Abstract
The regulators of complement activation cluster at chromosome 1q32 contains the complement factor H (CFH) and five complement factor H–related (CFHR) genes. This area of the genome arose from several large genomic duplications, and these low-copy repeats can cause genome instability in this region. Genomic disorders affecting these genes have been described in atypical hemolytic uremic syndrome, arising commonly through nonallelic homologous recombination. We describe a novel CFH/CFHR3 hybrid gene secondary to a de novo 6.3-kb deletion that arose through microhomology–mediated end joining rather than nonallelic homologous recombination. We confirmed a transcript from this hybrid gene and showed a secreted protein product that lacks the recognition domain of factor H and exhibits impaired cell surface complement regulation. The fact that the formation of this hybrid gene arose as a de novo event suggests that this cluster is a dynamic area of the genome in which additional genomic disorders may arise.
Keywords: complement, hemolytic uremic syndrome, genetic renal disease
The complement factor H (CFH) and complement factor H–related (CFHR1–CFHR5) genes reside in a 360-kb region in the regulators of complement activation (RCA) cluster on chromosome 1q32 (Figure 1A).1 This is an area of the genome that arose from several large genomic duplications,2,3 and these low-copy repeats can cause genome instability in this region.
Figure 1.
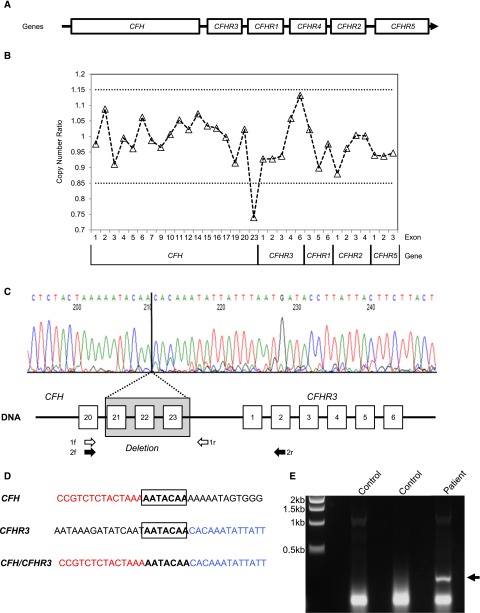
A 6.3-kb deletion in the RCA cluster results in a novel CFH/CFHR3 hybrid gene transcript. (A) Genomic organization of the RCA cluster region containing the CFH and CFHR genes. (B) MLPA of CFH, CFHR3, CFHR1, CFHR2, and CFHR5 showing the copy number ratio. The dotted lines represent ratios considered within the normal range. (C) Identification of breakpoint. Genomic DNA was amplified using a forward primer specific for CFH exon 20 (1f) and a reverse primer in the CFH 3′ region intergenic region (1r) and sequenced. The deletion of exons 21–23 of CFH is shown (shaded box), and the breakpoint is highlighted. (D) The CFH and CFHR3 sequence flanking the breakpoint. The 7-bp area of microhomology is shown (bold and boxed). (E) Confirmation of hybrid CFH/CFHR3 mRNA. A message for the hybrid CFH/CFHR3 gene was confirmed by amplifying patient and control cDNA with cross CFH and CFHR3 gene primers (black arrows 2f and 2r in C). The agarose gel shows amplified cDNA. The patient lane shows an amplified product consistent with a hybrid CFH/CFHR3 gene, which is not present in either control cDNA.
Mutations in CFH are the most common genetic predispositions to the thrombotic microangiopathy: atypical hemolytic uremic syndrome (aHUS).4 The majority of mutations in CFH occur at the C-terminal end responsible for host polyanion binding.5–7 Several of these CFH mutations (e.g., S1191L and V1197A) have been shown to be gene conversion events between CFH and CFHR1.8 Nonallelic homologous recombination involving CFH and CFHR1 can result in CFH/CFHR1 hybrid genes (Supplemental Figure 1).9,10 A single family with a hybrid CFH/CFHR3 gene (factor H [FH] protein complement control protein modules [CCPs] 1–19 and factor H–related 3 [FHR3] CCP1–CCP5) has been reported to arise through microhomology–mediated end joining (MMEJ)11 (Supplemental Figure 1). These genetic variants have all been shown to impair C3b/host polyanion binding on host cells, thus impairing local complement regulation.5,8,11
Recently, a reverse CFHR1/CFH hybrid gene arising through nonallelic homologous recombination has been described in aHUS. This encodes an FHR1, where the C-terminal CCPs are replaced by the C-terminal CCPs of FH.12,13 Unlike previously reported hybrid proteins, this does not impair FH cell surface binding but instead, acts as a competitive inhibitor of FH.13
In this study, we report a novel CFH/CFHR3 hybrid gene arising through MMEJ that impairs cell surface complement regulation.
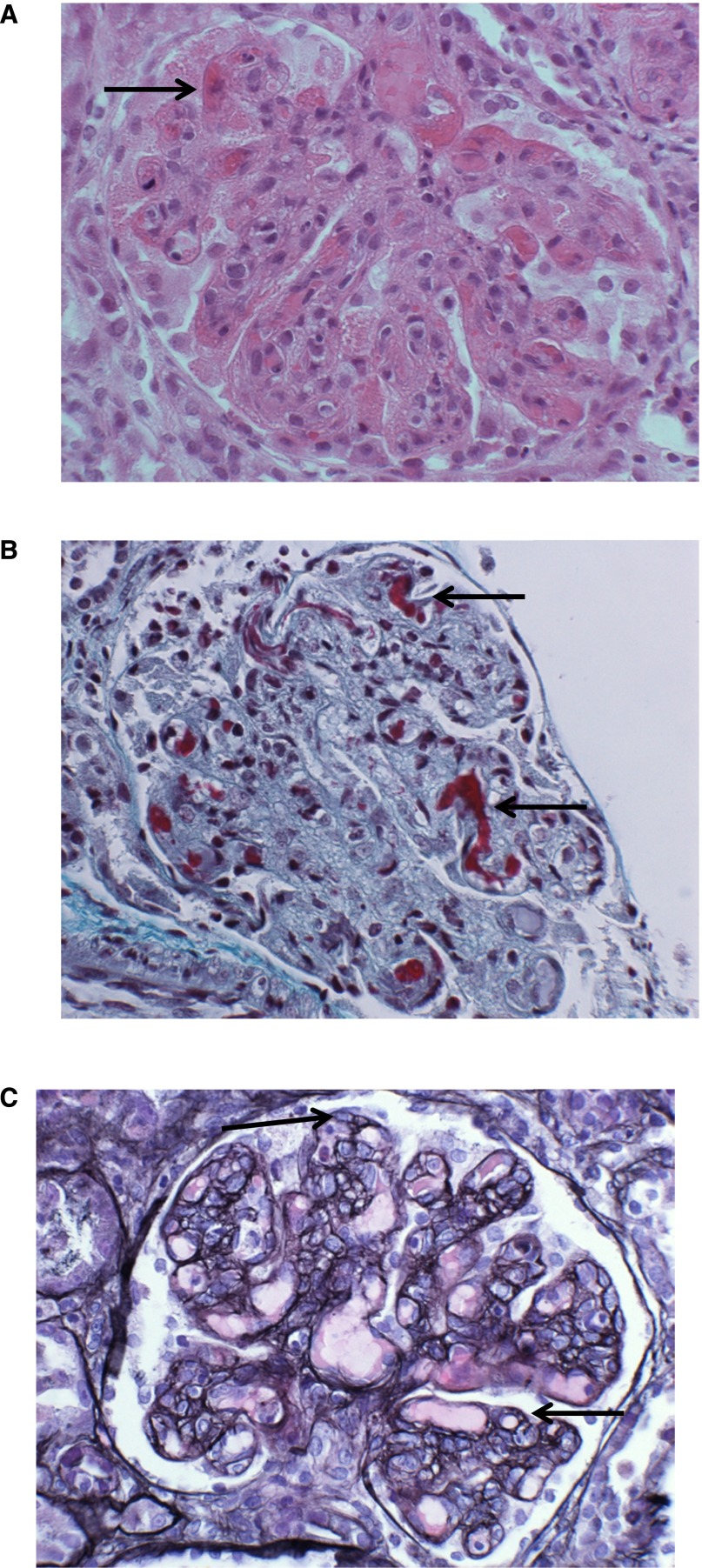
An 8-month-old boy presented with a diarrheal illness; on admission, creatinine was 52 μmol/L, and urea was 11.1 μmol/L. A blood film showed microangiopathic hemolytic anemia, and lactate dehydrogenase was 5747 units/L. Stool culture was positive for Escherichia coli O157:H7, and a diagnosis of Shiga toxin–producing E. coli (STEC) hemolytic uremic syndrome (HUS) was made. He did not require RRT and did not receive plasma exchange. He was discharged 2 weeks later with a creatinine of 107 μmol/L. He was readmitted 2 weeks later with an upper respiratory tract infection. Creatinine on readmission was 141 μmol/L, urea was 20.6 μmol/L, lactate dehydrogenase was 2398 units/L, and platelets were 128×109/L, with evidence of microangiopathic hemolytic anemia on blood film. This relapse suggested a diagnosis of aHUS rather than STEC HUS. Serum complement levels were within the normal range: C3, 1.05 g/L (0.68–1.80 g/L); C4, 0.30 g/L (0.18–0.60 g/L); and FH, 0.51 g/L (0.35–0.59 g/L). He commenced plasma exchange and required three sessions of hemofiltration. A renal biopsy confirmed a thrombotic microangiopathy (Figure 2). Creatinine returned to a baseline of approximately 100 μmol/L. He was treated with weekly plasma exchange for 3.5 years before starting Eculizumab, on which he has been maintained for 3 years. Creatinine is currently 200 μmol/L, and there have been no additional episodes of aHUS while on treatment with Eculizumab (Supplemental Figure 2).
Figure 2.
A renal biopsy from the patient demonstrating haemolytic uraemic syndrome. Renal biopsy of the patient showing thrombi (arrows) on capillary loops. (A) hematoxylin and eosin. (B) Masson trichrome. A developing membranoproliferative pattern of injury can be seen in (C) characterized by capillary loop double contours (arrows). Silver counterstained H&E, ×400.
Sanger sequencing of aHUS-associated genes (CFH, CFI, CFB, CD46, C3, THBD, and DGKE) did not reveal any mutations, although heterozygosity was found for the common Y402H polymorphism (rs1061170) in CFH. Multiplex ligation–dependent probe amplification (MLPA) of CFH and the CFHRs revealed a deletion in the CFH gene (Figure 1B). To define the extent of the deletion and the breakpoint, Sanger sequencing of genomic DNA was undertaken. This showed a 6.3-kb deletion extending from CFH intron 20 to the CFH 3′ intergenic region incorporating exons 21–23 of CFH (Figure 1C). Directly flanking the breakpoint was a 7-bp region of microhomology (Figure 1D). The deletion was not detected in either parent, suggesting that this was a de novo event.
We hypothesized that the loss of CFH exons 21–23 and the 3′ UTR of CFH would lead to aberrant splicing. In silico analysis showed that the next acceptor splice site following the CFH exon 20 donor site was 5′ of CFHR3 exon 2, thus potentially producing a transcript for a hybrid CFH/CFHR3 gene (Supplemental Figure 3). mRNA for this putative hybrid CFH/CFHR3 gene was confirmed by amplifying patient and control cDNA with CFH– and CFHR3–specific primers. This showed a product in the patient and not in healthy controls (Figure 1E).
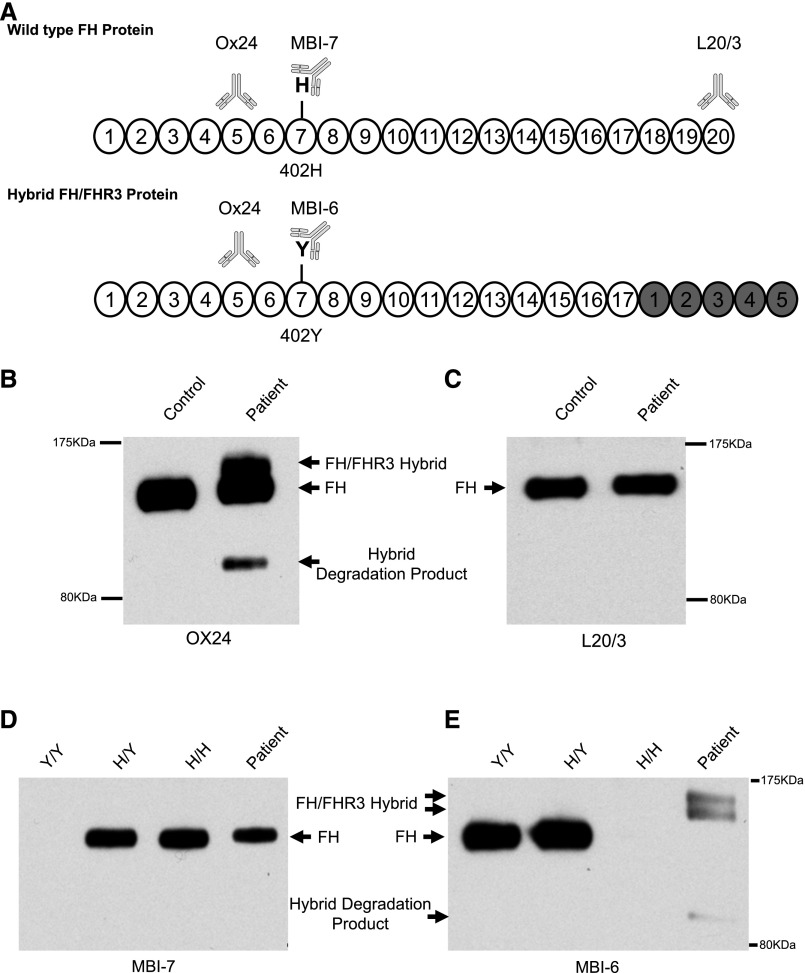
To confirm that this hybrid transcript led to the synthesis and secretion of a hybrid protein, Western blotting was performed using a series of epitope–defined anti–FH mAbs (Figure 3A). An initial blot probed with OX24, an FH CCP5–specific antibody, showed, in addition to FH, two additional bands only in the patient (Figure 3B). The species at approximately 160 kD was consistent with the predicted intact hybrid FH/FHR3. A species at approximately 120 kD was hypothesized to be a degradation product. No abnormal bands were detected in either parent (Supplemental Figure 4A). An FH C terminus–specific antibody (L20/3) did not reveal additional aberrant bands, indicating that these protein species lacked FH CCP20 (Figure 3C). Genotyping showed the patient to be heterozygous for the Y402H polymorphism in FH. This allowed the use of mAbs specific for histidine or tyrosine in CCP7 of FH.14 The normal allele’s product reacted to the histidine-specific mAb (MBI7) and gave a single band of approximately 150 kD, consistent with FH. The hybrid allele reacted to the tyrosine-specific mAb (MBI6), showing a doublet at approximately 160 kD (Figure 3D). FHR3 is known to be alternately glycosylated,15–17 and we hypothesize that the doublet is caused by an alternately glycosylated hybrid protein. Additionally, it can be seen that the presumed degradation product at 120 kD is only seen using the tyrosine-specific mAb, consistent with this only arising from the hybrid protein.
Figure 3.
Identification of a novel FH/FHR3 hybrid protein in patient serum. (A) The protein product of wild-type FH and the predicted FH/FHR3 hybrid protein consisting of CCP1–17 of FH and CCP1–5 of FHR3. The CCPs originating from FHR3 are highlighted in gray. The patient is heterozygous for a common polymorphism in CCP7 of FH (Y402H). Binding epitopes for the mAbs are shown: Ox24-CCP5, MBI7-CCP7 amino acid 402 histidine variant, MBI6-CCP7 amino acid 402 tyrosine variant, and L20/3-CCP20. (B) Serum Western blot using OX24 showing two additional bands in the patient in addition to FH. The upper band is consistent with the predicted size of the FH/FHR3 hybrid. (C) Serum Western blot using L20/3. When using this FH C terminus–specific antibody, no additional bands are seen, consistent with the hybrid protein lacking CCP18–20. (D) Serum Western blot using MBI7. This shows that the patient has a normal allele producing FH with the histidine at amino acid 402. (E) Serum Western blot using MBI6. The patient has three additional bands and no wild–type FH band. This is consistent with the FH/FHR3 hybrid protein carrying the tyrosine amino acid at position 402. The two additional upper bands represent differentially glycosylated hybrid protein, consistent with that seen in the native FHR3. There is a faint degradation product only detected with the MBI6 antibody. This is consistent with the breakdown product being generated only from the hybrid protein and suggests a less stable protein product. The controls were unaffected, unrelated, genotyped samples. In B and C, a sample from an individual heterozygous at Y402H was used. In D and E, samples from individuals homozygous for Y402 (Y/Y), homozygous for H402 (H/H), or heterozygous (Y/H) were used.
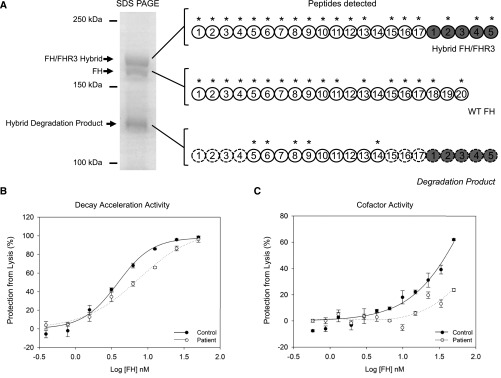
To confirm these findings, FH species were purified from serum using affinity chromatography with an OX24 mAb. These were separated by a 6% SDS-PAGE before trypsin digestion. Mass spectrometry was then carried out on these three purified bands (Figure 4A). Peptide species identified from the approximately 160-kD band confirmed that this was a hybrid FH/FHR3 protein. Protein fragments from the 150-kD band were consistent with FH. Fragments from CCP5, CCP6, CCP8, CCP9, and CCP14 of FH were seen in the band at approximately 120 kD. Coverage at CCP7 was insufficient to determine whether there was a tyrosine or histidine at position 402. The Western blot analysis and mass spectrometry data together show that breakdown products from only the hybrid protein are seen in serum.
Figure 4.
Impaired cell surface co-factor and decay acceleration activity of hybrid FH/FHR3 hybrid protein. (A) Mass spectrometry of purified proteins. The FH, FH/FHR3 hybrid, and degradation product were purified using affinity chromatography with an OX24 column. These FH species were separated by 6% SDS-PAGE, stained using Coomassie, cut from the gel (left panel), and submitted for trypsin digest and mass spectrometry. Peptides sequences identified using mass spectrometry are indicated with asterisks. The peptides detected in the hybrid degradation product by mass spectrometry are annotated on a full–length FH/FHR3 hybrid protein. CCPs that cannot be directly inferred from mass spectrometry data are outlined with a dashed line. A molecular mass of approximately 120 kD is consistent with an approximately 17 CCP protein. (B) Decay acceleration assays on sheep erythrocytes. The purified FH/FHR3 hybrid from the patient showed impaired cell surface complement regulation compared with wild type (WT) FH purified from control. Alternative pathway convertase (C3bBb) was formed on sheep erythrocytes. Cells were incubated for 15 minutes with dilutions of purified FH/FHR3 hybrid and WT FH before triggering lysis with NHSΔBΔH. Maximal lysis occurs in buffer-only (0 mM FH) conditions. Addition of WT FH caused decay of the C3 convertase and decay of convertase resulting in inhibition of lysis. The FH/FHR3 hybrid was up to 2-fold less efficient at inhibiting lysis. (C) C3b cofactor activity on sheep erythrocytes. WT FH and purified FH/FHR3 hybrid were tested for the ability to act as a cofactor for factor I–catalyzed inactivation of C3b deposited on the surfaces of sheep erythrocytes. The C3 convertase (C3bBb) was formed on residual C3b, and lysis was triggered by adding NHSΔBΔH. Maximal lysis occurs in the presence of buffer only (0 mM FH). The addition of WT FH and factor I produces iC3b, which decreases convertase formation and subsequent lysis, and this is shown as increasing amounts of inhibition of lysis (expressed as percentage of maximal lysis) after incubation with factor I and WT FH (black circles) or FH/FHR3 hybrid (white circles). The FH/FHR3 hybrid can be seen to be markedly less active than WT.
To examine the functional significance, the FH/FHR3 hybrid protein was purified from serum using affinity chromatography with an MBI6 mAb followed by gel filtration (Supplemental Figure 4B). The FH/FHR3 hybrid protein displayed both impaired cell surface decay acceleration (Figure 4B) and cofactor activity (Figure 4C, Supplemental Figure 5).
Thus, in this study, we show a deletion in the RCA cluster resulting in a novel de novo CFH/CFHR3 hybrid gene. Microhomology in the sequence flanking the breakpoint suggests that the deletion has occurred through MMEJ. Genomic disorders affecting CFH and the CFHRs as a result of nonallelic homologous recombination are found in approximately 4.5% of patients with aHUS. In contrast, only one patient with a genomic disorder secondary to MMEJ has been described.11
We have shown that the product of this gene, a 22 CCP domain protein, is secreted, albeit with degradation fragments present in the serum, suggesting impaired stability of the protein. Functional analysis of the purified FH/FHR3 protein showed impaired cell surface complement regulation.
This FH/FHR3 hybrid protein is similar to that described by Francis et al.,11 in that in both, the C-terminal end of FH is replaced by all five CCPs of FHR3 (Supplemental Figure 1). Although both hybrids lack CCP20 of FH responsible for cell surface protection, the hybrid reported here also lacks CCP18 and CCP19.
The functional role of FHR3 is unclear, with various regulatory activities being suggested.16,18 Unsurprisingly, given its high homology with CCP7 of FH, FHR3 binds to heparin.16 FHR3 also binds to C3b and C3d through CCP4 and CCP5.16,19 Competition between FHR3 and FH for surface-bound C3b has been described.18 The hybrid described by Francis et al.11 showed normal fluid–phase complement regulatory activity but a profoundly reduced cell surface complement regulatory activity.
The fact that such an FH/FHR3 hybrid should lack cell surface regulatory activity is not surprising. Both of these hybrids lack the CCP20 domain of FH responsible for cell surface localization.6 Additionally, structural analysis has revealed a specific hairpin structure suggesting a model whereby cell surface–bound C3b is engaged by both CCP1–4 and CCP19 and CCP20 of FH concurrently.20 The elongation of FH by the addition of extra CCPs would prevent such an orientation.
Although the initial presentation of disease was seen in association with STEC, the rapid relapse suggested aHUS rather than STEC HUS, and this was confirmed by the finding of the CFH/CFHR3 hybrid gene. In individuals carrying mutations in complement genes, additional triggers (e.g., pregnancy21 and infection22) are required for disease to manifest.4,22 In this case, STEC served to unmask the genetic predisposition to disease, such as in two previously reported patients with STEC-triggered aHUS.23
In summary, we describe a deletion in the RCA cluster arising through MMEJ that results in a novel CFH/CFHR3 hybrid gene. The fact that this arose as a de novo event suggests that this is a dynamic area of the genome where we should expect additional genomic disorders.
Concise Methods
The study was approved by Newcastle and North Tyneside 1 Research Ethics Committee, and informed consent was obtained in accordance with the Declaration of Helsinki.
Complement Assays
C3 and C4 levels were measured by rate nephelometry (Beckman Coulter Array 360). FH levels were measured by radial immunodiffusion (Binding Site). Screening for FH autoantibodies was undertaken using ELISA as described previously.24
Genetic Analyses and MLPA
Mutation screening of CFH,25 CFI,26 CFB,27 MCP,28 C3,29 and DGKE30 was undertaken using Sanger sequencing as previously described. Screening for genomic disorders affecting CFH, CFHR1, CFHR2, CFHR3, and CFHR5 was undertaken using MLPA in both the affected individual and 500 normal controls as previously described.11 A proprietary kit from MRC Holland (SALSA MLPA Kit P236-A1 ARMD; www.mlpa.com) was supplemented by specific in–house CFH probes (Supplemental Material). GeneMarker software (Version 1.90) was used to calculate dosage quotients.
Genomic Breakpoint Analyses
To identify the breakpoint of the deletion that resulted in the CFH/CFHR3 hybrid gene, genomic DNA was amplified using a forward primer specific for CFH in exon 20 and a reverse primer in the CFH 3′ region (Figure 1C). The sequence of the forward primer was GTAACTGTTATCAGTTGATTTGC, and the reverse was ACGGATTGCATGTATAAGTG. The product was then sequenced using direct fluorescent sequencing.
Confirmation of CFH/CFHR3 RNA Product
mRNA was extracted from peripheral blood lymphocytes of the patient, and cDNA was prepared. This was amplified using a forward primer in CFH exon 20 (TGGATGGAGCCAGTAATGTAACATGCAT) and a reverse primer in CFHR3 exon 2 (GAAATAGACCTCCATGTTTAATGTCTG) (Figure 1C). The PCR product was detected in an ethidium bromide–stained 1.5% agarose gel.
Western Blotting
Detection of abnormal protein products in serum arising as a consequence of the deletion was undertaken by Western blotting. Sera were diluted 1:100, and under nonreducing conditions, they were electrophoresed on 6% SDS-PAGE gel and transferred onto nitrocellulose. mAbs of known specificity to FH (OX24, CCP5, L20/3, CCP20, MBI6, CCP7 402Y, MBI7 CCP7, and 402H) were used with sheep anti–mouse Ig HRP (Jackson Immuno Research) at dilutions of 1:2000 (primary antibody) and 1:4000 (secondary antibody) (Supplemental Material). After washes in 1× TBST, blots were developed using Pierce ECL Western Blotting Substrate (Thermo Scientific).
Purification of FH Species
FH species (wild-type FH from normal allele, FH/FHR3 hybrid, and FH/FHR3 breakdown product all with CCP5 of FH) were purified from serum with affinity chromatography using immobilized mAb to FH (OX24) on a 1-ml HiTrap NHS HP Column (GE Healthcare). After washing with PBS, the bound proteins were then eluted using 0.1 M glycine (pH 2.7). The eluted material was pooled and concentrated for analysis by mass spectrometry.
The FH/FHR3 hybrid protein was purified from patient serum with affinity chromatography using immobilized mAb to FH CCP7 402Y (MBI6)31 on a 1-ml HiTrap NHS HP Column (GE Healthcare). After washing with PBS, the bound proteins were eluted using PBS/diethylamine (50 mM). Subsequently, the FH/FHR3 protein was purified from its degradation product and FHL1 using a Superdex 200 Size Exclusion Column. We also purified wild–type FH protein from a healthy control. FH or FH/FHR3 hybrid protein purified using the MBI6 column was used for cofactor and decay acceleration assays.
Mass Spectrometry
A 6% SDS-PAGE was run, and the three bands identified by Coomassie staining were excised from the gel as indicated in Figure 4A. Trypsin digest and mass spectrometry were then undertaken (Supplemental Material).
Cell Surface Decay Acceleration Assays
Decay acceleration on sheep erythrocytes was performed as previously described32 and in Supplemental Material. Briefly, FH/FHR3 hybrid from the patient and FH from controls were purified by immunoaffinity chromatography as above and gel filtered into PBS. Alternative pathway convertase (C3bBb) was formed on sheep erythrocytes. Cells were incubated for 15 minutes with 1.24–50 nM patient FH/FHR3 hybrid or control FH. The molar concentration of the purified patient FH/FHR3 was estimated using the extinction coefficient (272,170 M cm−1), whereas the extinction coefficient of FH (246,800 M cm−1) was used for the control. Lysis was initiated with 4% normal human serum depleted of factor B and FH (NHSΔBΔH). Maximal lysis was achieved by adding NHSΔBΔH to no FH wells (buffer only). To determine the amount of lysis, cells were pelleted by centrifugation, and hemoglobin release was measured at 420 nm (A420). Percentage of inhibition of lysis in the presence of increasing concentrations was defined as (A420[buffer only] − A420[FH])/A420[buffer only] ×100%.
Cell Surface Cofactor Assays
Cofactor activity on sheep erythrocytes was performed as previously described.32 Briefly washed C3b–coated sheep erythrocyte cells were resuspended in AP buffer and incubated with an equal volume of a range of concentrations of FH/FHR3 hybrid and wild-type FH and 2.5 μg/ml factor I (CompTech) for 8 minutes at 25°C. After three washes in AP buffer, AP convertase was formed on the remaining C3b. Lysis was initiated with 4% NHSΔBΔH. Again, cells were pelleted, and hemoglobin release was measured at 420 nm. Percentage of inhibition from lysis was calculated by the formula (A420[buffer only] − A420[FH])/A420[buffer only] ×100%.
Disclosures
A.A. has received fees from Alexion Pharmaceuticals for lectures. C.L.H. is also employed by GlaxoSmithKline and has shares in this company. Newcastle University has received fees from Alexion Pharmaceuticals for lectures and consultancy undertaken by T.H.J.G. and D.K.
Supplementary Material
Acknowledgments
We thank Achim Treumann for technical help with mass spectrometry.
The research leading to these results has received funding from European Union’s Seventh Framework Programme FP7/2007-2013 Grant 305608 (EURenOmics). Funding for this study was provided by United Kingdom Medical Research Council Grant G0701325. G.S.R.A. is funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico. E.K.S.W. is a Medical Research Council Clinical Training Fellow. D.K. is a Wellcome Trust Intermediate Clinical Fellow.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2015010100/-/DCSupplemental.
References
- 1.Díaz-Guillén MA, Rodríguez de Córdoba S, Heine-Suñer D: A radiation hybrid map of complement factor H and factor H-related genes. Immunogenetics 49: 549–552, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL: Complement factor H: Sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 37: 41–52, 2000 [DOI] [PubMed] [Google Scholar]
- 3.Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S: The factor H protein family. Immunopharmacology 42: 53–60, 1999 [DOI] [PubMed] [Google Scholar]
- 4.Kavanagh D, Goodship TH, Richards A: Atypical hemolytic uremic syndrome. Semin Nephrol 33: 508–530, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira VP, Herbert AP, Cortés C, McKee KA, Blaum BS, Esswein ST, Uhrín D, Barlow PN, Pangburn MK, Kavanagh D: The binding of factor H to a complex of physiological polyanions and C3b on cells is impaired in atypical hemolytic uremic syndrome. J Immunol 182: 7009–7018, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt CQ, Herbert AP, Kavanagh D, Gandy C, Fenton CJ, Blaum BS, Lyon M, Uhrín D, Barlow PN: A new map of glycosaminoglycan and C3b binding sites on factor H. J Immunol 181: 2610–2619, 2008 [DOI] [PubMed] [Google Scholar]
- 7.Blaum BS, Hannan JP, Herbert AP, Kavanagh D, Uhrín D, Stehle T: Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat Chem Biol 11: 77–82, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Heinen S, Sanchez-Corral P, Jackson MS, Strain L, Goodship JA, Kemp EJ, Skerka C, Jokiranta TS, Meyers K, Wagner E, Robitaille P, Esparza-Gordillo J, Rodriguez de Cordoba S, Zipfel PF, Goodship TH: De novo gene conversion in the RCA gene cluster (1q32) causes mutations in complement factor H associated with atypical hemolytic uremic syndrome. Hum Mutat 27: 292–293, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, Pirson Y, Jackson MS, Hughes A, Wood KM, Goodship JA, Goodship TH: Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med 3: e431, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maga TK, Meyer NC, Belsha C, Nishimura CJ, Zhang Y, Smith RJ: A novel deletion in the RCA gene cluster causes atypical hemolytic uremic syndrome. Nephrol Dial Transplant 26: 739–741, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis NJ, McNicholas B, Awan A, Waldron M, Reddan D, Sadlier D, Kavanagh D, Strain L, Marchbank KJ, Harris CL, Goodship TH: A novel hybrid CFH/CFHR3 gene generated by a microhomology-mediated deletion in familial atypical hemolytic uremic syndrome. Blood 119: 591–601, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Eyler SJ, Meyer NC, Zhang Y, Xiao X, Nester CM, Smith RJ: A novel hybrid CFHR1/CFH gene causes atypical hemolytic uremic syndrome. Pediatr Nephrol 28: 2221–2225, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valoti E, Alberti M, Tortajada A, Garcia-Fernandez J, Gastoldi S, Besso L, Bresin E, Remuzzi G, Rodriguez de Cordoba S, Noris M: A novel atypical hemolytic uremic syndrome-associated hybrid CFHR1/CFH gene encoding a fusion protein that antagonizes factor H-dependent complement regulation. J Am Soc Nephrol 26: 209–219, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakobyan S, Tortajada A, Harris CL, de Córdoba SR, Morgan BP: Variant-specific quantification of factor H in plasma identifies null alleles associated with atypical hemolytic uremic syndrome. Kidney Int 78: 782–788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skerka C, Kühn S, Günther K, Lingelbach K, Zipfel PF: A novel short consensus repeat-containing molecule is related to human complement factor H. J Biol Chem 268: 2904–2908, 1993 [PubMed] [Google Scholar]
- 16.Hellwage J, Jokiranta TS, Koistinen V, Vaarala O, Meri S, Zipfel PF: Functional properties of complement factor H-related proteins FHR-3 and FHR-4: Binding to the C3d region of C3b and differential regulation by heparin. FEBS Lett 462: 345–352, 1999 [DOI] [PubMed] [Google Scholar]
- 17.Skerka C, Chen Q, Fremeaux-Bacchi V, Roumenina LT: Complement factor H related proteins (CFHRs). Mol Immunol 56: 170–180, 2013 [DOI] [PubMed] [Google Scholar]
- 18.Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Köhl J, Zipfel PF, Weber BH, Skerka C: An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet 19: 4694–4704, 2010 [DOI] [PubMed] [Google Scholar]
- 19.Hellwage J, Jokiranta TS, Friese MA, Wolk TU, Kampen E, Zipfel PF, Meri S: Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol 169: 6935–6944, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Morgan HP, Schmidt CQ, Guariento M, Blaum BS, Gillespie D, Herbert AP, Kavanagh D, Mertens HD, Svergun DI, Johansson CM, Uhrín D, Barlow PN, Hannan JP: Structural basis for engagement by complement factor H of C3b on a self surface. Nat Struct Mol Biol 18: 463–470, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fakhouri F, Roumenina L, Provot F, Sallée M, Caillard S, Couzi L, Essig M, Ribes D, Dragon-Durey MA, Bridoux F, Rondeau E, Frémeaux-Bacchi V: Pregnancy-associated hemolytic uremic syndrome revisited in the era of complement gene mutations. J Am Soc Nephrol 21: 859–867, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caprioli J, Noris M, Brioschi S, Pianetti G, Castelletti F, Bettinaglio P, Mele C, Bresin E, Cassis L, Gamba S, Porrati F, Bucchioni S, Monteferrante G, Fang CJ, Liszewski MK, Kavanagh D, Atkinson JP, Remuzzi G International Registry of Recurrent and Familial HUS/TTP : Genetics of HUS: The impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood 108: 1267–1279, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberti M, Valoti E, Piras R, Bresin E, Galbusera M, Tripodo C, Thaiss F, Remuzzi G, Noris M: Two patients with history of STEC-HUS, posttransplant recurrence and complement gene mutations. Am J Transplant 13: 2201–2206, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth SJ, Holmes LV, Ward R, Morgan L, Goodship TH, Marchbank KJ: Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 115: 379–387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH: Factor H mutations in hemolytic uremic syndrome cluster in exons 18-20, a domain important for host cell recognition. Am J Hum Genet 68: 485–490, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship TH: Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 2150–2155, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Kavanagh D, Kemp EJ, Richards A, Burgess RM, Mayland E, Goodship JA, Goodship TH: Does complement factor B have a role in the pathogenesis of atypical HUS? Mol Immunol 43: 856–859, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Müslümanoğlu MH, Kavukcu S, Filler G, Pirson Y, Wen LS, Atkinson JP, Goodship TH: Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc Natl Acad Sci U S A 100: 12966–12971, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frémeaux-Bacchi V, Miller EC, Liszewski MK, Strain L, Blouin J, Brown AL, Moghal N, Kaplan BS, Weiss RA, Lhotta K, Kapur G, Mattoo T, Nivet H, Wong W, Gie S, Hurault de Ligny B, Fischbach M, Gupta R, Hauhart R, Meunier V, Loirat C, Dragon-Durey MA, Fridman WH, Janssen BJ, Goodship TH, Atkinson JP: Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood 112: 4948–4952, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaire M, Frémeaux-Bacchi V, Schaefer F, Choi M, Tang WH, Le Quintrec M, Fakhouri F, Taque S, Nobili F, Martinez F, Ji W, Overton JD, Mane SM, Nürnberg G, Altmüller J, Thiele H, Morin D, Deschenes G, Baudouin V, Llanas B, Collard L, Majid MA, Simkova E, Nürnberg P, Rioux-Leclerc N, Moeckel GW, Gubler MC, Hwa J, Loirat C, Lifton RP: Recessive mutations in DGKE cause atypical hemolytic-uremic syndrome. Nat Genet 45: 531–536, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hakobyan S, Harris CL, Tortajada A, Goicochea de Jorge E, García-Layana A, Fernández-Robredo P, Rodríguez de Córdoba S, Morgan BP: Measurement of factor H variants in plasma using variant-specific monoclonal antibodies: Application to assessing risk of age-related macular degeneration. Invest Ophthalmol Vis Sci 49: 1983–1990, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Wong EK, Anderson HE, Herbert AP, Challis RC, Brown P, Reis GS, Tellez JO, Strain L, Fluck N, Humphrey A, Macleod A, Richards A, Ahlert D, Santibanez-Koref M, Barlow PN, Marchbank KJ, Harris CL, Goodship TH, Kavanagh D: Characterization of a factor H mutation that perturbs the alternative pathway of complement in a family with membranoproliferative GN. J Am Soc Nephrol 25: 2425–2433, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.