in the top. (D) GFP and Glu tubulin immunostaining of NIH3T3 fibroblasts depleted of INF2 and expressing GFP-WT INF2 or GFP-INF2 E184K and R218Q variants that do not bind to mDia1. Arrows indicate transfected cells. (E) Quantification of cells with Glu MTs treated as in D. (F) Quantification of Glu MTs in cells depleted of mDia1 and transfected with GFP, GFP mDia1, or GFP mDia1ΔDAD, which lacks the region that binds to INF2. Data are mean ± SD from three independent experiments (>200 cells/experiment). *p < 0.05, **p < 0.001; ns, not significant; calculated by chi-square test. Bars, 20 μm.
We also examined whether the DAD domains could rescue the increased MT dynamicity observed in cells depleted of each formin (Figure 1E). Neither DAD domain rescued the increased MT growth or shrinkage rates or decreased pausing in cells depleted of their parent molecules (Figure 4B). Nonetheless, INF2-DAD significantly suppressed these parameters in mDia1-depleted cells, and mDia1-DAD failed to suppress them (Figure 4B, Supplemental Table S2, and Supplemental Movies S4–S12). These results lend further support to the idea that INF2 functions downstream of mDia1 in regulating MTs.
To test the relationship between mDia1 and INF2 further, we examined the localization of each formin in the absence of the other. We detected no obvious alteration in the localization of endogenous mDia1 in INF2-depleted cells (Supplemental Figure S6). However, the accumulation of INF2 along the length of MTs as detected by TIRF was lost in cells deprived of mDia1 (Figure 4C). Thus the localization of INF2 on MTs requires mDia1.
The DID of INF2 has been shown to interact directly with the DAD of mDia1 (Sun et al., 2011), potentially providing an explanation for how activated mDia1 may function upstream to activate INF2. To test this idea, we took advantage of variants in INF2’s DID that cause focal segmental glomerulosclerosis (FSGS) and disrupt this interaction (Brown et al., 2010; Sun et al., 2011). We attempted to rescue Glu MT formation in INF2-depleted cells by expressing wild-type INF2 or these disease variants. Unlike their wild-type counterpart, neither E184K nor R218Q INF2 restored levels of Glu MTs in INF2-depleted cells (Figure 4, D and E). Consistent with this result, mDia1Δ DAD, which cannot bind to INF2 (Sun et al., 2011), failed to rescue MT stability in cells depleted of mDia1 (Figure 4F). Together these results strongly support a role for mDia1 in the regulation of INF2 activity toward MTs and suggest that the interaction between mDia1-DAD and INF2-DID is required for this regulation.
IQGAP1 is required for formin-induced Glu MTs
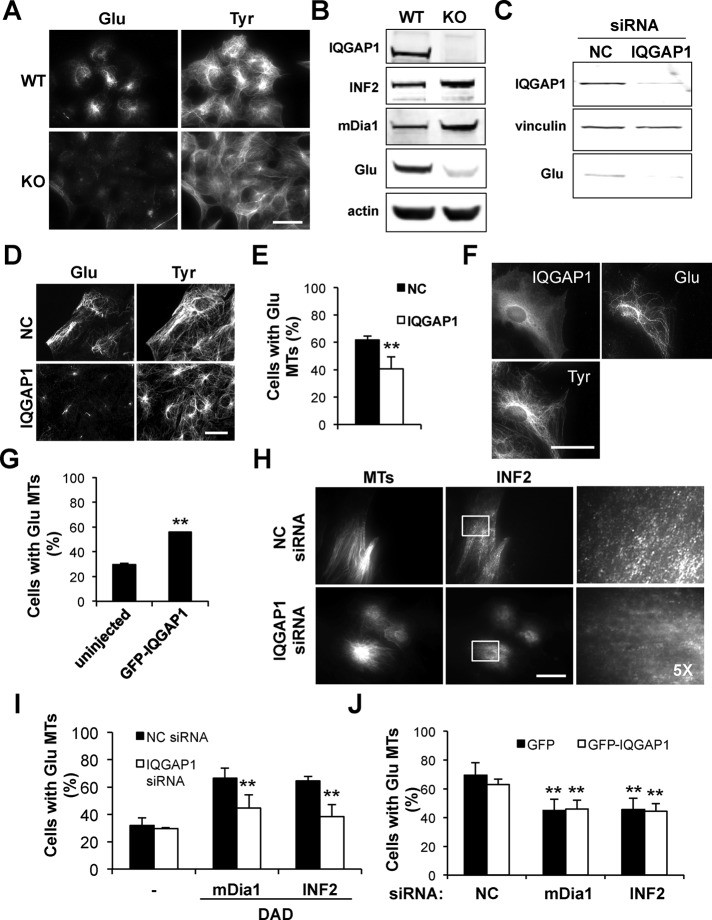
Despite the fact that mDia1’s DAD interacts directly with INF2’s DID in vitro (Sun et al., 2011), its expression did not induce Glu MTs in serum-starved cells in the absence of mDia1 (Figure 4B). This may reflect the low affinity of interaction between mDia1-DAD and INF2-DID (Sun et al., 2011). We hypothesized that in vivo full-length mDia1 was necessary to bring its DAD into the proximity of INF2, perhaps by binding a protein that bridges the two formins. IQGAP1 is such a candidate bridging molecule: it interacts specifically with activated mDia1, whose DAD is freed from interaction with its DID (Brandt et al., 2007), and it has been implicated in regulating MT capture and stability (Fukata et al., 2003; Watanabe et al., 2004; Wickstrom et al., 2010). IQGAP1 was also reported to coimmunoprecipitate with INF2, although the nature of the interaction was not characterized (Boyer et al., 2011). To examine a possible role for IQGAP1 in stimulating INF2’s MT activity by active mDia1, we first determined whether IQGAP1 was required to generate stable Glu MTs. Compared to wild-type (WT) mouse embryonic fibroblasts (MEFs), MEFs from IQGAP1-knockout (KO) mice exhibited almost no detectable Glu MTs by immunofluorescence or Glu tubulin by Western blot (Figure 5, A and B). This phenotype represented defective MT stabilization, as shown by lack of acetylated MTs, another tubulin posttranslational modification associated with MT stability, and the finding that Glu MT levels could be rescued with the MT stabilizer Taxol (Supplemental Figure S7). Lack of Glu MTs was not due to reduced levels of mDia1 or INF2, which in fact were up-regulated compared with WT MEFs (Figure 5B). Of importance, reexpression of IQGAP1 in IQGAP1 KO cells rescued Glu MT levels in these cells (Supplemental Figure S8, A and B). Acute depletion of IQGAP1 by siRNA oligonucleotides also reduced Glu tubulin levels biochemically and Glu MTs by immunofluorescence (Figure 5, C and E). Conversely, overexpression of IQGAP1 in serum-starved cells induced formation of Glu MTs (Figure 5, F and G). Thus IQGAP1 is required for LPA-stimulated Glu MT formation and is sufficient to induce Glu MTs when overexpressed.
FIGURE 5:
IQGAP1 is required for formin-mediated induction of Glu MTs. (A) Immunostaining of Glu and Tyr tubulin in IQGAP1 WT and KO MEFs. (B) Immunoblot of IQGAP1, INF2, mDia1, and Glu tubulin in IQGAP1 WT and KO MEFs. Actin is a loading control. (C) Immunoblot of IQGAP1 and Glu tubulin in NIH3T3 fibroblasts treated with noncoding (NC) or IQGAP1 siRNA. Vinculin is a loading control. (D) Immunostaining of IQGAP1, Glu, and Tyr tubulin in NIH3T3 fibroblasts treated with NC or IQGAP1 siRNA. (E) Quantification of Glu MTs in cells treated as in D. (F) Immunostaining of GFP, Glu, and Tyr tubulin in starved NIH3T3 fibroblasts expressing GFP-IQGAP1. (G) Quantification of cells with Glu MTs treated as in F. (H) TIRF microscopy of NIH3T3 fibroblasts treated with NC or IQGAP1 siRNA and immunostained for INF2 and Tyr tubulin (MTs). Far right, higher- magnifications of INF2 staining of the regions shown in the boxes. (I) Quantification of cells with Glu MTs after depleting IQGAP1 and transfecting either mDia1-DAD or INF2-DAD to activate endogenous formins. (J) Quantification of cells with Glu MTs in cells transfected with GFP or GFP-IQGAP1 after treatment with NC, mDia1, or INF2 siRNAs. Data are mean ± SD from three independent experiments (>200 cells/experiment). **p < 0.001 calculated by chi-square test. Bars, 20 μm.
As in cells depleted of mDia1, INF2 failed to localize to MTs in cells depleted of IQGAP1 (Figure 5H). However, whereas we detected INF2 along the length of stable Glu MTs, endogenous IQGAP1 only localized to cortical dynamic MTs in NIH3T3 cells (Supplemental Figure S9, A–C). Reexpression of IQGAP1 in IQGAP1 KO MEFs induced INF2 localization to Glu MTs (Supplemental Figure S8C). This result suggests that IQGAP1 may be necessary to activate INF2’s MT-stabilizing activity. To test this, we determined whether IQGAP1 was required for the induction of Glu MTs by mDia1’s DAD domain. mDia1-DAD domain failed to induce Glu MTs when expressed in cells depleted of IQGAP1 (Figure 5I). In addition, IQGAP1 overexpression failed to induce Glu MTs in mDia1- or INF2-depleted cells (Figure 5J). These observations suggest that IQGAP1 is necessary for mDia1’s activation of INF2 and that IQGAP1 cannot stimulate MT stability without the formins.
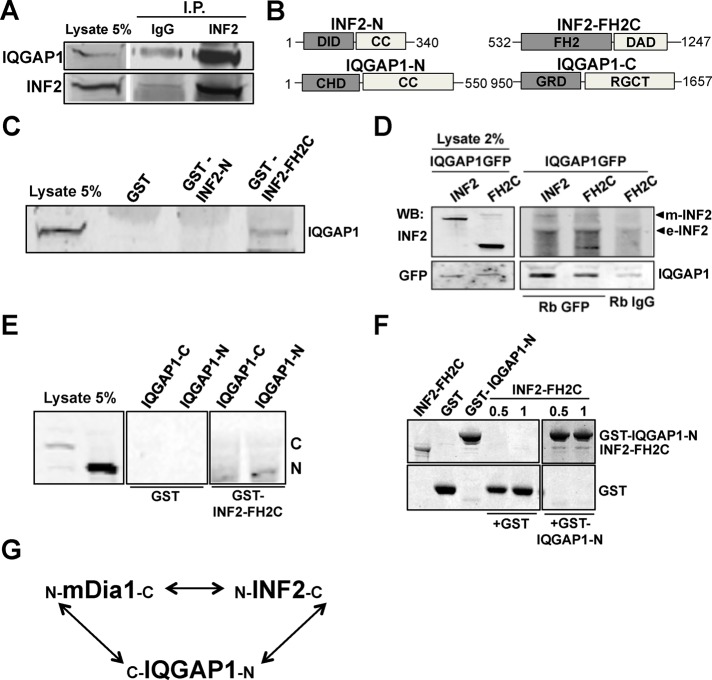
We confirmed that endogenous INF2 and IQGAP1 interacted by coimmunoprecipitation (Figure 6A). Active mDia1 interacts with the C-terminus of IQGAP1 (Brandt et al., 2007), so it was of interest to determine whether INF2 interacted with the same or a different site on IQGAP1. We found that full-length INF2 and INF2-FH2C, but not INF2-N, interacted with IQGAP1 by pull-down assays using recombinant or overexpressed proteins (Figure 6B-D). INF2-FH2C interacted with an N-terminal fragment of IQGAP1 but not a C-fragment (Figure 6E). Of importance, using recombinant proteins, we found that the interaction between INF2-FH2C and IQGAP1-N was direct (Figure 6F). Together these data show that each of the three components, mDia1, INF2, and IQGAP1, associate via nonoverlapping sites, potentially forming a trimeric complex (Figure 6G).
FIGURE 6:
IQGAP1 interacts directly with INF2. (A) Coimmunoprecipitation of INF2 and IQGAP1 from whole-cell lysates of NIH3T3 fibroblasts using nonimmune IgG or INF2 antibody. (B) Schematic of the fragments of INF2 and IQGAP1 used in C–F. (C) Immunoblot analysis of endogenous IQGAP1 in pull downs from whole-cell lysates using GST-INF2-N, GST-INF2-FH2C, and GST alone. (D) Coimmunoprecipitation of IQGAP1-GFP and Myc-INF2 (INF2) or Myc-INF2-FH2C using nonimmune IgG (Rb IgG) or GFP antibody. Both Myc-tagged (m-INF2) and endogenous INF2 (e-INF2) were detected by Western blotting using INF2 antibody. (E) Immunoblot analysis of pull downs from whole-cell lysates expressing GFP-IQGAP1-N or -C using GST-INF2-FH2C or GST alone. (F) Coomassie staining of binding reactions with GST-IQGAP1-N or GST alone (2 μM) with indicated concentrations (μM) of untagged INF2-FH2C. (G) Schematic of the interactions between mDia1, INF2, and IQGAP1.
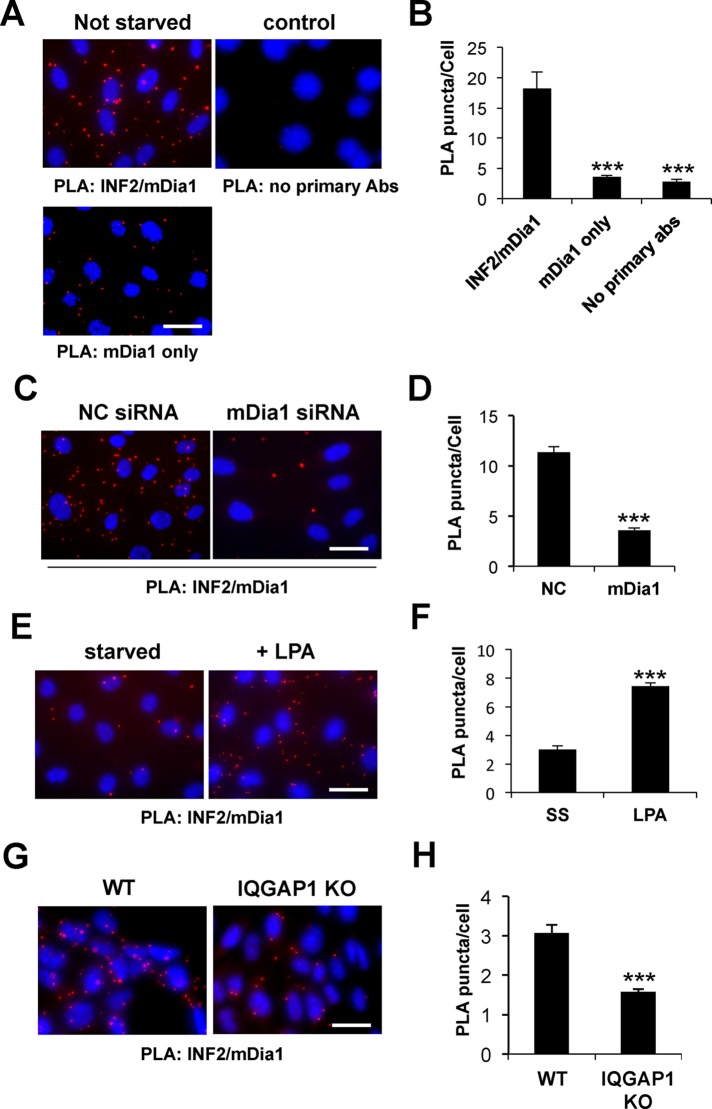
We attempted to isolate a complex by sucrose gradient fractionation and coimmunoprecipitation but did not detect one, suggesting that it may form transiently or may not resist cell lysis. To assess whether this complex might form in cells, we used in situ proximity ligation (PLA) assays that generate punctate signals when antibodies to the two interacting species are within 30–40 nm (Soderberg et al., 2006). First, we confirmed that the PLA signal (measured as number of puncta per cell) depended on the presence of mDia1 and INF2 primary antibodies and mDia1 expression (Figure 7, A–D). Next we observed that INF2/mDia1 PLA puncta increased upon LPA addition (Figure 7, E and F) and that INF2/mDia1 puncta were significantly reduced in immortalized MEFs from IQGAP1 KO mice (Figure 7, G and H). These results indicate that LPA induces the association of INF2 and mDia1 in cells and that this depends on IQGAP1.
FIGURE 7:
IQGAP1 is required for the interaction of mDia1 and INF2 stimulated by LPA. (A) Images from PLA assays of serum-grown NIH3T3 fibroblasts using antibodies against mDia1 and INF2 or as controls, leaving out mDia1 antibody or both antibodies. Red puncta indicate interaction between mDia1 and INF2; nuclei are stained with DAPI. (B) Quantification of PLA puncta/cell (±SEM) in samples prepared as in A; >50 cells/sample. (C) PLA puncta in NIH3T3 fibroblasts depleted of mDia1 by siRNA. (D) Quantification of PLA puncta/cell (±SEM) in samples prepared as in C; >140 cells/sample. (E) PLA puncta in starved NIH3T3 fibroblasts stimulated with LPA. (F) Quantification of PLA puncta/cell (±SEM) in samples prepared as in E; >80 cells/sample. (G) PLA puncta in IQGAP1 WT and KO MEFs. (H) Quantification of PLA puncta/cell (±SEM) in samples prepared as in G; >80 cells/sample. ***p < 0.0001 by Mann–Whitney test. Bars, 20 μm.
DISCUSSION
One of the crucial questions for how formins regulate MT stability is whether their action is direct or is mediated by downstream events. A number of formins, including mDia1, mDia2, and INF2, have been shown to bind to MTs and enhance their stability in vitro (Bartolini et al., 2008; Gaillard et al., 2011; Thurston et al., 2012) and in vivo (Palazzo et al., 2001a; Wen et al., 2004; Bartolini et al., 2008; Andres-Delgado et al., 2012; Thurston et al., 2012; Daou et al., 2014). Previously mDia1 was shown to interact with the MT plus end–tracking proteins EB1 and APC, suggesting that the formation of a complex might be involved in generating stable MTs (Gundersen et al., 2004; Wen et al., 2004). Here we found that a second formin, INF2, implicated in MT stability in T-cells, was also required for MT stability and regulation of MT dynamics in NIH3T3 fibroblasts and, critically, functioned downstream of mDia1. Either formin was necessary to reduce MT growth and shrinkage rates while increasing pausing, suggesting that at steady state, Glu MT stability is critically dependent on reduction of MT dynamics as if stabilized by a “capping” mechanism indicated by previous observations (Cook et al., 1998; Infante et al., 2000; Palazzo et al., 2001a). Consistent with a more proximal effect on MTs, INF2 localized to stable MTs, and this localization required mDia1 and IQGAP1. These data suggest a revised model for MT stabilization by formins in which one formin (mDia1) activates a second formin (INF2), allowing it to bind to and stabilize MTs through its FH2 domain.
In addition to functional and localization data, this sequential model for formin action on MTs is supported by interaction data on mDia1, INF2, and IQGAP1. A previous study showed that the DAD domain of mDia1 interacted with the DID domain of INF2 (Sun et al., 2011), and we found that this interaction was essential for inducing stable Glu MTs. Of importance, mDia1’s DAD was unable to suppress MT dynamicity or induce Glu MTs in the absence of endogenous mDia1, indicating that in vivo mDia1’s DAD needed to be presented to INF2 in the context of full-length mDia1. Given the previously described interactions of INF2 and active mDia1 with IQGAP1 (Brandt et al., 2007; Boyer et al., 2011), we suspected that IQGAP1 may act to bring mDia1 and INF2 together to facilitate interaction between their DAD and DID. Indeed, our data showing that IQGAP1 is required for induction of stable Glu MTs by LPA or the DAD domains of either formin strongly support such a model. In addition, our studies show that INF2 and IQGAP1 interact through sites that would allow simultaneous binding of mDia1. Despite this, we were unable to isolate a stable complex containing the three proteins. Nonetheless, we used PLA to show that mDia1 and INF2 interacted in vivo and that this interaction was stimulated by LPA and required IQGAP1. Together these results suggest that the interaction between mDia1, INF2, and IQGAP1 is transient and may release INF2 once it has been activated.
To our knowledge, these data are the first to suggest that formins function in series to regulate the formation of a cytoskeletal array. Previous studies implicated multiple formins in regulating both the actin and MT cytoskeletons but did not show that two formins functioned sequentially to induce a single cytoskeletal change. This result raises the question of whether INF2 is a downstream effector of mDia1 only or whether other formins can converge in the activation of INF2 to regulate MT stability, thus allowing activation of MT stability downstream of multiple stimuli. It is also possible that the mDia1-INF2 pair can be activated to regulate actin structures in addition to stable MT arrays. For example, INF2’s DID interaction with mDia1’s DAD was shown to inhibit the polymerization of actin by mDia1 (Sun et al., 2011, 2013). Formins are known to interact with other regulators of the cytoskeleton, including MT plus end–tracking proteins EB1 and APC (Gundersen et al., 2004), MT-modifying enzyme HDAC6 (Destaing et al., 2005), and actin regulators APC (Moseley et al., 2007; Webb et al., 2009; Okada et al., 2010) and spire (Rosales-Nieves et al., 2006; Dahlgaard et al., 2007; Quinlan, 2013). Perhaps the selected recruitment of these cytoskeletal regulators downstream of distinct signaling pathways contributes to the formation of distinct cytoskeletal arrays.
Unlike loss of MT stability, no effect on centrosome reorientation was detected in either mDia1- or INF2-silenced cells, suggesting that, unlike T-cells, the activity of these formins on Glu MTs has no functional consequence on the polarization of the centrosome/nuclear axis in NIH3T3 cells at the edge of a wounded monolayer. T-cells seem to depend on Glu MTs for centrosome orientation, but repeated studies show that this not the case in fibroblasts (Palazzo et al., 2001a; Bartolini and Gundersen, 2006). We cannot fully explain this discrepancy, but it may reflect differences in the mechanisms of centrosome orientation in the two systems: in NIH3T3 fibroblasts, the nucleus is moved rearward to orient the centrosome (Gomes et al., 2005; Luxton et al., 2010; Luxton and Gundersen, 2011), whereas in T-cells, the centrosome is moved (Ritter et al., 2013).
Our work is not the first to implicate IQGAP1 as a scaffolding protein for regulators of MTs. For example, CLIP-170 and APC interact with IQGAP1 to mediate the transient capture of MTs at cortical regions (Watanabe et al., 2004). Similarly, CLASP2, which also interacts with IQGAP1, is involved in MT stability in migrating cells (Drabek et al., 2006) and might regulate the association of IQGAP1 with MTs and EB1 (Watanabe et al., 2009). Induction of Glu MTs in starved cells was shown to occur upon GSK3β inhibition caused by mDia-mediated novel protein kinase C activation, suggesting positive regulation of CLASP2/IQGAP1 complex formation by formin function (Eng et al., 2006). Comprehensive in vitro and in vivo analysis will be required to understand the sequence of the association of all these proteins and whether they assemble into a MT “stabilosome.”
Mutations in INF2’s DID domain that disrupt the interaction between INF2 and mDia1 have been implicated in the etiology of FSGS and Charcot–Marie–Tooth (CMT) diseases (Brown et al., 2010; Boyer et al., 2011; Sun et al., 2011, 2013). Our results show that these mutations also prevent the formation of stable Glu MTs, raising the possibility that lack of MT stability may contribute to these diseases. CMT hereditary neuropathy refers to a group of disorders caused by a variety of mutations affecting axonal function, the insulating myelin coating, or both, whereas FSGS is primarily a glomerular disorder that causes renal dysfunction and in which the affected genes encode regulators of the actin cytoskeleton in podocytes. Although loss of INF2-mediated inhibition of Rho/mDia-driven actin polymerization in the foot process of podocytes is one mechanism proposed for mutant INF2 in the pathogenesis of FSGS (Sun et al., 2011, 2013), the precise role of INF2 mutations in the etiology of CMT and FSGS is largely unexplained. MTs are known to maintain the shape and integrity of podocyte major processes, and disruption of MT stability is predicted to indirectly affect the delivery of regulators of actin dynamics in the foot processes. In addition, loss of MT stability by mutant INF2 might equally be crucial for maintenance of axonal integrity at the onset of CMT. Examination of MT stability in tissue or cells isolated from patients may reveal whether loss of MT stability by mutated INF2 correlates with the onset of neuropathy and/or kidney dysfunction.
MATERIALS AND METHODS
Plasmids and reagents
DNA constructs expressing the following INF2 fragments tagged with c-myc or glutathione S-transferase (GST) at their amino terminus were described previously (Andres-Delgado et al., 2010, 2012; Madrid et al., 2010): myc-FH2DAD (amino acids [aa] 532–1010); myc-FH1FH2 (aa 266–955); myc–WT FH2 (aa 532–955) or with the Lys792Ala or double Lys792Ala and Ile643Ala mutations; myc–C-terminus of INF2-1 (splicing variant 1; aa 1013–1249); GST-DID (aa 1–265); GST-FH2DAD (aa 532–1010); and GST-C1 (aa 1013–1249). DNA constructs expressing GFP fused to INF2-1 or to INF2 proteins with the single Lys792Ala, triple Leu976, Leu977, Leu986Ala, or combined mutations (Lys792Ala/Leu976, Leu977, Leu986Ala) were made by cloning INF2 sequences from the corresponding constructs in pSport6 (Madrid et al., 2010) in the pEGFP-C1 expression vector. DNA constructs expressing FH2DADC fragments with mutations in the CAAX box of INF2-1 (FH2DADC-AIVQ and FH2DADC-CIVL) or with the carboxyl-terminal end of INF2-1 substituted by that of INF2-2 (splicing variant 2 without CAAX box; FH2DAD-N) were achieved by PCR using a perfectly matched 5′ oligonucleotide primer and a 3′ oligonucleotide primer containing the appropriate modification and cloning the amplification fragment in the plasmid pCR3.1 (ThermoFisher Scientific, Waltham, MA). GFP- or myc-tagged DAD of INF2 was made by cloning the amplified fragment containing the DAD domain of INF2 (aa 956–1010) in the pCR3.1 or pGFP-C1 vectors. GST-FH2DADC1 (aa 532–1249) was made by cloning the myc-tagged FH2DADC1 fragment of INF2-1 in pGEX-4T-1. His-tagged-mDia1-FH2DAD (aa 748-1203) was a kind gift of Robert Grosse (University of Heidelberg, Heidelberg, Germany; Brandt et al., 2007). GFP mDia1 lacking the DAD domain was generated using standard PCR procedures using a template kindly provided by S. Narumiya (Kyoto University, Kyoto, Japan). GFP-INF2-E184K and -R218Q mutants were constructed using a QuikChange Site-Directed Mutagenesis kit (Stratagene, San Diego, CA) according to the manufacturer’s protocols. Cherry-mDia1FH1COOH was generated as described (Okada et al., 2010). GFP-IQGAP1, GFP-IQGAP1-C (1950–1657), and -N (1–550), as well as GST-IQGAP1-N and -C plasmids, were kindly provided by Geri Kreitzer (Cornell University, Ithaca, NY) and George Bloom (University of Virginia, Charlottesville, VA). All constructs were verified by sequencing. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise noted. LPA was obtained from Avanti (Alabaster, AL).
Antibodies
Antibodies used include polyclonal rabbit 301 generated against INF2 peptide SVKEGAQRKWAALKEKLGPC (amino acids 2–19), mouse monoclonal supernatant (1E4) against the same sequence, and rabbit polyclonal anti-INF2 (Proteintech, Rosemont, IL). The INF2 peptide was synthesized in an automated peptide synthesizer (Abimed, Langerfeld, Germany) coupled to keyhole limpet hemocyanin (ThermoFisher Scientific) and injected in rabbits and mice. Spleen cells from immunized mice were fused to myeloma cells and a hybridoma clone (E4) selected that produced antibody that recognized INF2 in extracts from Cos-7 cells transiently expressing myc-tagged INF2. The specificity of rabbit antiserum and the E4 monoclonal antibody was assayed by immunoblot (IB) and immunofluorescence (IF) analyses. Other antibodies used include mDia1 (clone 51; 1:100 for IF, 1:500 for WB; BD Biosciences, San Jose, CA), rat anti–tyrosinated tubulin (YL-1/2; 1:10 for IF, 1:1000 for IB), rabbit anti-Glu tubulin (1:500 for IF, 1:10,000 for IB; Gundersen et al., 1984), mouse (1:8000; ThermoFisher Scientific) or rabbit (FL-335; 1:1000; Santa Cruz Biotechnology, Dallas, TX) anti–glyceraldehyde 3-phosphate dehydrogenase, mouse anti–β-actin (C4; 1:1000), mouse anti-pericentrin (1:100; BD Biosciences), mouse anti-PDI (1:1000 for IB; Stressgen, Victoria, Canada), mouse anti-Myc (9E10; 1:100; Santa Cruz Biotechnology), mouse anti–acetylated tubulin (6-11B-1; 1:100), mouse anti–α-tubulin (DM1A; 1:1000), rabbit anti-GFP (Millipore, Billerica, MA), mouse (10 μg/ml for IF, 1 μg/ml for IB) and rabbit (1:250 for IF, 1:1000 for IB) anti-IQGAP1 antibodies (kind gifts of George Bloom), and rabbit anti-IQGAP1 (H-109; 1:100 for IF, 1:500 for IB; Santa Cruz Biotechnology). For endogenous immunolocalization, a mouse anti-IQGAP1 from BD Biosciences was used (1:100).
Protein purification, GST pull-down, and direct protein-binding assays
GST-tagged proteins (GST, GST-INF2-FH2DADC, GST-INF2-FH2DAD, GST-INF2-C, and GST-INF2-N) were expressed in Escherichia coli BL-21 and purified on agarose-coupled glutathione (GE Healthcare, Little Chalfont, United Kingdom) according to the manufacturer’s protocols. Histidine-mDia1-FH2DAD was purified as described (Bartolini et al., 2008). Purified proteins were dialyzed into in HKCL buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH.7.4, and 140 mM KCl) using Slide-A-Lyzer dialysis cassettes (10,000 MWCO; ThermoFisher Scientific) overnight at 4°C. GST-pull-down assays were carried out by incubating glutathione-agarose–bound purified GST-tagged proteins (20 μg) with precleared whole-cell lysates prepared using NP-40 lysis buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol [DTT], phenylmethylsulfonyl fluoride [PMSF], and protease and phosphatase inhibitor cocktails from ThermoFisher Scientific) from cells transfected with the indicated GFP constructs. Complexes were allowed to form for 2 h at 4°C and eluted from beads with SDS loading buffer after three washes with NP-40 lysis buffer. Direct protein-binding assays were performed in HKCL buffer plus 1 mM DTT by incubating the indicated amounts of GST or GST-IQGAP1-N with untagged INF2-FH2C plus or minus untagged INF2-DID for 30 min at room temperature. Untagged proteins were generated by overnight incubation with thrombin at 4°C, followed by elution from benzoamidine agarose to eliminate thrombin from the final protein solution. Protein complexes were isolated by adding glutathione-agarose for 1 h at 4°C, followed by several washes in HKCL before final elution in SDS loading buffer.
Coimmunoprecipitation assays
NIH3T3 fibroblasts grown in 10-cm dishes (at least two dishes per sample) were lysed at confluence by scraping with EBC buffer (50 mM Tris-HCl, pH 8, 120 mM NaCl, 0.5% NP-40, 1 mM ethylene glycol tetraacetic acid, 1 mM NaF, 500 μM Na3VO4, 1 mM PMSF, and proteasome and phosphatase inhibitor cocktails) for 30 min on ice, and cell lysates were precleared by centrifugation at 130,000 × g at 4°C. Each precleared sample was incubated with the primary antibody or preimmune immunoglobulin G (IgG) serum (∼10 μg/sample) for 4 h at 4°C. A premade mixture of protein A–plus protein G–Sepharose 4B beads equilibrated in EBC buffer (50/50 slurry) was then added to each sample and incubated for 2 h at 4°C. Immunocomplexes were washed 3× with EBC buffer using spin filter tubes (ThermoFisher Scientific) and eluted in hot SDS loading buffer before boiling and loading onto SDS–PAGE for Western blot analysis.
Cell culture and microinjection
NIH3T3 fibroblasts were grown in DMEM and 10% calf serum as previously described (Cook et al., 1998; Palazzo et al., 2001a). WT and KO IQGAP1MEFs (Kim et al., 2011) were grown in DMEM plus 10% fetal calf serum. For drug or microinjection experiments, cells were grown to confluence on acid-washed coverslips, serum starved or not for 48 h, and wounded 30 min before treatment or cDNA injection. For plasmid microinjection, cDNAs were resuspended in HKCL buffer at 50–200 μg/ml, precleared at 10,000 × g for 30 min at 4°C, and microinjected into nuclei of cells selected randomly from the edges of a wounded monolayer 2 h before fixation and immunostaining.
siRNA, cDNA transfection, and Western blot analysis
NIH3T3 fibroblasts were transfected with siRNA oligonucleotides (Shangai GenePharma, Shanghai, China) using RNAiMax according to manufacturer’s protocols (Invitrogen). siRNA duplexes targeting mDia1 and INF2 were based on previously published sequences (Bartolini and Gundersen, 2006; Madrid et al., 2010). IQGAP1 siRNA (5′-GGAUAAAUUUAACGUGGAUdTdT-3′) was generated through the Dharmacon (Lafayette, CO) siRNA design algorithm (www.thermoscientificbio.com/design-center/), and a scrambled noncoding sequence provided by the manufacturer was used as a negative control. Knockdown efficiency and percentage of cells with Glu MTs or oriented centrosomes were analyzed 48 or 72 h after transfection as indicated in the figures. For cDNA expression for immunoprecipitation studies, NIH3T3 fibroblasts were transfected at 30% confluence in growth medium with Lipofectamine Plus (ThermoFisher Scientific) according to the manufacturer’s protocols and allowed to express for 24 h. For cDNA expression in cells previously silenced by siRNA transfection, cells were transfected 48 h after siRNA transfection and allowed to express an additional 24 h before fixation. For Western blot analysis, unless otherwise stated, cells were lysed in RIPA buffer (1% Triton X-100, 50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1 mM PMSF, and protease and phosphatase inhibitors), normalized for protein concentration by bicinchoninic acid assay, boiled in SDS sample buffer, separated by SDS–PAGE, and Western blotted. Incubation with primary antibodies was followed by incubation with the appropriate IR680- or IR800-conjugated secondary antibodies (1:10,000; Rockland Immunochemicals, Pottstown, PA), and final data acquisition and quantification were performed using an Odyssey imaging system (Li-COR Biosciences, Lincoln, NE).
Epifluorescence and TIRF microscopy
NIH3T3 fibroblasts grown on coverslips were typically fixed in methanol at −20°C for 10 min to preserve MT integrity and rehydrated in TBS (10 mM Tris-buffered saline, pH 7.4). Secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA) and were preabsorbed to minimize cross-reaction with other species. Immunostained samples were observed using a Nikon TE2000 microscope with a 60×/1.45 objective and an Orca II ER charge-coupled device (CCD) camera (Hamamatsu, Hamamatsu, Japan) controlled by MetaMorph software. In experiments assessing Glu MT levels, injected cells or 200 cells randomly chosen at the edge of an artificial wound were scored as positive if >10 MTs were brightly stained by the Glu antibody. This cutoff was selected based on previous considerations (Cook et al., 1998; Palazzo et al., 2001a). For centrosome orientation, cells at the edge of an artificial wound were randomly selected and scored as previously described (Palazzo et al., 2001b). Both counts were preformed blinded. Data were quantified and expressed as means ± SEM from at least three independent experiments (>200 cells/experiment).
MT dynamics
NIH3T3 fibroblasts were infected with pMSCV-puro-tagRFP-C4 α-tubulin retrovirus to generate a red fluorescent protein (RFP)–tubulin stably expressing cell line. RFP-tubulin NIH3T3 cells were transfected with noncoding control siRNA (NC) or either siRNA to silence Dia1 (simDia1) or INF2 (siINF2) for 72 or 48 h and then transfected with enhanced GFP control, mDia1 DAD-EGFP (aa 1180–1255), or INF2 DAD-EGFP (aa 955–1010) for another 24 h. Live imaging of MT dynamics in transfected cells was performed at 37°C and 5% CO2 for 5 min (5 s/frame) with a 100× PlanApo objective (numerical aperture 1.45) and an iXon X3 CCD camera (Andor, Belfast, United Kingdom) on a Nikon Eclipse Ti microscope controlled by Nikon’s NIS-Elements software (Nikon, Tokyo, Japan). Movies were analyzed by ImageJ using a manual tracking plug-in. Statistics among groups were performed by two-way analysis of variance (ANOVA test). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Preparation of detergent-resistant cell fraction
Confluent starved cells before and after treatment with LPA for 2 h were scraped and washed twice in ice-cold phosphate-buffered saline before lysis in ice-cold fractionation buffer (10 mM 1,4-piperazinediethanesulfonic acid, pH 6.8, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% Triton X-100, 1 mM DTT, and protease inhibitors from ThermoFisher Scientific) for 20 min on ice. The cell lysate was centrifuged at 4°C for 3 min at 1500 × g to isolate the soluble from the Triton X-100–insoluble fraction. Denaturing SDS loading buffer was added and matching aliquots of each protein boiled and loaded onto SDS–PAGE for Western blot analysis.
In situ ligation proximity assays
Samples were fixed in methanol at −20°C and processed for Duolink in situ amplification as suggested by the manufacturer’s protocol (Onlink Bioscience, Uppsala, Sweden) using mouse anti-mDia1 (clone 51, 1:100; BD Biosciences) and rabbit ant-INF2 (301). Random images of stained samples were captured by epifluorescence microscopy using a 60× oil immersion objective, and PLA signals were quantified using custom software. Briefly, cell boundaries were drawn, and the number of PLA puncta and their sizes (in pixels) were determined by the software using thresholded images. Data were exported to Microsoft Excel or GraphPad Prism 4.0 for statistical analysis and plotting. All PLA experiments were performed at least three times, and for each experiment, 10 images were taken per condition using identical microscope and camera settings. In each experiment, a minimum of 50 cells were counted per condition and used for analysis.
Other methods
See Supplemental Materials and Methods for MT binding and stability assays and confocal microscopy.
Supplementary Material
Acknowledgments
We thank Feng Ning Yuan for excellent technical assistance and members of the Gundersen lab for very helpful discussions and critical comments on the manuscript. We are particularly grateful to Wakam Chang for writing the software to count PLA puncta and David Sacks (National Institutes of Health, Bethesda, MD) for sharing IQGAP1-null MEFs. Our gratitude goes to George Bloom and Geri Kreitzer for sharing IQGAP1 reagents. This work was supported by National Institutes of Health Grant GM105536 to G.G.G. and Grants BFU2012-32532 and CONSOLIDER COAT CSD2009-00016 to M.A.A. from the Ministerio de Economía y Competitividad, Spain.
Abbreviations used:
- CMT
Charcot–Marie–Tooth
- DAD
diaphanous autoregulatory domain
- DID
diaphanous inhibitory domain
- DRF
diaphanous-related formin
- FH1/2
formin homology 1/2
- FSGS
focal segmental glomerulosclerosis
- GBD
GTPase-binding domain
- GFP
green fluorescent protein
- Glu MT
detyrosinated microtubule; IgG, immunoglobulin G
- LPA
lysophosphatidic acid
- MT
microtubule
- PLA
in situ proximity ligation
- siRNA
small interfering RNA
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0489) on March 30, 2016.
REFERENCES
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Amano M, Kaneko T, Maeda A, Nakayama M, Ito M, Yamauchi T, Goto H, Fukata Y, Oshiro N, Shinohara A, et al. Identification of Tau and MAP2 as novel substrates of Rho-kinase and myosin phosphatase. J Neurochem. 2003;87:780–790. doi: 10.1046/j.1471-4159.2003.02054.x. [DOI] [PubMed] [Google Scholar]
- Andres-Delgado L, Anton OM, Bartolini F, Ruiz-Saenz A, Correas I, Gundersen GG, Alonso MA. INF2 promotes the formation of detyrosinated microtubules necessary for centrosome reorientation in T cells. J Cell Biol. 2012;198:1025–1037. doi: 10.1083/jcb.201202137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres-Delgado L, Anton OM, Madrid R, Byrne JA, Alonso MA. Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood. 2010;116:5919–5929. doi: 10.1182/blood-2010-08-300665. [DOI] [PubMed] [Google Scholar]
- Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–867. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Formins and microtubules. Biochim Biophys Acta. 2010;1803:164–173. doi: 10.1016/j.bbamcr.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, Gundersen GG. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer O, Nevo F, Plaisier E, Funalot B, Gribouval O, Benoit G, Huynh Cong E, Arrondel C, Tete MJ, Montjean R, et al. INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med. 2011;365:2377–2388. doi: 10.1056/NEJMoa1109122. [DOI] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Goode BL. Formins at a glance. J Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Schlondorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, Higgs HN, Henderson JM, Pollak MR. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42:72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Cook TA, Nagasaki T, Gundersen GG. Rho guanosine triphosphatase mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J Cell Biol. 1998;141:175–185. doi: 10.1083/jcb.141.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13:539–553. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou P, Hasan S, Breitsprecher D, Baudelet E, Camoin L, Audebert S, Goode BL, Badache A. Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol Biol Cell. 2014;25:658–668. doi: 10.1091/mbc.E13-08-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destaing O, Saltel F, Gilquin B, Chabadel A, Khochbin S, Ory S, Jurdic P. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- Drabek K, van Ham M, Stepanova T, Draegestein K, van Horssen R, Sayas CL, Akhmanova A, Ten Hagen T, Smits R, Fodde R, et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr Biol. 2006;16:2259–2264. doi: 10.1016/j.cub.2006.09.065. [DOI] [PubMed] [Google Scholar]
- Eng CH, Huckaba TM, Gundersen GG. The formin mDia regulates GSK3beta through novel PKCs to promote microtubule stabilization but not MTOC reorientation in migrating fibroblasts. Mol Biol Cell. 2006;17:5004–5016. doi: 10.1091/mbc.E05-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukata M, Nakagawa M, Kaibuchi K. Roles of Rho-family GTPases in cell polarisation and directional migration. Curr Opin Cell Biol. 2003;15:590–597. doi: 10.1016/s0955-0674(03)00097-8. [DOI] [PubMed] [Google Scholar]
- Gaillard J, Ramabhadran V, Neumanne E, Gurel P, Blanchoin L, Vantard M, Higgs HN. Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol Biol Cell. 2011;22:4575–4587. doi: 10.1091/mbc.E11-07-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Goulimari P, Kitzing TM, Knieling H, Brandt DT, Offermanns S, Grosse R. Galpha12/13 is essential for directed cell migration and localized Rho-Dia1 function. J Biol Chem. 2005;280:42242–42251. doi: 10.1074/jbc.M508690200. [DOI] [PubMed] [Google Scholar]
- Goulimari P, Knieling H, Engel U, Grosse R. LARG and mDia1 link Galpha12/13 to cell polarity and microtubule dynamics. Mol Biol Cell. 2008;19:30–40. doi: 10.1091/mbc.E06-11-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Gomes ER, Wen Y. Cortical control of microtubule stability and polarization. Curr Opin Cell Biol. 2004;16:106–112. doi: 10.1016/j.ceb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- Infante AS, Stein MS, Zhai Y, Borisy GG, Gundersen GG. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113:3907–3919. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
- Kim H, White CD, Li Z, Sacks DB. Salmonella enterica serotype Typhimurium usurps the scaffold protein IQGAP1 to manipulate Rac1 and MAPK signalling. Biochem J. 2011;440:309–318. doi: 10.1042/BJ20110419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat Neurosci. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- Kreitzer G, Liao G, Gundersen GG. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol Biol Cell. 1999;10:1105–1118. doi: 10.1091/mbc.10.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Lin SX, Gundersen GG, Maxfield FR. Export from pericentriolar endocytic recycling compartment to cell surface depends on stable, detyrosinated (glu) microtubules and kinesin. Mol Biol Cell. 2002;13:96–109. doi: 10.1091/mbc.01-05-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid R, Aranda JF, Rodriguez-Fraticelli AE, Ventimiglia L, Andres-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gomez S, Jimenez A, et al. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Maiti S, Michelot A, Gould C, Blanchoin L, Sokolova O, Goode BL. Structure and activity of full-length formin mDia1. Cytoskeleton. 2012;69:393–405. doi: 10.1002/cm.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Bartolini F, Okada K, Wen Y, Gundersen GG, Goode BL. Regulated binding of adenomatous polyposis coli protein to actin. J Biol Chem. 2007;282:12661–12668. doi: 10.1074/jbc.M610615200. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Bartolini F, Deaconescu AM, Moseley JB, Dogic Z, Grigorieff N, Gundersen GG, Goode BL. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–1096. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Machius M, Rosen MK. Crystal structure of the Formin mDia1 in autoinhibited conformation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012896. e12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol. 2001a;3:723–729. doi: 10.1038/35087035. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Eng CH, Schlaepfer DD, Marcantonio EE, Gundersen GG. Localized stabilization of microtubules by integrin- and FAK-facilitated Rho signaling. Science. 2004;303:836–839. doi: 10.1126/science.1091325. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, Gundersen GG. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001b;11:1536–1541. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- Quinlan ME. Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development. 2013;140:4417–4425. doi: 10.1242/dev.097337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V, Gurel PS, Higgs HN. Mutations to the formin homology 2 domain of INF2 protein have unexpected effects on actin polymerization and severing. J Biol Chem. 2012;287:34234–34245. doi: 10.1074/jbc.M112.365122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Ritter AT, Angus KL, Griffiths GM. The role of the cytoskeleton at the immunological synapse. Immunol Rev. 2013;256:107–117. doi: 10.1111/imr.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Nieves AE, Johndrow JE, Keller LC, Magie CR, Pinto-Santini DM, Parkhurst SM. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nat Cell Biol. 2006;8:367–376. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Johnson EA, Vizcarra CL, Bois JS, Quinlan ME. Interaction between microtubules and the Drosophila formin Cappuccino and its effect on actin assembly. J Biol Chem. 2014;289:4395–4404. doi: 10.1074/jbc.M113.499921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sun H, Schlondorff J, Higgs HN, Pollak MR. Inverted formin 2 regulates actin dynamics by antagonizing Rho/diaphanous-related formin signaling. J Am Soc Nephrol. 2013;24:917–929. doi: 10.1681/ASN.2012080834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Schlondorff JS, Brown EJ, Higgs HN, Pollak MR. Rho activation of mDia formins is modulated by an interaction with inverted formin 2 (INF2) Proc Natl Acad Sci USA. 2011;108:2933–2938. doi: 10.1073/pnas.1017010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston SF, Kulacz WA, Shaikh S, Lee JM, Copeland JW. The ability to induce microtubule acetylation is a general feature of formin proteins. PLoS One. 2012;7:e48041. doi: 10.1371/journal.pone.0048041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kakeno M, Matsui T, Harada T, Wang S, Itoh N, Sato K, Matsuzawa K, Iwamatsu A, et al. Phosphorylation of CLASP2 by GSK-3beta regulates its interaction with IQGAP1, EB1 and microtubules. J Cell Sci. 2009;122:2969–2979. doi: 10.1242/jcs.046649. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Webb RL, Zhou MN, McCartney BM. A novel role for an APC2-Diaphanous complex in regulating actin organization in Drosophila. Development. 2009;136:1283–1293. doi: 10.1242/dev.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- Wickstrom SA, Lange A, Hess MW, Polleux J, Spatz JP, Kruger M, Pfaller K, Lambacher A, Bloch W, Mann M, et al. Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev Cell. 2010;19:574–588. doi: 10.1016/j.devcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang F, Niu YJ, Liu HL, Rui R, Cui XS, Kim NH, Sun SC. Formin mDia1, a downstream molecule of FMNL1, regulates Profilin1 for actin assembly and spindle organization during mouse oocyte meiosis. Biochim Biophys Acta. 2015;1853:317–327. doi: 10.1016/j.bbamcr.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Formin-induced nucleation of actin filaments. Curr Opin Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.