Abstract
Myeloid-derived suppressor cells (MDSCs) play critical roles in primary and metastatic cancer progression. While MDSC regulation is widely variable even within patients harboring the same type of malignancy, the mechanisms governing such heterogeneity are largely unknown. Here, integrating human tumor genomics and syngeneic mammary tumor models, we demonstrate that mTOR signaling in cancer cells dictates a mammary tumor’s ability to stimulate MDSC accumulation through regulating G-CSF. Inhibiting this pathway or its activators (e.g., FGFR) impairs tumor progression, which is partially rescued by restoring MDSCs or G-CSF. Tumor-initiating cells (TICs) exhibit elevated G-CSF. MDSCs reciprocally increase TIC frequency through activating Notch in tumor cells, forming a feed-forward loop. Analyses of primary breast cancers and patient-derived xenografts (PDXs) corroborate these mechanisms in patients. These findings establish a non-canonical oncogenic role of mTOR signaling in recruiting pro-tumorigenic MDSCs and show how defined cancer subsets may evolve to promote and depend upon a distinct immune microenvironment.
Myeloid derived suppressor cells (MDSCs) are a heterogeneous population defined as CD11b+Gr1+ cells. They can be roughly divided into granulocytic and monocytic subsets using Ly6G and Ly6C as markers, respectively. Both CD11b+Ly6G+ and CD11b+Ly6C+ cells have immunosuppressive activities, although different mechanisms may be utilized. The granulocytic subset is more often found expanded in tumor models and is involved in promoting tumor progression1–4, although anti-tumor effects have also been observed5. In the clinic, MDSCs were first identified in the peripheral blood of cancer patients as non-lymphoid hematopoietic suppressor cells6 that have been subsequently shown to increase during progression in many cancer types7, 8. Similar to mouse, most human MDSCs carry markers of immature myeloid lineage cells and qualify as either granulocytic (CD11b+CD33+CD15+HDLAlow) or monocytic (CD11b+CD14+HDLAlow) subsets3, 4, 9.
Considerable information has been obtained about the biogenesis and functions of MDSCs. The cytokines responsible for MDSC accumulation include G-CSF10–14, GM-CSF15, IL1β16, 17, IL618, PGE219, IFNγ20, IL421, and VEGF22. The immunosuppressive mechanisms utilized by MDSCs involve secretion of TGFβ, generation of nitric oxide and reactive oxygen species, and metabolic depletion of L-arginine by arginase 11, 2. These activities can blunt cytotoxicity, block proliferation, or induce apoptosis of cytotoxic T lymphocytes and natural killer cells. Other MDSC functions include formation of a pre-metastatic niche23, enhancement of tumor invasion24, 25 and stimulation of angiogenesis25.
Despite this knowledge, we have a limited understanding of why and how individual tumors vary widely in their propensity to induce MDSCs. Here, we demonstrate that this propensity is determined by an oncogenic signaling pathway and linked to the subpopulation of tumor initiating cells (TICs).
RESULTS
Inter-tumoral heterogeneity of MDSC infiltration
We examined myeloid cells in a variety of syngeneic mammary tumor models of diverse genetic backgrounds and tumorigenic drivers. MMTV-WNT1, MMTV-WNT1-iFGFR, and P53-PTEN double-knockout (DKO) are genetically engineered mouse models in the FVB background. MMTV-WNT1 is a widely used model of basal-like tumors. WNT1-iFGFR is a bigenic model based on MMTV-WNT1, in which FGFR signaling can be inducibly activated26, 27. The P53-PTEN DKO was generated by conditional deletion of these tumor suppressors using a MMTV-driven Cre. P53N tumor lines initially arose from transplanted P53-null mammary gland tissues in Balb/c mice, and are maintained through mouse-to-mouse orthotopic transplantation. Despite the common loss of P53, P53N lines exhibited remarkable inter-tumoral heterogeneity in genomic copy number, gene expression profiles, and TIC frequencies28. The 67NR-4TO7-4T1 series are established cell lines derived from a spontaneous tumor in Balb/c mice. Taken together, these reagents provide an unbiased representation of currently available syngeneic models.
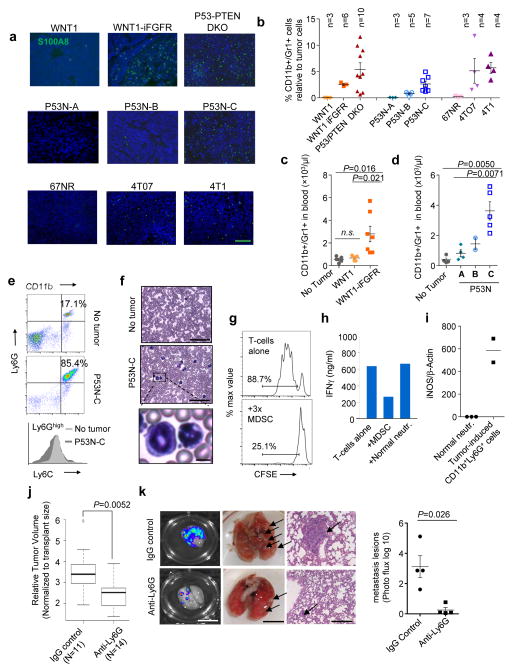
Mammary tumors were generated either by spontaneous tumorigenesis (MMTV-WNT1, WNT1-iFGFR, and P53-PTEN DKO), or orthotopic transplantation of primary tumor tissues (P53N series) or cell suspensions (67NR, 4TO7, and 4T1). When tumors reached 1 cm3, we examined myeloid cell infiltration by immunofluorescence staining of S100A8, which was predominantly expressed by CD11b+Gr1+ cells rather than tumor cells (Supplementary Fig. 1b). Significant inter-tumoral heterogeneity was discovered (Fig. 1a and Supplementary Fig. 1a). Quantification of CD11b+Gr1+ cells in dissociated tumors was largely consistent with S100A8 staining (Fig. 1b). The accumulation of CD11b+Gr1+ cells is systemic, as they were also found in peripheral blood and the frequencies closely correlated with the tumor-infiltrating myeloid cells (Fig. 1c,d). Overall, the inter-model variations of CD11b+Gr1+ cells were as high as 50- to 100-fold. In contrast, tumors of the same model exhibited only a 2- to 5-fold variation (Fig. 1b). Thus, the local and systemic accumulation of CD11b+Gr1+ cells appears to be a stable trait of each tumor line.
Figure 1. Inter-tumoral heterogeneity of MDSC infiltration.
a. Representative immunofluorescence staining of S100A8 in indicated tumors. Green=S100A8, Blue=DAPI (nucleus), scale bar: 100μm.
b. Flow cytometry quantification of tumor-infiltrating CD11b+Gr1+ cells. Animal numbers in each model are indicated. Five independent experiments were performed with consistent results and one representative one is shown.
c–d. Flow cytometry quantification of CD11b+Gr1+ cells in peripheral blood. Animal numbers: c. tumor-free (TF): n=4, MMTV-Wnt1: n=5, MMTV-Wnt1 iFGFR: n=7. d. TF: n=4, P53N-A: n=4, P53N-B: n=2, P53N-C: n=5. Three independent experiments were performed with consistent results and a representative experiment is shown.
e. FACS analyses show that CD11b+ cells in blood of P53N-C tumor-bearing mice express Ly6C at a lower and more variable level compared to normal neutrophils. Upper: Representative data of Ly6GhighCD11b+ leukocytes in TF and P53N-C tumor-bearing mice. Lower: Ly6C histogram of CD11b+Ly6G+ cells in P53N-C tumor-bearing (dark) or TF (light) mice. Each panel is representative of 6 mice per group.
f. H&E staining of blood cells in P53N-C tumor-bearing mice compared to TF control, scale bars, 50μm. Bottom: Representative abnormal granulocytic cells. Scale bar, 5μm.
g. In vitro T-cell proliferation, measured by CFSE assay, is inhibited by MDSCs. Left: histograms of CFSE levels in CD3- and IL-2 stimulated T-cells with or without co-culture of MDSCs at 1:3 ratio. See Supplementary Fig. 1 for quantification.
h. IFNγ concentration in conditioned medium of T-cells cultured with and without MDSCs or normal neutrophils.
In g–h, triplicate wells of each group were analyzed. Two independent experiments were conducted with representative results shown.
i. iNOS expression in MDSCs and normal neutrophils by real-time qPCR. Data derived from 3 and 2 animals, respectively.
j. Box-whisker plot of mammary tumor size in animals treated with IgG control and Ly6G depletion-antibody, respectively.
k. Pulmonary metastasis. From left: representative IVIS images, visible lesions, and H&E staining of lung metastases in control and anti-Ly6G groups and bioluminescence quantification. n=4 animals per group, selected from larger group to have similar orthotopic tumor sizes. Scale bars, 10μm, 5μm, and 50μm from left to right.
In j–k, experiment was performed twice with similar consistent results. One representative experiment is shown.
Error bars indicate S.E.M. P values are determined by two-tailed Student’s t tests. Statistics source data of Fig. 1c, 1d and 1k are provided in Supplementary Table 4.
The vast majority of CD11b+Gr1+ cells expressed a high level of Ly6G and a variable level of Ly6C (Fig. 1e), and exhibited a granulocytic morphology (Fig. 1f). Neutrophils isolated from tumor-free (TF) mice also express CD11b and Ly6G. However, P53N-C tumor-induced CD11b+Ly6G+ cells differ from normal neutrophils by their heterogeneous expression of Ly6C (Fig. 1e), which typically indicates an immature status29. Co-culture of CD11b+Gr1+ cells with T cells inhibited CD3- and IL2-induced T-cell proliferation and IFNγ production (Fig. 1g,h and Supplementary Fig. 1c), whereas normal neutrophils did not exhibit these effects (Fig. 1h and Supplementary Fig. 1c). Moreover, iNOS expression is over 500-fold higher in CD11b+Ly6G+ cells than in normal neutrophils (Fig. 1i). Thus, these CD11b+Gr1+ cells are immunosuppressive and, therefore, MDSCs by definition.
MDSCs can be depleted by administration of a monoclonal antibody against Gr112. The depletion inhibited orthotopic growth of P53N-C tumors (Fig. 1j). Moreover, treatment following orthotopic tumor resection significantly reduced distant metastasis to lungs (Fig. 1k) demonstrating the tumor-promoting role of MDSCs.
The oncogenic mTOR pathway dictates MDSC accumulation
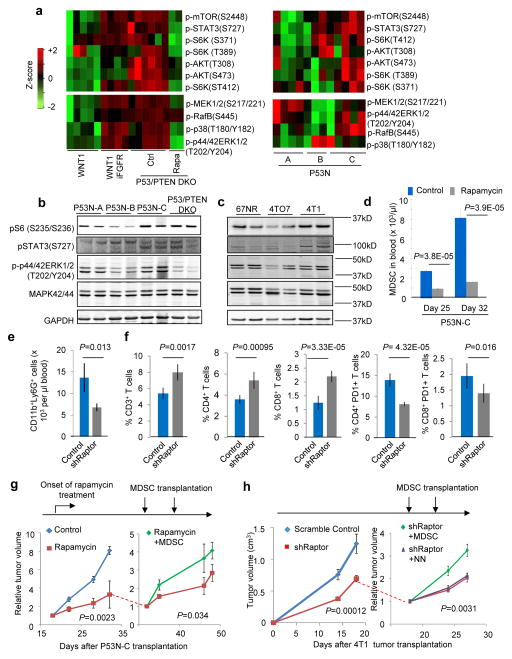
To identify the determinant of inter-tumoral heterogeneity of MDSCs, we performed reverse phase protein array (RPPA) analysis of 200 (phos-)proteins covering major signaling pathways (Supplementary Table 1). MDSC accumulation correlated with the AKT-mTOR and the MAPK pathways across the WNT1, WNT1-iFGFR, and P53-PTEN DKO models, and only the AKT-mTOR pathway in the P53N series (Fig. 2a), suggesting that the mTOR pathway is a likely driver of MDSC accumulation. This was largely confirmed by Western blot analysis in P53N tumors (Fig. 2b) as well as in the 67NR-4TO7-4T1 series (Fig. 2c).
Figure 2. mTOR pathway drives tumor-induced MDSC accumulation.
a. Reverse Phase Protein Array (RPPA) profiling in indicated mammary tumor models. Signaling molecules involved in mTOR and MAPK pathways are selected to make heat maps. Each column represents a biological replicate. Each biological replicate is the mean of technical triplicate. See Supplementary Table 1 for raw data.
b–c. Western blotting with indicated antibodies to validate RPPA results in indicated tumor models. Each lane shows one independent tumor.
d. MDSC quantification in blood of P53N-C tumor-bearing mice at indicated time points and rapamycin treatment. Three independent experiments were performed and yielded similar results. One representative experiment with n=5 animals per group is shown.
e. MDSC quantification in blood of animals carrying control 4T1 tumors or 4T1 expressing short-hairpin RNA against mTOR complex 1 protein Raptor (shRaptor). n=8 and 24 animals for control and shRaptor groups, respectively.
f. T-cell quantification in blood of the animals in e. From left: %CD3+ T cells, %CD4+ T cells and %CD8+ T-cells in white blood cells, % CD4+PD1+ cells and % CD8+PD1+ cells in CD3+ cells. n=8 and 24 animals for control and shRaptor groups, respectively.
g. P53N-C tumor growth curves under different treatment conditions. Horizontal arrow on the left: onset of mTOR inhibitor rapamycin treatment. On day 32, rapamycin-treated group was randomized into two sub-groups as indicated by dotted line, one of which received transplantation of 4×106 MDSCs for two times (indicated by vertical arrows). Tumor size was normalized for each sample relative to tumor size at onset of differential treatments. n=6 animals per group. P value calculation: general linear model accounting for repeated measurements at different time points.
h. Tumor growth curves of 4T1 cells expressing shRaptor (n=24 animals) or scrambled shRNA control (n=8 animals). On Day 15 after tumor transplantation, shRaptor group was randomized into three groups with or without adoptive transfer of MDSCs or normal neutrophils (NN) (arrows indicate time points of cell-transfers). n=8 per group after randomization.
Experiments in 2e–h were performed twice with similar consistent results.
Error bars indicate S.E.M., and P values are calculated by two-tailed Student’s t tests unless otherwise noted.
Treatment with rapamycin, an mTOR inhibitor, reduced MDSCs in P53N-C tumor-bearing mice (Fig. 2d). Rapamycin may affect non-cancer cells including MDSCs themselves. To verify that the mTOR activity responsible for MDSC accumulation is cancer cell-intrinsic, we employed a short hairpin RNA (shRNA) to deplete Raptor, an essential protein of the mTOR Complex 1, in 4T1 cancer cells (Supplementary Fig. 2). This depletion resulted in a significant reduction of MDSCs in vivo (Fig. 2e). We also observed an increase of overall T cells and a decrease of PD1+ T cells (Fig. 2f). These data strongly support that the oncogenic mTOR signaling may mediate this systemic immunosuppression.
To further distinguish cancer cell-autonomous functions of mTOR from its systemic impact through MDSCs, we performed a “rescue” experiment by adoptively transferring MDSCs into rapamycin-treated animals bearing P53N-C tumors (Fig. 2g), or into animals transplanted with 4T1 cells expressing the shRNA against Raptor (Fig. 2h). In both cases, tumor progression was partially restored (Fig. 2g,h). In contrast, normal neutrophils were unable to rescue tumor progression (Fig. 2h). Thus, at least part of the tumor-promoting effects of the mTOR pathway is through MDSC induction.
The mTOR pathway drives expression of G-CSF
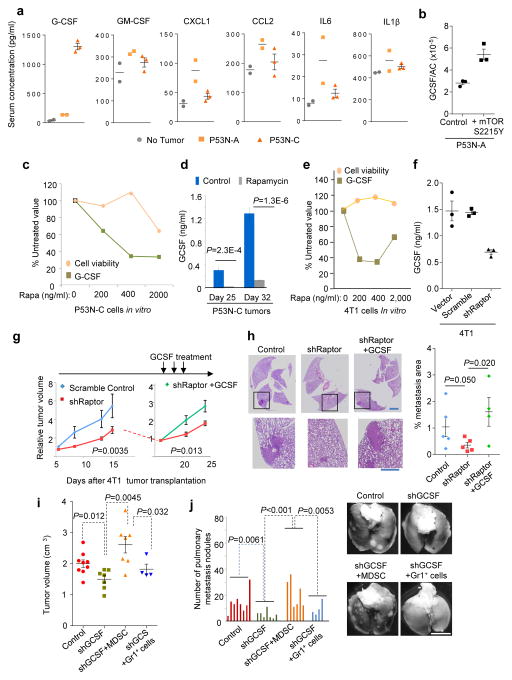
We surveyed over 20 cytokines using multiplexed luminex assays in some P53N models. The serum G-CSF concentration was higher in the P53N-C model than in TF or P53N-A. Other cytokines did not exhibit this trend (Fig. 3a). Expression of a constitutively active mTOR mutant30 in P53N-A tumors led to increased expression of G-CSF (Fig. 3b). Conversely, rapamycin treatment on P53N-C cells/tumors resulted in the reduction of G-CSF in conditioned medium (CM) or in serum (Fig. 3c,d). For 4T1 cells, either rapamycin treatment or genetic depletion of Raptor also decreased G-CSF expression (Fig. 3e,f). Taken together, these data support that G-CSF is a downstream target of the mTOR pathway in tumor cells.
Figure 3. Oncogenic mTOR pathway drives MDSC accumulation through G-CSF.
a. Bioplex assays of indicated cytokines in sera. Data are from two TF, two P53N-A tumor-bearing and three P53N-C tumor-bearing mice.
b. Quantification of G-CSF expression by real-time qPCR in P53N-A tumor cells expressing constitutively active mTOR mutant (S2215Y) or wild type control.
c. Cell viability (by WST-1 assay) and G-CSF quantity (by ELISA) in P53N-C cells as a function of rapamycin concentration.
d. Quantification of serum G-CSF by ELISA in P53N-C tumor-bearing mice with rapamycin or vehicle treatment. Two experiments were performed with consistent results. One representative experiment with n=5 animals per group is shown.
e. Rapamycin’s effect on 4T1 cell viability and GCSF expression.
f. ELISA quantification of G-CSF in CM of 4T1 cells expressing shRaptor, the scrambled shRNA control or empty vector.
In Fig. 3b, 3c, 3e and 3f, two independent experiments were conducted each with technical triplicates. One representative experiment is shown.
g. Growth curves of 4T1 cells expressing shRaptor (n=12 animals) or scrambled shRNA (n=6 animals). On Day 17 after tumor transplantation, shRaptor group was randomized into two groups (n=6 animals each), one of which received G-CSF. The P values were determined using general linear models accounting for repeated measurements at different time-points.
h. Representative H&E staining and pulmonary metastasis quantification from experiments in g, day 28 post tumor transplantation. n= 5 animals for the first two groups, and 4 animals for the third group. 10 fields were randomly chosen from each animal and averaged. P values are calculated by one-tailed Student’s t tests. Scale bars, 200μm and 50μm in the upper and lower panels, respectively.
i–j. Tumor volume (i) and lung metastasis (j) of 4T1 cells expressing shGCSF or vector control (n=8 animal). On Day 17 shGCSF group was randomized into three groups of untreated (shGCSF, n=7 animals), treated with MDSCs (shGCSF+MDSC, n=7 animals) or treated with Gr1+ splenocytes of TF mice (shGCSF+Gr1+, n=4 animals). Tumor volumes on Day 24 and lung metastases on Day 35 are shown. The lung metastasis burden was graded as described in Methods. P values are calculated by two-tailed Student’s t tests. Scale bar, 5mm.
Error bars indicate S.E.M. Statistics source data of Fig. 3c, 3i and 3j are provided in Supplementary Table 4.
We asked if systemic restoration of G-CSF could rescue delayed tumor progression caused by mTOR inhibition. We administered recombinant G-CSF protein to mice bearing 4T1 tumors expressing the shRNA against Raptor. This treatment restored orthotopic tumor growth and pulmonary metastasis (Fig. 3g,h). Thus, G-CSF functionally mediates at least part of the tumor-promoting functions of mTOR.
G-CSF’s role in MDSC formation and tumor progression has been investigated in previous studies10–14, 31. To confirm these effects in our models, we depleted G-CSF either by a neutralizing antibody in P53N-C model or by shRNA-mediated knockdown in the 4T1 model. These approaches lowered serum G-CSF by 50% or 70%, respectively (Supplementary Fig. 3a,b), resulted in a decrease of MDSCs (Supplementary Fig. 3c,d), and retarded tumor progression and metastasis (Fig. 3i,j). Importantly, this retardation was reversed by adoptive transfer of MDSCs but not splenic Gr1+ cells from TF mice (Fig. 3i,j). The depletion of G-CSF did not generate discernable effects on cancer cells in vitro (Supplementary Fig. 3e), arguing against a simple autocrine effect of G-CSF. Taken together, these data further demonstrate that G-CSF promoted tumor progression is mediated by MDSCs.
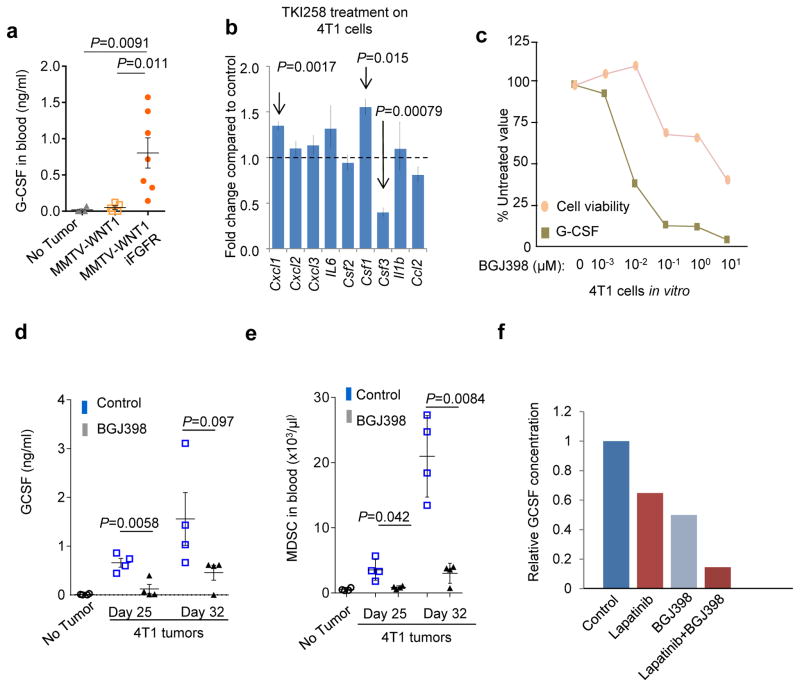
Pharmacological inhibition of mTOR may have confounding effects due to its roles in non-cancer cells. Diverse upstream signals may stimulate mTOR activity, some of which are more cancer-specific, such as activation of FGFR and loss of PTEN (Fig. 2a). Indeed, the induction of FGFR signaling in WNT1-iFGFR bi-genic tumors led to a 7-fold increase of serum G-CSF (Fig. 4a). 4T1 cells are known to possess autocrine FGFR signaling32. Reanalysis of a published dataset revealed that treatment of 4T1 cells by TKI258, a potent FGFR inhibitor, reduces Csf3 expression, the gene encoding G-CSF. Other MDSC-inducing cytokines did not exhibit the same trend (Fig. 4b). Furthermore, inhibition of the upstream FGFR signaling by BGJ398 also reduced G-CSF both in vitro (Fig. 4c) and in vivo (Fig. 4d). Accordingly, this inhibition also led to a significant reduction of MDSCs (Fig. 4e). Simultaneously inhibition of both EGFR and FGFR significantly reduced G-CSF expression in the P53N-C model (Fig. 4f), although targeting FGFR alone was ineffective, suggesting a redundant role of these receptor tyrosine kinases. Thus, G-CSF is a mediator of mTOR-driven MDSC accumulation. Targeting upstream activators of the mTOR pathway such as FGFR, therefore, may represent viable therapeutic strategies to mitigate MDSC-mediated tumor progression, in addition to their direct effects on cancer cells.
Figure 4. FGFR, an upstream activator of the mTOR pathway, can be targeted to reduce G-CSF production.
a. Quantification of serum G-CSF by ELISA in mice bearing indicated tumors. Number of animals per group: No tumor: n=4, MMTV-Wnt1: n=5, MMTV-Wnt1 iFGFR: n=7. Two independent experiments performed with similar results. One representative experiment is shown.
b. Gene expression alteration of several cytokines related to MDSC expansion in 4T1 cells upon treatment of TKI258 (n=3 biological replicates, raw data available from GSE19222).
c. 4T1 cell viability (measured by WST-1 assay) and G-CSF quantity (by ELISA) are plotted as a function of concentration of BGJ398. All results are presented as percent of control (untreated). For each data point, cells were cultured in three wells. The experiment was done two times with consistent results. One representative experiment is shown.
d. Quantification of serum G-CSF by ELISA in 4T1 tumor-bearing mice and TF mice at indicated time points and treatment.
e. Quantification of MDSCs in the blood of 4T1 tumor-bearing mice at indicated time-points with or without BGJ398 treatment.
For d–e, three independent experiments were performed with similar results. A representative one with n=4 animals per group is shown.
f. Relative G-CSF concentration as measured by ELISA in the supernatant of P53N-C cells treated with indicated drugs for 6 hours. For each data point, cells were cultured in three wells. The experiments were repeated twice with consistent results.
Error bars indicate S.E.M., and P values are calculated by two-tailed Student’s t tests. Statistics source data of Fig. 4a, 4d and 4e are provided in Supplementary Table 4.
Genomic analyses support the role of G-CSF and MDSCs in mTOR-driven immunosuppression and tumor progression
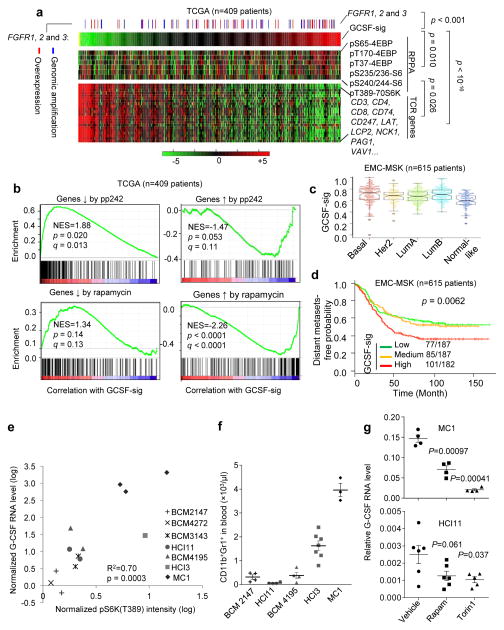
We examined the Cancer Genome Atlas (TCGA)33 datasets comprised of genomic, transcriptomic and proteomic data of human breast cancers. To assess the G-CSF/MDSC status, we obtained a gene expression signature derived from the hematopoietic cells of human patients subjected to G-CSF treatment34. This signature (designated as GCSF-sig) encompasses genes overexpressed by G-CSF-induced human hematopoietic cells. In TCGA datasets, the GCSF-sig positively correlates with mTOR pathway activity as indicated by the RPPA data, and inversely correlates with T-cell receptor (TCR) pathway genes (Fig. 5a). As one of the upstream activation signals, the genomic amplification and expression of FGFR positively correlated with the GCSF-sig (Fig. 5a). The correlation among FGFR, the GCSF-sig, and the TCR genes was also confirmed in other clinical datasets (Supplementary Fig. 4a–d)35, but not in a panel of breast cancer cell lines in vitro (Supplementary Fig. 4e), suggesting that this association is not due to intrinsic overlap between the gene sets, rather it appears to be dictated by non-tumor cells.
Figure 5. A G-CSF responsive gene signature (GCSF-sig) links the mTOR pathway to MDSC infiltration and immunosuppression in human breast cancer.
a. Heat maps show the expression of G-CSF signature as a single score (top), RPPA data of phospho-proteins reflecting the mTOR activity (middle), and TCR pathway components (bottom). The red or blue sticks above the heat maps indicate tumor samples whose FGFR-1 or -2 or -3 expression is higher than the population means by >2 standard deviations at the levels of transcription (red) or genomic copy number (blue). The P values are based on Student’s t-test on the Pearson correlation coefficients among different variables.
b. GSEA shows the correlation between GCSF-sig and gene sets up- or down-regulated by rapamycin and pp242. NES: normalized enrichment score. q: false discovery rate. P values were computed based on comparison against random simulation as implemented in GSEA.
c. Box-whisker plots of GCSF-sig scores in different subtypes of breast cancer.
d. Kaplan-Meier curves show distant metastasis-free survival (DMSF) in the EMC-MSK dataset divided into tertiles based on GCSF-sig score. P values were calculated using the log rank test.
e. Scatter plots show the correlation between G-CSF expression and the mTOR activity as indicated by the direct substrate pS6K(T389) determined by Western blotting. Each point represents an independent tumor. Seven different PDXs with 1–3 independent tumors were examined (n=13 tumors). The p value was computed using Student’s t-test on Pearson correlation coefficient.
f. Flow cytometry quantification of MDSCs in peripheral blood in animals carrying indicated PDX lines. n=4, 4, 4, 7, and 3 animals for the five PDX lines (from left to right), respectively. Error bars indicate S.E.M. P = 9.8E–10 based on one-way ANOVA across different PDX lines, and 0.00045 by two-sided Student’s t test between low-pS6K(T389) PDXs (BCM2147, HCI11 and BCM4195, see Fig. 5e) and high-pS6K(T389) PDXs (HCI3 and MC1, see Fig. 5e).
g. Two PDX lines, MC1 and HCI11, were treated with Torin 1 or rapamycin in 3D suspension cultures. The expression of G-CSF was determined by qPCR. The experiment was repeated twice with similar results. One representative experiment is shown. Upper panel: n = 4biological replicates for each group. Lower panel: n=6, 6, and 5 biological replicates for the three groups, respectively. Error bars indicate S.E.M. P values are calculated by two-tailed Student’s t tests.
Statistics source data of Fig. 5g and 5f are provided in Supplementary Table 4.
To further evaluate the mTOR pathway, we utilized two gene expression signatures denoting the responses of cancer cells to mTOR inhibitors, rapamycin36 and pp24237. The G-CSF-sig inversely correlates with these signatures in Gene Set Enrichment Analysis (GSEA) (Fig. 5b), suggesting a positive association with mTOR activity. The GCSF-sig is expressed at a higher level in both the Basal and Luminal B (LumB) breast cancer subtypes (Fig. 5c), and correlates with the risk of distant metastasis (Fig. 5d). The GCSF-sig is associated with poor prognosis in multivariate prognosis analysis in two different datasets with long-term follow-up35, 38 (Supplementary Fig. 4f).
Tumor-induced MDSC accumulation appears independent of the adaptive immune system as it was also observed in immune-deficient mice (Supplementary Fig. 4g). Therefore, it was possible to test human xenografts for their ability to induce MDSCs. A strong connection between mTOR activity and G-CSF expression was observed in a panel of patient-derived xenografts (PDXs)39, 40 (Fig. 5e). Specifically, we assayed pS6K(T389) as an indicator of mTOR activity by Western blotting, and G-CSF expression by quantitative real-time PCR (Supplementary Fig. 4h). PDXs expressing a higher level of pS6K(T389) and G-CSF also induced more MDSCs in vivo (Fig. 5f). Moreover, treatment of mTOR inhibitors, rapamycin or Torin 1, reduced G-CSF in two of the PDX lines cultured in 3D suspension medium (Fig. 5g).
Collectively, these data suggest that human tumors with an activated mTOR pathway are more likely to have an increased infiltration of MDSCs, a decreased infiltration of T cells and a worse prognosis.
TICs exhibit enhanced G-CSF production
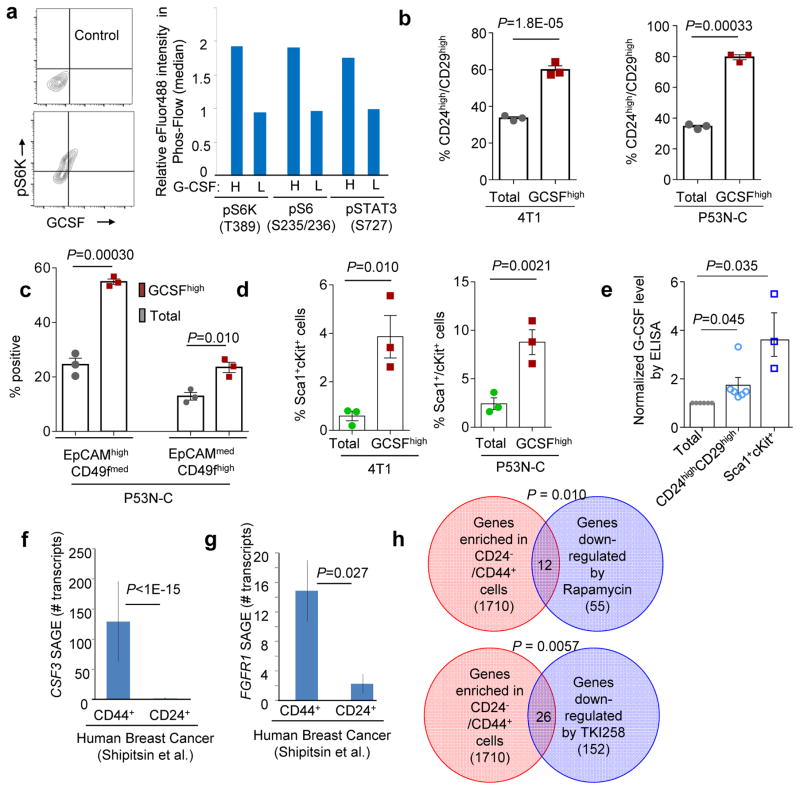
In addition to inter-tumoral heterogeneity, we also observed an intra-tumoral heterogeneity of G-CSF production. Intracellular staining of G-CSF, pS6K(T389), pS6(S235-S236) and pSTAT3(S727) followed by FACS (Supplementary Fig. 5a,b) revealed a correlation between G-CSF and mTOR activity at the single cell level (Fig. 6a). Among the top 5% of G-CSFhigh cells (>2 × mean) in 4T1 and P53N-C models approximately 60–80% are CD24highCD29high, a widely used definition of TICs, as opposed to 30% in the entire population (Fig. 6b and Supplementary Fig. 5c,d). We then examined a different set of TIC markers, EpCAM and CD49f. Among P53N-C cells, the top 5% G-CSFhigh cells fell into two distinguishable populations, both expressing to different degrees EpCAM and CD49f. Together they account for over 80% of G-CSFhigh cells (Fig. 6c). Sca-1 and cKit have also been used as TIC markers in the 4T1 model41, and Sca-1highcKithigh cells were also enriched in G-CSFhigh subpopulation of both 4T1 and P53N-C tumors (Fig. 6d). Taken together, these data support the conclusion that G-CSFhigh cells enrich for TICs.
Figure 6. G-CSFhigh cells enrich for TICs.
a. G-CSF and mTOR substrates were analyzed by intracellular FACS staining. Left: Isotype-control antibody and representative staining of GCSF and phos-S6K(T389) in P53N-C. Right: Median fluorescence intensity (relative to total population) of indicated phos-proteins in G-CSFhigh (H) vs G-CSFlow (L) subpopulations. The average values of three technical replicates are shown. Two independent experiments were performed and one representative experiment is shown.
b. 4T1 (left) or P53N-C (right) cells with G-CSF expression (top 5.3%, designated as G-CSFhigh) were compared to total population in CD24 and CD29.
c. G-CSFhigh P53N-C were analyzed for CD49f and EpCAM expression relative to total population. Two populations with slightly different EpCAM/CD49f expression were identified and assigned as EpCAMhighCD49fmed and EpCAMmedCD49fhigh.
d. 4T1 and P53N-C cells were analyzed for Sca1 and cKit. Graph shows percentages of Sca1+cKit+ subgroup.
For b–d, n=3 independent experiments each with technical replicates. Mean values of all three experiments are used to generate the plot and calculate p values.
e. ELISA validation: CD24highCD29high and Sca1+cKit+ cells were isolated by FACS and analyzed for GCSF (ELISA) and compared to mock-sorted total population. n= 6 (CD24 and CD29) and 3 (ScaI and cKit) independent FACS experiments.
f–g. SAGE libraries of CD44+ and CD24+ cell populations were analyzed for CSF3 (f) and FGFR1 (g) expression. n=5 independent tumors/specimens. For each, a P value was computed based on Poisson distribution to test the hypothesis that CD44+ and CD24+ populations have the same frequencies of the transcripts. Bonferroni correction was used to adjust for multiple comparisons. Fisher’s method was then used to combine the P values. Raw data are in Supplementary Table 6 of ref43.
h. Overlap between genes preferentially expressed in human CD44+ cell fraction and genes down-regulated by mTOR inhibitors Rapamycin and TKI258 is shown in Venn diagrams. Numbers of overlapping genes are indicated. P value was computed using Fisher’s Exact Tests.
Error bars indicate S.E.M., and P values are calculated by two-tailed Student’s t tests unless otherwise noted. Statistics source data of Fig. 6 b–e are provided in Supplementary Table 4.
The converse hypothesis that TICs overexpress G-CSF is supported by the elevated G-CSF level in CD24highCD29high cells and Sca1+cKit+ subpopulations as indicated by FACS (Supplementary Fig. 5e,f), or by ELISA (Fig. 6e). A similar conclusion was also reached using EpCAM and CD49 as markers (Supplementary Fig. 5g,h).
To ask if TICs have enhanced ability to induce MDSC differentiation as compared to non-TICs, we treated primary bone marrow cells of naïve mice with tumor cell CM. On a per-cell basis, tumor cells grown under a TIC-enriching condition (mammospheres in suspension medium42) secreted more G-CSF than under the condition favoring differentiation (2D cultures). Accordingly, tumor CM obtained under the TIC-enriching condition induced increased MDSCs (CD11b+Ly6G+Ly6Cvariable). This increase was reversed by G-CSF neutralizing antibodies (Supplementary Fig. 5i).
To test the above conclusions in human cancer, we analyzed a dataset of Serial Analysis of Gene Expression on purified CD44+ and CD24+ cancer cells43. CSF3 transcripts were exclusively found in CD44+ cells but not CD24+ cells, suggesting that cancer cells with TIC-like phenotype overexpress G-CSF (Fig. 6f). CD44+ cells also express a significantly higher level of FGFR1 transcripts (Fig. 6g). To test if CD44+ cells may possess increased activity of FGFR and mTOR pathway, we intersected the CD44+-associated genes43 with gene expression signatures that reflect mTOR inhibition37 by rapamycin or FGFR inhibition by TKI29832, and discovered statistically significant overlaps (Fig. 6h). Finally, we treated human primary bone marrow cells with CM from PDX cells. The TIC-enriched CM induced more differentiation of CD11b+CD33+CD15+CD16− cells (Supplementary Fig. 5j), which are known to be immature granulocytic cells9, 44. Taken together, these results provide evidence for the clinical relevance of the connection between TICs, G-CSF, and MDSCs.
MDSCs enhance TIC features through Notch signaling
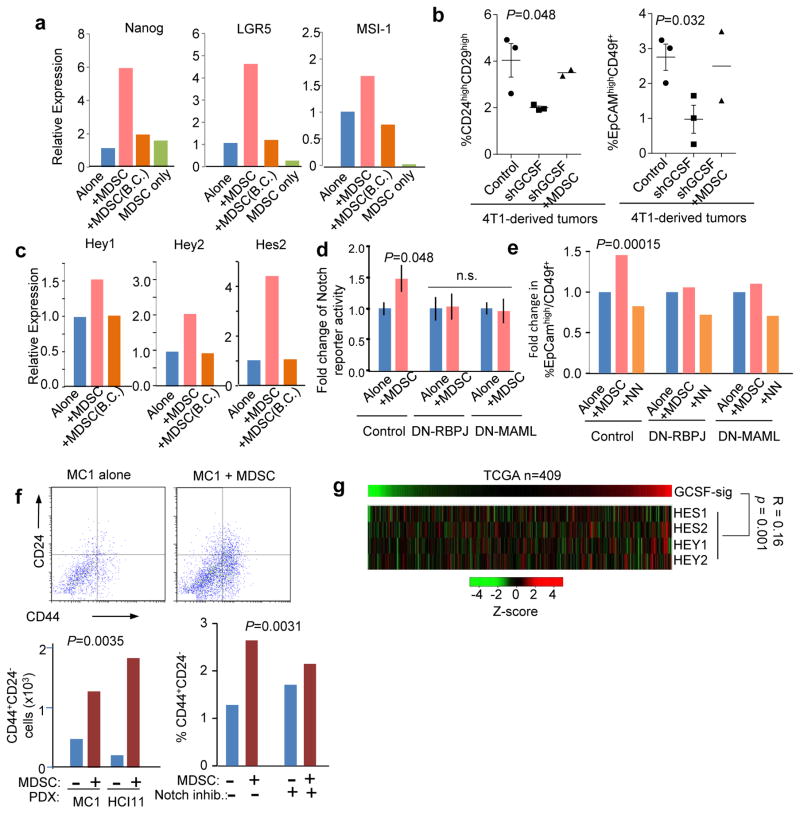
We asked how MDSCs contribute to tumor initiation. MDSCs have been shown to promote angiogenesis24, 45. The perturbation of the mTOR-G-CSF axis did not appear to affect tumor angiogenesis, as suggested by immunofluorescence staining of CD31 (Supplementary Fig. 6). Monocytes and macrophages have been shown to enhance TIC features46. We asked if granulocytic MDSCs have similar functions. The presence of MDSCs increased the number of mammospheres formed by 4T1 cells (Supplementary Fig. 7a). Moreover, MDSCs also disproportionally increased number of cancer cells expressing murine mammary TIC markers including CD24, CD29, CD49f, Sca-1, and c-Kit (Supplementary Fig. 7b). Co-culturing with MDSCs stimulated several stemness-related genes in cancer cells, including Nanog, LGR5, and MSI-147 (Fig. 7a). Cancer cells extracted from 4T1 tumors with G-CSF knockdown exhibited reduced frequencies of CD24highCD29high cells and EpCAMhighCD49f+ cells (Fig. 7b), and a decreased ability to form mammospheres (Supplementary Fig. 7c). These alterations could be rescued by adoptive MDSC transfer (Fig. 7b), suggesting that MDSCs contribute to the expansion of TICs in vivo. Taken together, these data indicate that MDSCs enhance the tumor initiation.
Figure 7. MDSCs promote breast cancer TIC features.
a. Gene expression of Nanog, Lgr5 and Msi-1 in 4T1 mammosphere cultures with or without MDSCs. 4T1 and MDSCs were co-cultured in direct contact, or separated [0.4μm pore size, Boyden Chamber (B.C.)]. Cells were cultured in different wells to form 3–6 technical replicates. Three experiments yielding consistent results were performed.
b. Percentage of CD24highCD29high cells (left) or EpCAMhighCD49f+ cells (right) in mammary tumors formed by 4T1 cells expressing short-hairpin RNA against G-CSF (shGCSF) or vector control. Part of the mice bearing shGCSF tumors are subjected to MDSC transplantation (shGCSF+MDSC). n=3, 3, and 2 tumors for the three groups, respectively.
c. Gene expression of three canonical Notch target genes in co-culture experiments described in a. 3–6 technical replicates were used. Two experiments yielding consistent results were performed.
d. Relative bioluminescence intensity of cancer cells expressing a luciferase Notch reporter, either cultured alone (Alone) or together with MDSCs (+MDSC). Dominant negative RBPJ (DN-RBPJ) or MAML (DN-MAML), or control vector was co-expressed with notch reporter in cancer cells. n=13 wells of cells that were independently set up, transfected with plasmids, and co-cultured with MDSCs/NNs.
e. Quantification of EpCAMhighCD49f+ cell frequencies in cancer cells cultured in 3D suspension medium either alone (Alone), with MDSCs (+MDSC) or normal neutrophils (+NN). DN-RBPJ or DN-MAML, or control vectors were expressed in cancer cells. The P value was determined by two-way ANOVA across various treatment groups with different genetic perturbations as one factor (three levels: control, DN-RBPJ, and DN-MAML) and co-culturing conditions as the second factor (three levels: control, +MDSC, and +NN). Data were derived from two independent experiments..
f. MC1 and HCI11 cells were maintained in 3D suspension cultures with or without MDSCs. CD24 and CD44 expression was assessed by FACS. The P values were determined using two-way ANOVA with different co-culturing conditions as one factor (two levels: −MDSC and +MDSC), and different PDX lines (two levels: HCI1 and MC1, left) or Semagacestat treatment (two levels: “−” and “+”, right) as the second factor. Data were obtained from two (left) or three (right) independent experiments.
g. Heat maps show expression of GCSF sig and indicated Notch target genes. Pearson correlation coefficient between GCSF-sig and the sum of the four genes, and the corresponding P value (by two-sided Student’s t test on correlation coefficients) is shown.
Error bars indicate S.E.M., and P values are calculated by two-tailed Student’s t tests unless otherwise noted. Statistic source data for Fig. 7b and 7d–f are provided in Supplementary Table 4.
Direct cell-cell interaction appeared to be critical for this function, as MDSCs separated in the Boyden Chambers were unable to enhance the stemness-related genes (Fig. 7a). This points to pathways that require cell juxtaposition and dictate stem or progenitor cell properties. The Notch pathway meets these criteria48. Indeed, MDSC co-culturing significantly increased expression of multiple canonical Notch target genes in a contact-dependent fashion (Fig. 7c). To determine if this increase is restricted to cancer cells, we transduced a Notch reporter49 into cancer cells, and observed that the reporter activity was enhanced by MDSCs. This enhancement was abolished by co-expression of a dominant negative (DN) mutant of Rbpj50 or MAML51 in cancer cells (Fig. 7d). Consistently, the MDSC-mediated increase of CD49+EpCAMhigh cells was reduced by DN-MAML or DN-Rbpj (Fig. 7e). Normal neutrophils were unable to induce the same effects (Fig. 7e). In human models, co-culturing of MDSCs and MC1 or HCI11 cells (isolated from the respective PDX models) significantly increased the proportion of CD44+CD24− cells (Fig. 7f). This increase was almost completely abolished by a Notch inhibitor, Semagacestat (Fig. 7f). Finally, the GCSF-sig correlates with Notch target genes in the TCGA dataset (Fig. 7g). Taken together, these results strongly support that MDSCs enrich TICs through Notch signaling in cancer cells.
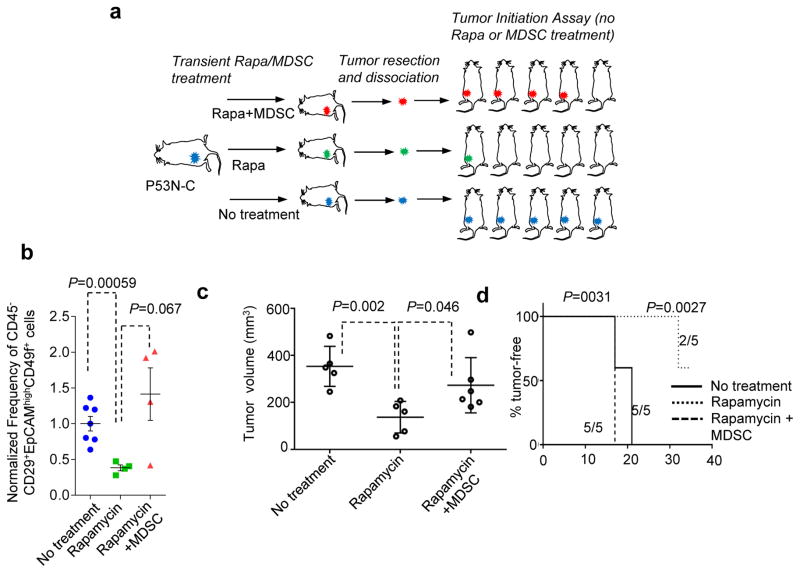
To test the impact of mTOR-MDSC cascade on tumor initiation in vivo (Fig. 8a), we transiently treated P53N-C tumor-bearing mice with rapamycin. A subset of the rapamycin-treated mice also received transplantation of exogenous MDSCs to selectively rescue the MDSC-mediated effects downstream of the mTOR pathway. The tumors were then subjected to tumor initiation assays without further rapamycin or MDSC treatment. As expected, compared to control tumors, rapamycin-treated tumors exhibited a significant decrease of CD29+EpCAMhighCD49f+ cells (Fig. 8b). Provision of exogenous MDSCs reversed this decrease (Fig. 8b). When transplanted into untreated mice at high cell numbers (5,000 and 1,000 cells per mouse), the rapamycin-treated group exhibited slower tumor progression (Fig. 8c, Supplementary Fig. 7d). At low cell numbers (200 cells and 40 cells per mouse) tumor initiation is significantly delayed in this group (Fig. 8d, Supplementary Fig. 7e). There was also a statistically significant decrease of TIC frequency compared to the other groups (p = 0.016 by Extreme Limiting Dilution Analysis52). The effects of rapamycin on tumor size, latency and TIC frequency were all partially reversed by concomitant transfer of MDSCs (Fig. 8c,d; Supplementary Fig. 7d,e). These data show that the transient inhibition of mTOR signaling reduced TIC frequency, and, more importantly, this reduction is partially mediated by MDSCs.
Figure 8. mTOR/MDSC cascade increases TIC frequency in vivo.
a. Schematics of experiments to test the MDSC-mediated effects of the mTOR pathway on tumorigenesis capacity of P53N-C cells. P53N-C tumors were allowed to grow to about 0.5 cm3 before being randomized into three groups: untreated, Rapa (rapamycin 5mg/kg, three times per week for two weeks), and Rapa + MDSC (4 × 106 cells transplanted twice a week for two weeks). After the treatment, tumors were extracted and subjected to assays shown in b–d.
b. Quantification of frequencies of CD29+EpCAMhighCD49f+ cells of P53N-C tumors subjected to the treatment shown in a. n=7,4, and 4 for the three groups, respectively. Error bars, S.E.M.
c. Results of tumor initiation assays. Tumor formation after injection of equal numbers of tumor-cells was compared. Tumor size on day 17 post orthotopic tumor cell injection (5,000 cells per mouse). n=5 mice per group. Error bars, S.D.
d. Kaplan Meier curve of TF survival. Low tumor cell number injection (40 cells per mouse) reveals increased latency of tumor cells derived from rapamycin-treated mice. Number of mice with tumors at end of observation time and total numbers of mice are also indicated for each condition. P values were computed by log-rank test.
Statistics source data of Fig. 8b is provided in Supplementary Table 4.
DISCUSSION
Our results indicate that mammary tumors relying on different oncogenic pathways may also differ in their ability to alter the immune system. Recent genomic studies have suggested enormous heterogeneity of tumor-infiltrating immune cells53–55. However, why and how different tumors evolve to enrich distinct immune cells remains poorly understood. Here, our data demonstrate how a tumor cell intrinsic oncogenic pathway, the mTOR pathway, determines a tumor’s capacity to accumulate granulocytic MDSCs, providing one partial answer to these questions.
Our data demonstrate that G-CSF is a downstream target gene of the mTOR pathway, and mediates MDSC accumulation. The role of MDSCs and G-CSF in tumor progression and therapeutic responses have been elucidated12, 13, 16, 18, 31. However, the role of mTOR-G-CSF cascade has not been reported, and these results extend the oncogenic functions of mTOR pathway from a cancer cell intrinsic to a systemic level. Our data suggest a therapeutic strategy to combine current immunotherapies with mTOR inhibitors, which has already been applied to endocrine-therapy-resistant ER+ tumors in the clinic56. A recent study utilized a PI3K inhibitor and demonstrated synergistic effects with immune checkpoint blockade57, supporting that inhibition of the PI3K-mTOR pathway may help overcome immunosuppression and enhance immunotherapies. A potential caveat of this strategy is that the PI3K-mTOR pathway also plays an important role in T cells58. As a result, PI3K and mTOR inhibitors themselves are expected to be immunosuppressive. Here, our data provided mechanistic insights into potential upstream targets including FGFR and G-CSF. Drugs blocking FGFR or G-CSF may inhibit MDSC accumulation without potentially detrimental effects on the adaptive immune system.
MDSCs enhance TIC features. This is consistent with the previously reported role of MDSCs in ovarian cancer59. The mechanism here, however, appears to be distinct - through the Notch pathway in tumor cells. In addition, our data also indicate that TICs enrich the ability to produce G-CSF, and hence contribute more significantly to MDSC accumulation. Taken together we identified a feed-forward loop between TICs and MDSCs, suggesting that the tumorigenic potential may evolve hand-in-hand with tumor-induced immunosuppression.
Among the variety of tumor models we examined, only a subset resulted in granulocytic MDSC accumulation. Thus, these cells are not universally required, and there may be different immune cells playing distinct roles in other tumor contexts. For instance, another major type of MDSCs, monocytic MDSCs, also possess potent immunosuppressive activities37. Further studies will be necessary to determine if monocytic MDSCs are involved in different breast subsets of breast cancers, and are regulated by different oncogenic pathways.
METHODS
Animal studies and tumor models
All animal experiments were carried out in accordance with a protocol approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Female animals of 5–6 weeks are used as the recipients of tumor tissue or cell line transplantation. BALB/C and SCID/Beige were used as recipients of syngeneic tumors (P53-null series and 4T1-4TO7-67NR cell line series) or PDXs, respectively. MMTV-WNT1, MMTV-WNT1-iFGFR, and P53-PTEN DKO mice are in FVB background.
The mouse breast cancer models were as follows: P53N-A (formerly assigned T11), P53N-B (formerly assigned 2225L) and P53N-C (formerly assigned 2208L) from our P53-null mouse mammary tumor bank described in ref 28 are on BALB/C background and were used in syngeneic BALB/C mice unless stated otherwise. Models on the FvB background and maintained in FvB strain of mice: MMTV Wnt1 (termed Wnt1 for brevity) and MMTV Wnt1 tumors transduced with inducible FGFR (termed Wnt1-iFGFR) have been described elsewhere 27. The P53-PTEN DKO was generated by conditional deletion of these tumor suppressors using a MMTV-driven Cre via nipple-injection of adenoviral Cre into transgenic mice carrying respective Flox alleles of P53 and PTEN.
Breast cancer PDXs were maintained by injecting 1–2mm size tumor-pieces into fat pad-cleared mammary glands of SCID Beige mice. The development of PDX lines was conducted under the Institutional Review Board-approved protocols, and was documented in previous studies 39,40. The current study utilized already-established PDXs that had been de-identified, and therefore, has been granted protocol exemption by the Institutional Review Board of Baylor College of Medicine for not involving human subjects. PDXs used in this study: HCI11 (ER+, FGFR+), HCI3 (ER+), BCM3143 (Her2+) and triple negative subtype MC1, BCM2147, BCM4272 and BCM4195, which were obtained from two independent sources 39,40.
Cell lines
None of the cell lines used in this study is listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample. 4T1, 4TO7, and 67NR cell lines were purchased from Karmanos Cancer Institute. No cell line authentication was performed. Cell lines were subjected to bimonthly tests for mycoplasma contamination.
Lentivirus-transduction of tumor cells
The expression vector for a firefly luciferase-GFP fusion protein (FLUC-PWIPZ) was packaged into lentivirus in 293T cells with pMD2 and pSpAX2 packaging system. Lentiviral stocks were filtered and concentrated by ultracentrifugation. To label P53N-C tumor cells, tumors were digested into single cell suspensions by collagenase 3 treatment, incubated with FLUC-PWIPZ lentivirus and polybrene for 24h under mammosphere assay conditions to preserve tumor characteristics. At 48h culture period, successfully labeled cells were isolated by FACS sorting of GFP positive cells and injected to mammary glands of BALB/C mice for tumor development. Tumors that retained GFP and luciferase signal (about 40% of tumors) were collected and comprised the tumor stock used in experiments.
4T1 cells were transduced with GCSF-specific shRNA (shGCSF), Raptor-specific shRNA (shRaptor) or a scramble-control non-specific shRNA (NS) using the lentiviral GIPZ vector system (Thermo Scientific Open Biosystems) which allows puromycin selection to obtain pure transduced cell population. Knockdown efficiencies were determined by gene specific Realtime-qPCR and by ELISA (R&D Systems) for GCSF or by quantitative Western blotting for Raptor (Odyssey System, rabbit anti-raptor, Cell Signaling Technology).
P53N-A model derived tumor cells were transfected using Xtreme Gene HP DNA Transfection Reagent (Version 08, Roche) with expression plasmid for a constitutively active mTOR mutant, pcDNA3-AU1-mTOR-S2215Y, which was a gift from F. Tamanoi (Addgene plasmid # 26037).
In vivo treatments
1–2mm size tumor pieces were implanted orthotopically, in the fourth mammary gland on the left side of the animal. When tumor size reached an average of 80mm3, treatments were initiated as indicated in figure legends. In all experiments, the initial implantation was conducted to animals at the age of 5–6 weeks.
BGJ398 was dissolved at 10mg/ml in DMSO, and mixed with PEG300/5%glucose (3/1) to administer by oral gavage (15 mg kg −1, daily, 5 d per week).
Rapamycin was administered by intraperitoneal (IP) injection (100μg in 200μl of 10%PEG400, 10% TWEEN80 and 4% Ethanol, dosage was at 5mg kg −1, three times per week).
Anti-Ly6G treatments comprised of two IP injections per week of 100 μg anti-Ly6G (BioXCell) in PBS. Anti-GCSF (R&D Systems) was IP-injected daily, five times per week, at 10 μg injection dose. Control animals received equal amount of isotype control antibody (BioXCell).
Recombinant human GCSF (Novoprotein, specific activity: 6×107 IU/mg) was injected subcutaneously in the flank at 1.5μg per injection in 150μl PBS every two days for the duration indicated in Figures.
Gr1+ cells of tumor-bearing mice (MDSCs), or for controls, from TF mice, were enriched via magnetic cell separation kit (Biotinylated anti-mouse Gr1, BD Pharmingen, Biotin Selection Kit, Stem Cell Technologies) yielding >95% purity. For in vivo transfer, cells were washed in PBS and immediately injected via retro-orbital route. Two consecutive injections of 4×106 cells spaced 5 d were applied.
Tumor and tissue analysis
Tumor length (L) and width (W) were measured with a caliper. Tumor volumes were calculated using the formula L*W^2*π/6. MDSCs in tumor sections were identified by immunohistochemistry with anti-S100A8 (clone 335806, R&D Systems). Immunofluorescence staining of 5μm tumor sections was carried out as previously described. To assess tumor vascularity, tumor sections were stained with CD31 antibody, followed by DAPI staining of nuclei. Images of stained sections were analyzed for number of S100A8+ cells, number of DAPI+ nuclei, and area covered by CD31 with the help of ImageJ and Photoshop. The images shown in Fig. 1a,f and Supplementary Fig. 6 are representative of at least 5 randomly picked fields from multiple tumors.
Lung metastasis was quantified by bioluminescence imaging (luciferase labeled tumor cells) or enumeration of visible lesions: Upon sacrifice, animals were perfused with PBS, lungs were excised and immediately subjected to bioluminescence measurement with IVIS Lumina II equipment. Then lungs were fixed in 4% PFA and visible lesions were counted with the aid of a magnifying stereoscope. Lung metastasis nodules were counted by two persons who were blinded with regard to a sample’s experimental-group origin. Severity of metastasis was scored based on these metastasis nodule counts. Paraffin embedded lung samples were also submitted to H&E staining to reveal size of lung metastases. For quantification, 10 stacked H&E stained sections per lung, spaced 200μm to cover representative portion of whole lung, were evaluated. Sections were scanned and total lung area and metastasis-covered area were measured using Photoshop. Fig. 1k, 3h and 3j show the average level of lung metastasis burdens in their corresponding group (group sizes indicated in the corresponding figure legends).
Blood was drawn and collected in EDTA coated tubes. To separate plasma from blood cells, a 15 min centrifugation at 4,000 rpm, 4°C, was performed. Plasma was stored at −80 °C before analysis by Bioplex (23-cytokines-kit, Biorad) or ELISA (mouse GCSF DuoSet, R&D Systems). Blood cells were subjected to red blood cell lysis, followed by FACS staining or other analyses.
Flowcytometry of blood and tumor samples
To characterize and quantify MDSCs by FACS, cells were incubated with FcR blocker (eBioscience), then stained with Ly6C-PE-CF594 (BD Biosciences), Ly6G-PerCPCy5.5 (BD Bioscience), CD11b-APC (eBioscience) and/or anti-GR1 (RB6, BD Biosciences). MDSCs were identified as GR1+CD11b+ cells, granulocytic MDSCs as Ly6GhighLy6Clow or −CD11b+, monocytic MDSCs as Ly6ChighLy6GlowCD11b+. To determine absolute cell numbers counting beads (BD Biosciences) were added before FACS acquisition. T-cells were analyzed with an antibody combination of CD3-PerCPCy5.5, CD4-APC, CD8-FITC (all eBioscience) and PD1-PE-CF594 (BD Biosciences). Please also see Supplementary Table 2 for antibody use information.
Tumor infiltrating MDSCs and T-cells were quantitated by enzymatic digestion of the dissected tumors (Mouse tumor cell dissociation kit, Miltenyi Biotec), followed by FACS staining with the above antibody combinations. Staining for the common leukocyte marker CD45 was also included to facilitate the distinction of leukocytes from tumor cells and other stromal cells.
Reverse phase protein array
Reverse phase protein array assays were carried out as described previously with minor modifications. Protein lysates were prepared from tissue samples with Tissue Protein Extraction Reagent (TPER) (Pierce, Rockford, IL) supplemented with 450 mM NaCl and a cocktail of protease and phosphatase inhibitors (Roche Life Science, Indianapolis, IN). Protein lysates at 0.5 mg/ml of total protein were denatured in SDS sample buffer (Life Technologies, Grand Island, NY) containing 2.5% 2-mercaptoethanol at 100°C for 8 min. The Aushon 2470 Arrayer (Aushon BioSystems, Billerica, MA) with a 40 pin (185 μm) configuration was used to spot lysates onto nitrocellulose-coated slides (Grace Bio-labs, Bend, OR) using an array format of 960 (experimental and controls) lysates/slide with each sample spotted as technical triplicates (2880 spots/slide). Slides were prepared for antibody labeling by blocking for 1 hr with I-Block reagent (Applied Biosystems, Foster City, CA) followed by 15 min incubation with Re-Blot reagent (Dako, Carpinteria, CA). Antibody labeling was performed at room temperature with an automated slide stainer Autolink 48 (Dako) using specific primary antibodies and appropriate biotinylated secondary antibody (Vector). An amplified fluorescent detection signal was achieved with Vectastain-ABC Streptavidin-Biotin Complex (Vector, PK-6100) followed by TSA-plus Biotin Amp Reagent diluted at 1:250 (Perkin Elmer, NEL749B001KT) and a 1:50 dilution of LI-COR IRDye 680 Streptavidin (Odyssey). Total protein content of each spotted lysate was assessed by fluorescent staining with Sypro Ruby Protein Blot Stain for selected subset of slides (Molecular Probes). Fluorescent-labeled slides were scanned on a GenePix AL4200 scanner, and the images were analyzed with GenePix Pro 7.0 (Molecular Devices). For normalization, raw image intensity of each spot is subtracted by that of negative control and then divided by total protein values. Tumors with different genetic backgrounds are analyzed separately.
Western Blotting
Tumor lysates described in RPPA section above were also subjected to gel electrophoresis using 25μg protein per lane of pre-cast 4–12% gradient minigels (Novex, Life Technologies), followed by transfer onto Nitrocellulose membrane (Novex, Life Technologies), blocking with 5% dry milk in TBST for 1h, shaking with supplier-recommended concentration of primary antibodies (all from Cell Signaling Technology) at room-temperature for 4h or at 4° C overnight, and developing with secondary antibodies that were labeled for detection with Odyssey system. Labeled molecular weight marker was also loaded on the gels. Protein bands were quantified using Odyssey features with scan times set to exclude overexposure. For representative images shown in Fig. 2b and Supplementary Fig. 2, at least three independent experiments were performed with similar images obtained.
In vitro T-cell activation assay
T-cells of naïve BALB/C mice were enriched from spleen by negative selection on magnetic beads (removal of TER119+, B220+ and CD11b+ cells) and labeled with CFSE (1uM, Molecular Probes). T-cells were cultured alone or admixed with MDSCs (at 1:0.33, 1:1 and 1:3 ratios) in 96well plates. T-cell activation was with anti-CD3 (coating of wells at 5μg/ml O/N, 4 °C) and IL-2 (huIL-2, 5ng/ml). At 5 days of culture cells were collected and CFSE dilution was determined by FACS. Samples were also stained for Ly6G, to be able to exclude non-T-cells (MDSCs) from the analysis.
Cell culture supernatants were also collected and analyzed by Bioplex assay of IFNγ.
In vitro drug treatment
4T1 cells were purchased from the Barbara Ann Karmanos Cancer Center. 2208L cells were derived from P53N-C tumors by continuous culture for more than one year. They were shown to express high E-cadherin levels suggesting the maintenance of luminal subtype of breast cancer-characteristics and were apparently free of stromal components (only p53N genotype detected) at the time of use. Routine mycoplasma inspection was performed bi-monthly to ensure negativity of infection. No STR profiling or other authentication procedures have recently been performed. For drug treatment experiments, cells were seeded at 100,000 cells per well in 12well format, or 10,000 cells per well of 96well format the day before the experiment. Drug treatments were done after one wash in FBS-free DMEM (high glucose) medium for 8h or 24h as indicated in figures. BGJ398 (Novartis), Torin-1 and Lapatinib stocks were prepared in DMSO at 1000x of final concentration. Rapamycin was dissolved at 1000x of final concentration in ethanol. Control cells received corresponding amounts of vehicle-only. Cell supernatants were collected and, after a brief spin, analyzed on GCSF-ELISA plates (mouse GCSF DuoSet, R&D Systems). In cell survival assays, WST-1 reagent (Roche) was added at end of drug treatment period following the supplier’s instructions and OD450nm was measured.
Tumor initiation assay
P53N-C tumors were extracted and dissociated into single-cell suspension. FACS was employed to remove lineage-positive cells and dead cells. The remaining cancer cells were serially diluted into 50,000, 10,000, 2,000, and 400 cells/ml. 100 μl cell suspension was transplanted into cleared mammary fat pads of recipient mice.
Mammosphere assay
Tumor cells (20,000 cells per well) were cultured in low-adhesion 24well plate in DMEM/F12 medium with murine bFGF (20 ng mL−1, Life Technologies), murine EGF (20 ng mL−1, Life Technologies) and B27 supplement (GIBCO). For co-culture, 80,000 Gr1+ MDSCs were added. As indicated, in some instances MDSCs and tumor cells were co-cultured in Boyden chambers, putting MDSCs on insets with 0.4 μm pore size membranes. Co-culture periods were 5 h for RNA expression study, 24 h for FACS of cancer stem cell surface markers, and 6 to 12 d to determine mammosphere forming units. As indicated in figures, specific control groups received Gr1+ cells of TF mice (NN, normal neutrophils) instead of Gr1+ MDSCs. These control cells were isolated from bone marrow with the same magnetic sorting procedure used for MDSCs (Biotinylated anti-mouse Gr1, BD Pharmingen, Biotin Selection Kit, Stem Cell Technologies). Mammospheres were enumerated by manual counting of low magnification images of the cultures (Cellcount). The images in Supplementary Fig. 7a are representative of over 20 images obtained from different experiments. For flow cytometry, mammospheres were dissociated by trypsin-EDTA digestion. Total tumor cell counts were obtained by FACS of the GFP-tagged tumor cells.
FACS of GCSF, phospho-S6K, breast cancer stem cell markers
Single cell suspensions of tumor cells were incubated on ice with FcR blocker (eBioscience) in staining buffer (1% FBS in PBS), followed by cell surface marker staining with the following antibodies: CD24-PE, Epcam-APC, ScaI-PE, CD29-PECy7, CD49f-eFLUOR450 and cKit-APC and CD45-eFLUOR450 as indicated in figures. When intracellular staining was done, cells were fixed and permeabilized using reagents of eBioscience’s FOXP3 staining kit. Intracellular staining was performed in presence of 2% normal mouse serum, 2% normal goat serum and 2% FBS to reduce non-specific binding. Antibodies used were GCSF-APC (eBioscience) and rabbit anti-pS6K(T389, Cell Signaling), rabbit anti-pS6 (S235/236, Cell Signaling) or rabbit anti-pStat3(S727, Cell Signaling) followed by goat anti-rabbit-eFluor488.
For analyses of CSCs in tumor tissues, we first performed tumor dissociation using gentleMACS dissociators and tumor dissociation kit manufactured by Miltenyi Biotech. The cell suspension was then subjected to staining described above. Due to technical variations, tumors harvested together and analyzed at the same time were considered as an experimental set, and the systematic differences between different sets were removed by normalization (set the mean frequencies of “No treatment” samples as one within each set).
Co-culture of human bone marrow cells with human breast cancer PDX cells
Cell suspensions of PDX tumor cells were prepared with Miltenyi Biotech tumor dissociation kit as described above. To obtain tumor-cell conditioned media, 1×106 tumor cells/ml were cultured for 3 days under mammosphere-condition (3D condition, low-adhesion plate, mammosphere medium described above) or on regular culture plates (2D condition, DMEM-high glucose with 10%FBS). Cell supernatant was collected, cleared by centrifugation and stored at −20deg until use.
Unprocessed human bone marrow was obtained from healthy volunteers (Poietics, consent agreement with vendor, Lonza). Red blood cells (RBCs) were removed by lysis.
In treatments with tumor-conditioned media, 1×106 bone marrow cells/well of 24well plate were cultured in IMDM-medium (IMDM, 10% heat-inactivated FBS, 0.01M HEPES, 55μM 2-mercapto-ethanol, 1% antibiotic/anti-micotic). To keep culture conditions consistent between groups, controls received 7.5% of 3D plus 7.5% of 2D non-conditioned tumor cell culture medium and experimental groups received 7.5% of 3D tumor cell conditioned medium plus 7.5% of non-conditioned 2D medium, or 7.5% of 2D tumor cell conditioned medium plus 7.5% of non-conditioned 3D medium, respectively. Similar experiments with mouse bone marrow cells and mouse breast cancer cell line 4T1 were carried out under same conditions. GCSF-neutralizing antibody was added as indicated in Figures, at 6μg/ml (approximately five times ED50 based on information provided by supplier). Recombinant human GCSF (Novoprotein) was used at 40ng/ml (specific activity: 6×10^7 IU/mg).
After 3-day culture, cells were analyzed by FACS. Cells with the phenotype of CD11b+CD33+CD15+CD16−HLA-DR−/low (human) or CD11b+Ly6G+ (mouse) and granulocytic characteristics in FSC-SSC profile were enumerated and assigned as granulocytic myeloid-derived suppressor cells.
Human bone marrow cells were also directly co-cultured with PDX tumor cells at 4:1 ratio under mammosphere conditions. At end of 8-day culture period, CD24 and CD44 profile of tumor cells was determined by FACS. Tumor cells and bone marrow cells were discriminated based on FSC and SSC properties and CD11b.
PDX cell co-culture with MDSCs
PDX MC1 cell suspensions were prepared as described above, cleared of mouse stromal cells using mouse cell removal kit (Miltenyi Biotec) and mixed with purified MDSCs (Gr1+) from blood of MC1 tumor bearing mouse and cultured under mammosphere conditions. The human cancer stem cell phenotype CD24−CD44+ was determined by flowcytometry with anti-human CD24-PECy7 (eBioscience) and anti-human CD44-PE (Biolegend) and included a mouse MDSC marker staining to facilitate the exclusion of this population by gating during data analysis.
Notch pathway studies
The γ-secretase inhibitor Segamacestat (Selleck Biochem) was dissolved in DMSO as 10mM stock and used at 10μM final concentration. Controls received same amount of DMSO. For assays of notch pathway activation, the notch reporter plasmid pCBFRE-luc, a gift from N. Gaiano (Addgene plasmid # 26897), was used. The notch reporter was co-transfected with a constitutive expression (EF1α promoter-driven) renilla-luciferase plasmid. Firefly luciferase (Notch reporter) and Renilla luciferase bioluminescence activity were measured by in vivo imaging system (IVIS) and determined consecutively by adding respective substrates (D-luciferin, Gold Biotechnology and RediJect Coelenterazine h, PerkinElmer, respectively) and included background measurements before and between reporter analyses. Notch reporter signal was normalized to the signal from constitutive plasmid. Dominant negative forms of RBPJ (DN-RBPJ) and MAML (DN-MAML) were applied to inhibit the intracellular notch pathway. This was done either by co-transfection in the notch reporter assays or using lentiviral transduction followed by GFP-based flow cytometric sorting to remove non-transduced cell population.
Bioinformatics analysis
TCGA dataset was obtained from the TCGA Data Portal in December 2012. Data of different platforms were matched based on patient identifiers. Patients with incomplete records of DNA-copy number, mRNA expression, or RPPA data were excluded. EMC-MSK dataset was derived as described in the previous study 62. METABRIC dataset 38 was described elsewhere and obtained from Cancer Research UK, Cambridge Research Institute. FGFR signature was derived from a previous study by comparing MMTV-WNT-iFGFR tumors to MMTV-WNT tumors 27. The genes that are significantly altered by inducible activation of FGFR1 were determined using a cutoff of FC (fold change) > 2 and p < 0.05 (by two-sided Student’s t tests). The signature was then applied to the gene expression profiles of p53-null tumors. Specifically, a single value score was computed using the formulas: ΣG↑−ΣG↓, where G↑ represents the expression values of up-regulated genes and G↓ represents the expression values of down-regulated genes. The G-CSF signature was derived based on a previous study using the same approach as for FGFR signature. In Figure 3b, we utilized GSE19222 to analyze the effect of an FGFR inhibitor, TKI258, on the expression of multiple cytokines. In Supplementary Fig. 4e, we used GSE12777 to gauge the correlation between GCSF-sig and TCR genes in cell lines.
Gene Set Enrichment Analyses (GSEA) software was downloaded from the website http://www.broadinstitute.org/gsea/index.jsp. Gene signatures reflecting the action of pp242, rapamycin and G-CSF were derived from previous studies 34. A continuous score of G-CSF signature was computed using a similar approach as described in the previous paragraph for the FGFR signature. GSEA was applied to the TCGA dataset using genes up- or down-regulated by pp242 and rapamycin as gene sets 36,37, and G-CSF signature scores as the continuous phenotype indices. P values of the enrichment were computed using built-in algorithms of GSEA based on random simulation.
Kaplan-Meier curve generation and tests (Fig. 4D), multivariate fitting of Cox Proportional Hazard Model (Supplementary Fig. 4F), gene expression comparison across different molecular subtypes (Fig. 4C), and other data mining procedures and statistical assessments were performed using R Statistical Software based on log-rank tests.
RT-qPCR
RNA was collected and processed in TRIZOL, reverse transcribed to cDNA with Applied Biosystems’ High Capacity Reverse Transcription Kit, and analyzed by Realtime-qPCR on an ABI7500 Fast Real-time PCR machine using SsoFast EvaGreen QPCR reagent (Biorad). Results were normalized to beta-actin and/or GFP (tumor cell label) and relative values were calculated as 2^[−(CTgene − CT reference)]. Gene-specific primers used are listed in Supplementary Table 3.
Statistics and Reproducibility
All results are presented in the form of Mean +/− S.E.M. unless otherwise specified. Sample sizes for in vivo experiments are noted in the corresponding figures or figure legends. Individual animals and independently prepared or treated primary tissue samples (e.g., PDXs and primary human bone marrows) are considered as biological replicates. Different wells in one experiment are considered as technical replicates. In each experiment the size is determined based on our prior knowledge of the variability of experimental output. Specifically, in animal experiments, a sample size of 4–8 mice per group allows us to detect a 50% difference in tumor size with power of 60–85% and α=0.05. Animals that suffered non cancer-related disease conditions (e.g., pathogen infection) were diagnosed by the Center of Comparative Medicine at Baylor College of Medicine based on routine inspection, and excluded from experiments per the recommendation of veterinarians based on pre-established protocols. Otherwise animals were included into analyses. For experiments shown in Fig. 2g, 2h, and 3g, a randomization process was performed by randomly assigning animals into two separate groups. After assignment, tumor sizes of the two resultant groups were compared to make sure there is no statistically significant different (by two-side Student’s t test). No randomization was performed in other experiments. The researchers were not blinded to allocation during experiments and outcome assessment except for experiment in Fig. 3j, in which the results were assessed by a researcher blinded to group allocation. For in vitro experiments including FACS analyses, in most cases two to three independent experiments were performed each with 3–6 technical replicates. Specific information is included in corresponding figure legends. No specific tests were conducted to test the assumption of normal distribution. The difference between means of different experimental groups was analyzed using two-tailed unpaired Student’s t test unless otherwise noted in respective legends.
F tests were performed to compare variation within different groups, and t tests were performed with or without the assumption of equal variation accordingly. In a few experiments (see Fig. 7e and 7f), we computed P values in the framework of two-way ANOVA to achieve more stable estimates of biological variations by collectively analyzing multiple biological conditions or perturbations. Specifically, in Fig. 7e, we treated different genetic perturbations (control, DN-Rbpj, and DN-Maml) as one factor, and different co-culture conditions (control, +MDSC, and +NN) as the second factor. In Fig. 7f, we used different co-culture condition (with or without MDSCs) as one factor, different PDX models (HCI11 and MC1, left panel) or different treatment (with or without Notch inhibitor, right panel) as the second factor. Two-way ANOVA tested the null hypotheses that there is no significant difference among different levels of the same factor and there is no interaction between the two factors. The analyses were performed using the built-in two-way ANOVA analysis tool in Excel. Detailed output including results of post hoc tests is provided in Supplementary Table 4.
Supplementary Material
Acknowledgments
Grant Support
We would like to thank Novartis for providing BGJ-398, and H.C. Lo and D. Weiss for helpful input. We also thank Antibody-Based Proteomics Shared Resource of the Dan L. Duncan Cancer Center supported by Cancer Center Support grant NCI P30CA125123, and Dr. Alana Welm for generously providing some of the PDX models. X. H.-F. Z. is supported by NCI CA183878, Breast Cancer Research Foundation, US Department of Defense DAMD W81XWH-13-1-0195, Susan G. Komen CCR14298445, and McNair Medical Institute. T.W. is supported by Helis Foundation. H.W. is supported by US Department of Defense DAMD W81XWH-13-1-0296. Studies with the p53 null tumors were supported by NIH grant CA148761 to J.M.R and with the WNT1-iFGFR tumors by NIH grant CA16303 to J.M.R. RRPA experiments were supported by Cancer Prevention and Research Institute of Texas (CPRIT) Core Facilities Support Award RP120092 to D.P.E. The authors acknowledge the joint participation by Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine.
Footnotes
The authors disclose no potential conflicts of interest.
Authors’ Contributions
Conception and design: X. H-F Zhang, J.M. Rosen, T. Welte
Development of methodology: T. Welte, D. Zheng, I.S. Kim, L. Tian, X. Gao, S. Huang, J. Xu, T. Wang. Q. Mo.
Acquisition of data: T. Welte, I.S. Kim, L. Tian, X. Gao, H. Wang, J. Li, X.B. Holdman, J.I. Herschkowitz, A. Pond, G. Xie, S. Kurley, T. Nguyen, L. Liao, D. Edward, S. Huang, J. Xu, Y. Li, M.T. Lewis, T. Wang, T.F. Westbrook, L. Xin.
Analysis and interpretation of data: T. Welte, X. H-F Zhang, J.M. Rosen, T. Wang, I.S. Kim, Q. Mo.
Writing, review of manuscript: X. H-F Zhang, T. Welte, T. Wang, J.M. Rosen
Study supervision: X. H-F Zhang, J.M. Rosen
Accession numbers and datasets
Referenced accessions: Previously published microarray data that were re-analysed here are available from GEO under accession codes GSE7400 34, GSE19222 32, GSE12777 61. The EMC-MSK dataset 62 is comprised of GSE2603, GSE5327, GSE2034, and GSE12276. METABRIC dataset 38 is deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/), which is hosted by the European Bioinformatics Institute, under accession number EGAS00000000083.
References
- 1.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunt SK, Clements VK, Hanson EM, Sinha P, Ostrand-Rosenberg S. Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through Toll-like receptor 4. J Leukoc Biol. 2009;85:996–1004. doi: 10.1189/jlb.0708446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marvel D, Gabrilovich DI. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J Clin Invest. 2015;125:3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granot Z, et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer Cell. 2011;20:300–314. doi: 10.1016/j.ccr.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strober S. Natural suppressor (NS) cells, neonatal tolerance, and total lymphoid irradiation: exploring obscure relationships. Annu Rev Immunol. 1984;2:219–237. doi: 10.1146/annurev.iy.02.040184.001251. [DOI] [PubMed] [Google Scholar]
- 7.Almand B, et al. Increased production of immature myeloid cells in cancer patients: a mechanism of immunosuppression in cancer. J Immunol. 2001;166:678–689. doi: 10.4049/jimmunol.166.1.678. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Montero CM, et al. Increased circulating myeloid-derived suppressor cells correlate with clinical cancer stage, metastatic tumor burden, and doxorubicin-cyclophosphamide chemotherapy. Cancer Immunol Immunother. 2009;58:49–59. doi: 10.1007/s00262-008-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solito S, et al. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann N Y Acad Sci. 2014;1319:47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, et al. BMP4 Inhibits Breast Cancer Metastasis by Blocking Myeloid-Derived Suppressor Cell Activity. Cancer Res. 2014;74:5091–5102. doi: 10.1158/0008-5472.CAN-13-3171. [DOI] [PubMed] [Google Scholar]
- 11.Donkor MK, et al. Mammary tumor heterogeneity in the expansion of myeloid-derived suppressor cells. Int Immunopharmacol. 2009;9:937–948. doi: 10.1016/j.intimp.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 12.Kowanetz M, et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci U S A. 2010;107:21248–21255. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shojaei F, et al. G-CSF-initiated myeloid cell mobilization and angiogenesis mediate tumor refractoriness to anti-VEGF therapy in mouse models. Proc Natl Acad Sci U S A. 2009;106:6742–6747. doi: 10.1073/pnas.0902280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PLoS One. 2011;6:e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serafini P, et al. High-dose granulocyte-macrophage colony-stimulating factor-producing vaccines impair the immune response through the recruitment of myeloid suppressor cells. Cancer Res. 2004;64:6337–6343. doi: 10.1158/0008-5472.CAN-04-0757. [DOI] [PubMed] [Google Scholar]
- 16.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–290. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 17.Song X, et al. CD11b+/Gr-1+ immature myeloid cells mediate suppression of T cells in mice bearing tumors of IL-1beta-secreting cells. J Immunol. 2005;175:8200–8208. doi: 10.4049/jimmunol.175.12.8200. [DOI] [PubMed] [Google Scholar]
- 18.Bunt SK, et al. Reduced inflammation in the tumor microenvironment delays the accumulation of myeloid-derived suppressor cells and limits tumor progression. Cancer Res. 2007;67:10019–10026. doi: 10.1158/0008-5472.CAN-07-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinha P, Clements VK, Fulton AM, Ostrand-Rosenberg S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007;67:4507–4513. doi: 10.1158/0008-5472.CAN-06-4174. [DOI] [PubMed] [Google Scholar]
- 20.Gallina G, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J Clin Invest. 2006;116:2777–2790. doi: 10.1172/JCI28828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronte V, et al. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170:270–278. doi: 10.4049/jimmunol.170.1.270. [DOI] [PubMed] [Google Scholar]
- 22.Gabrilovich D, et al. Vascular endothelial growth factor inhibits the development of dendritic cells and dramatically affects the differentiation of multiple hematopoietic lineages in vivo. Blood. 1998;92:4150–4166. [PubMed] [Google Scholar]
- 23.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell. 2008;13:23–35. doi: 10.1016/j.ccr.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du R, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welm BE, et al. Inducible dimerization of FGFR1: development of a mouse model to analyze progressive transformation of the mammary gland. J Cell Biol. 2002;157:703–714. doi: 10.1083/jcb.200107119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pond AC, et al. Fibroblast growth factor receptor signaling dramatically accelerates tumorigenesis and enhances oncoprotein translation in the mouse mammary tumor virus-Wnt-1 mouse model of breast cancer. Cancer Res. 2010;70:4868–4879. doi: 10.1158/0008-5472.CAN-09-4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herschkowitz JI, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2012;109:2778–2783. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Ji M, Klarmann KD, Keller JR. Repression of Id2 expression by Gfi-1 is required for B-cell and myeloid development. Blood. 2010;116:1060–1069. doi: 10.1182/blood-2009-11-255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coffelt SB, et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522:345–348. doi: 10.1038/nature14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dey JH, et al. Targeting fibroblast growth factor receptors blocks PI3K/AKT signaling, induces apoptosis, and impairs mammary tumor outgrowth and metastasis. Cancer Res. 2010;70:4151–4162. doi: 10.1158/0008-5472.CAN-09-4479. [DOI] [PubMed] [Google Scholar]
- 33.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzzeo MP, Yang J, Casella G, Reddy V. Hematopoietic stem cell mobilization with G-CSF induces innate inflammation yet suppresses adaptive immune gene expression as revealed by microarray analysis. Exp Hematol. 2007;35:1456–1465. doi: 10.1016/j.exphem.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XH, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei G, et al. Gene expression-based chemical genomics identifies rapamycin as a modulator of MCL1 and glucocorticoid resistance. Cancer Cell. 2006;10:331–342. doi: 10.1016/j.ccr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Wang BT, et al. The mammalian target of rapamycin regulates cholesterol biosynthetic gene expression and exhibits a rapamycin-resistant transcriptional profile. Proc Natl Acad Sci U S A. 2011;108:15201–15206. doi: 10.1073/pnas.1103746108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013;73:4885–4897. doi: 10.1158/0008-5472.CAN-12-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Movahedi K, et al. Identification of discrete tumor-induced myeloid-derived suppressor cell subpopulations with distinct T cell-suppressive activity. Blood. 2008;111:4233–4244. doi: 10.1182/blood-2007-07-099226. [DOI] [PubMed] [Google Scholar]
- 42.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Dumitru CA, Moses K, Trellakis S, Lang S, Brandau S. Neutrophils and granulocytic myeloid-derived suppressor cells: immunophenotyping, cell biology and clinical relevance in human oncology. Cancer Immunol Immunother. 2012;61:1155–1167. doi: 10.1007/s00262-012-1294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, et al. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–421. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 46.Lu H, et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oskarsson T, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 49.Yu X, et al. Notch signaling activation in human embryonic stem cells is required for embryonic, but not trophoblastic, lineage commitment. Cell Stem Cell. 2008;2:461–471. doi: 10.1016/j.stem.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blokzijl A, et al. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163:723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valdez JM, et al. Notch and TGFbeta form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell Stem Cell. 2012;11:676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Nagalla S, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gentles AJ, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iglesia MD, et al. Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res. 2014;20:3818–3829. doi: 10.1158/1078-0432.CCR-13-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baselga J, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med. 2012;366:520–529. doi: 10.1056/NEJMoa1109653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim K, et al. Eradication of metastatic mouse cancers resistant to immune checkpoint blockade by suppression of myeloid-derived cells. Proc Natl Acad Sci U S A. 2014;111:11774–11779. doi: 10.1073/pnas.1410626111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui TX, et al. Myeloid-derived suppressor cells enhance stemness of cancer cells by inducing microRNA101 and suppressing the corepressor CtBP2. Immunity. 2013;39:611–621. doi: 10.1016/j.immuni.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acharyya S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoeflich KP, et al. In vivo antitumor activity of MEK and phosphatidylinositol 3-kinase inhibitors in basal-like breast cancer models. Clin Cancer Res. 2009;15:4649–4664. doi: 10.1158/1078-0432.CCR-09-0317. [DOI] [PubMed] [Google Scholar]
- 62.Zhang XH, et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.