Abstract
We conducted genome-wide association studies of three phenotypes: subjective well-being (N = 298,420), depressive symptoms (N = 161,460), and neuroticism (N = 170,910). We identified three variants associated with subjective well-being, two with depressive symptoms, and eleven with neuroticism, including two inversion polymorphisms. The two depressive symptoms loci replicate in an independent depression sample. Joint analyses that exploit the high genetic correlations between the phenotypes (|ρ̂| ≈ 0.8) strengthen the overall credibility of the findings, and allow us to identify additional variants. Across our phenotypes, loci regulating expression in central nervous system and adrenal/pancreas tissues are strongly enriched for association.
INTRODUCTION
Subjective well-being—as measured by survey questions on life satisfaction, positive affect, or happiness—is a major topic of research within psychology, economics, and epidemiology. Twin studies have found that subjective well-being is genetically correlated with depression (characterized by negative affect, anxiety, low energy, bodily aches and pains, pessimism, and other symptoms) and neuroticism (a personality trait characterized by easily experiencing negative emotions such as anxiety and fear)1–3. Depression and neuroticism have received much more attention than subjective well-being in genetic-association studies, but the discovery of associated genetic variants with either of them has proven elusive4,5.
In this paper, we report a series of separate and joint analyses of subjective well-being, depressive symptoms, and neuroticism. Our primary analysis is a genome-wide association study (GWAS) of subjective well-being based on data from 59 cohorts (N = 298,420). This GWAS identifies three loci associated with subjective well-being at genome-wide significance (p < 5×10−8). We supplement this primary analysis with auxiliary GWAS meta-analyses of depressive symptoms (N = 180,866) and neuroticism (N = 170,910), performed by combining publicly available summary statistics from published studies with new genome-wide analyses of additional data. In these auxiliary analyses we identify two loci associated with depressive symptoms and eleven with neuroticism, including two inversion polymorphisms. In depression data from an independent sample (N = 368,890), both depressive symptoms associations replicate (p = 0.004 and p = 0.015).
In our two joint analyses, we exploit the high genetic correlation between subjective well-being, depressive symptoms, and neuroticism (i) to evaluate the credibility of the 16 genome-wide significant associations across the three phenotypes, and (ii) to identify novel associations (beyond those identified by the GWAS). For (i), we investigate whether our three subjective well-being-associated SNPs “quasi-replicate” by testing them for association with depressive symptoms and neuroticism. We similarly examine the quasi-replication record of the depressive symptoms and neuroticism loci by testing them for association with subjective well-being. We find that the quasi-replication record closely matches what would be expected given our statistical power if none of the genome-wide significant associations were chance findings. These results strengthen the credibility of (most of) the original associations. For (ii), we use a “proxy phenotype” approach6: we treat the set of loci associated with subjective well-being at p < 10−4 as candidates, and we test them for association with depressive symptoms and neuroticism. At the Bonferroni-adjusted 0.05 significance threshold, we identify two loci associated with both depressive symptoms and neuroticism and another two associated with neuroticism.
In designing our study, we faced a tradeoff between analyzing a smaller sample with a homogeneous phenotype measure versus attaining a larger sample by jointly analyzing data from multiple cohorts with heterogeneous measures. For example, in our analysis of subjective well-being, we included measures of both life satisfaction and positive affect, even though these constructs are conceptually distinct7. In Supplementary Note and Supplementary Figure 1, we present a theoretical framework for evaluating the costs and benefits of pooling heterogeneous measures. In our context, given the high genetic correlation across measures, the framework predicts that pooling increases statistical power to detect variants. This prediction is supported by our results.
RESULTS
GWAS of subjective well-being
Following a pre-specified analysis plan, we conducted a sample-size-weighted meta-analysis (N = 298,420) of cohort-level GWAS summary statistics. The phenotype measure was life satisfaction, positive affect, or (in some cohorts) a measure combining life satisfaction and positive affect. We confirmed previous findings9 of high pairwise genetic correlation between life satisfaction and positive affect using bivariate LD Score regression10 (ρ̂ = 0.981 (SE = 0.065); Supplementary Table 1). Details on the 59 participating cohorts, their phenotype measures, genotyping, quality-control filters, and association models are provided in Online Methods, Supplementary Note, and Supplementary Tables 2–6.
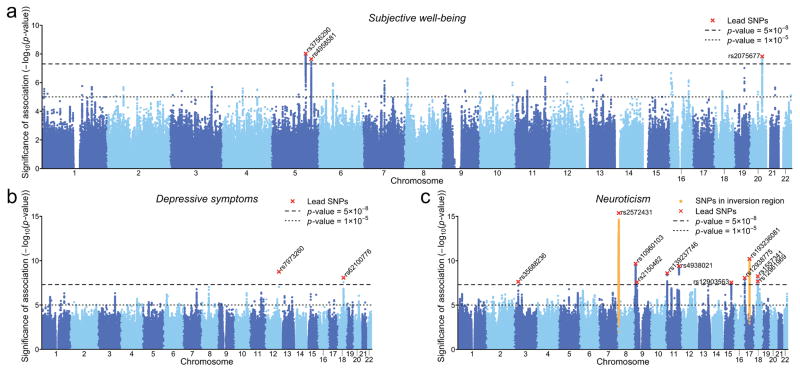
As expected under polygenicity11, we observe inflation of the median test statistic (λGC = 1.206). The estimated intercept from LD Score regression (1.012) suggests that nearly all of the inflation is due to polygenic signal rather than bias. We also performed family-based analyses that similarly suggest minimal confounding due to population stratification (Online Methods). Using a clumping procedure (Supplementary Note), we identified three approximately independent SNPs reaching genome-wide significance (“lead SNPs”). These three lead SNPs are indicated in the Manhattan plot (Figure 1a) and listed in Table 1. The SNPs have estimated effects in the range 0.015 to 0.018 standard deviations (SDs) per allele (each R2 ≈ 0.01%).
Fig. 1. Manhattan plots of GWAS results.
(a) Subjective well-being (N = 298,420), (b) Depressive symptoms (N = 180,866), (c) Neuroticism (N = 170,911). The x-axis is chromosomal position, and the y-axis is the significance on a −log10 scale. The upper dashed line marks the threshold for genome-wide significance (p = 5×10 8); the lower line marks the threshold for nominal significance (p = 10 5). Each approximately independent genome-wide significant association (“lead SNP”) is marked by ×. Each lead SNP is the lowest p-value SNP within the locus, as defined by our clumping algorithm (Supplementary Note).
Table 1.
Summary of polymorphisms identified across analyses.
| Panel A. Genome-Wide Significant Associations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| Subjective Well-Being (SWB, N = 298,420) | |||||||||||
|
| |||||||||||
| SNPID | CHR | BP | EA | EAF | Beta | SE | R2 | p-value | N | Quasi-Repl | |
| rs3756290 | 5 | 130,951,750 | A | 0.24 | −0.0177 | 0.0031 | 0.011% | 9.6×10−9 | 286,851 | ||
| rs2075677 | 20 | 47,701,024 | A | 0.76 | 0.0175 | 0.0031 | 0.011% | 1.5×10−8 | 288,454 | DS** | |
| rs4958581 | 5 | 152,187,729 | T | 0.66 | 0.0153 | 0.0027 | 0.011% | 2.3×10−8 | 294,043 | DS*** | |
|
| |||||||||||
| Neuroticism (N = 170,908) | |||||||||||
|
| |||||||||||
| SNPID | CHR | BP | EA | EAF | Beta | SE | R2 | p-value | N | Quasi-Repl | |
|
| |||||||||||
| rs2572431# | 8 | 11,105,077 | T | 0.41 | 0.0283 | 0.0035 | 0.039% | 4.2×10−16 | 170,908 | SWB* | |
| rs193236081## | 17 | 44,142,332 | T | 0.77 | −0.0284 | 0.0045 | 0.028% | 6.3×10−11 | 151,297 | ||
| rs10960103 | 9 | 11,699,270 | C | 0.77 | 0.0264 | 0.0038 | 0.024% | 2.1×10−10 | 165,380 |
|
|
| rs4938021 | 11 | 113,364,803 | T | 0.34 | 0.0233 | 0.0037 | 0.024% | 4.0×10−10 | 159,900 | ,SWB* | |
| rs139237746 | 11 | 10,253,183 | T | 0.51 | −0.0204 | 0.0034 | 0.021% | 2.6×10−9 | 170,908 | ||
| rs1557341 | 18 | 35,127,427 | A | 0.34 | 0.0213 | 0.0036 | 0.021% | 5.6×10−9 | 165,579 |
|
|
| rs12938775 | 17 | 2,574,821 | A | 0.47 | −0.0202 | 0.0035 | 0.020% | 8.5×10−9 | 163,283 | SWB* | |
| rs12961969 | 18 | 35,364,098 | A | 0.2 | 0.0250 | 0.0045 | 0.020% | 2.2×10−8 | 156,758 | ||
| rs35688236 | 3 | 34,582,993 | A | 0.69 | 0.0213 | 0.0037 | 0.019% | 2.4×10−8 | 161,636 | ||
| rs2150462 | 9 | 23,316,330 | C | 0.26 | −0.0217 | 0.0038 | 0.018% | 2.7×10−8 | 170,907 | ||
| rs12903563 | 15 | 78,033,735 | T | 0.50 | 0.0198 | 0.0036 | 0.020% | 2.9×10−8 | 157,562 | ,SWB* | |
|
| |||||||||||
| Depressive Symptoms (DS, N = 180,866) | |||||||||||
|
| |||||||||||
| SNPID | CHR | BP | EA | EAF | Beta | SE | R2 | p-value | N | Quasi- Repl/Repl | |
|
| |||||||||||
| rs7973260 | 12 | 118,375,486 | A | 0.19 | 0.0306 | 0.0051 | 0.029% | 1.8×10−9 | 124,498 |
|
|
| rs62100776 | 18 | 50,754,633 | A | 0.56 | −0.0252 | 0.0044 | 0.031% | 8.5×10−9 | 105,739 | ,SWB* | |
|
| |||||||||||
| Panel B. SNPs Identified via Proxy-Phenotype Analyses of SWB Loci with p-value<10−4 | |||||||||||
|
|
|||||||||||
| Depressive Symptoms in Non-Overlapping Cohorts | |||||||||||
|
| |||||||||||
| SNPID | CHR | BP | EA | EAF | BetaDS | SEDS | R2 | pDS | Bonferroni | NDS | |
|
| |||||||||||
| rs4346787† | 6 | 27,491,299 | A | 0.113 | −0.023 | 0.0059 | 0.011% | 9.8×10−5 | 0.0160 | 142,265 | |
| rs4481363 | 5 | 164,483,794 | A | 0.524 | 0.014 | 0.0038 | 0.009% | 3.1×10−4 | 0.0499 | 142,265 | |
|
| |||||||||||
| Neuroticism in Non-Overlapping Cohorts | |||||||||||
|
| |||||||||||
| SNPID | CHR | BP | EA | EAF | Betaneuro | SEneuro | R2 | pneuro | Bonferroni | Nneuro | |
|
| |||||||||||
| rs10838738 | 11 | 47,663,049 | A | 0.49 | 0.0178 | 0.0039 | 0.016% | 5.0×10−6 | 0.0009 | 131,864 | |
| rs10774909 | 12 | 117,674,129 | C | 0.52 | −0.0150 | 0.0039 | 0.011% | 1.2×10−4 | 0.0203 | 131,235 | |
| rs6904596 | 6 | 27,491,299 | A | 0.09 | −0.0264 | 0.0072 | 0.012% | 2.5×10−4 | 0.0423 | 116,335 | |
| rs4481363 | 5 | 164,474,719 | A | 0.49 | 0.0151 | 0.0040 | 0.011% | 1.9×10−4 | 0.0316 | 122,592 | |
EA: effect allele. EAF: effect allele frequency. All effect sizes are reported in units of SDs per allele. “Quasi-Repl.”: phenotypes for which SNP was found to be nominally associated in quasi-replication analyses conducted in independent samples.
significant at the 5%-level,
significant at the 1%-level,
significant at the 0.1%-level.
inversion-tagging polymorphism on chromosome 8.
inversion-tagging polymorphism on chromosome 17.
proxy for rs6904596 (R2 = 0.98).
We also conducted separate meta-analyses of the components of our subjective well-being measure, life satisfaction (N = 166,205) and positive affect (N = 180,281) (Online Methods). Consistent with our theoretical conclusion that pooling heterogeneous measures increased power in our context, the life satisfaction and positive affect analyses yielded fewer signals across a range of p-value thresholds than our meta-analysis of subjective well-being (Supplementary Table 7).
GWAS of depressive symptoms and neuroticism
We conducted auxiliary GWAS of depressive symptoms and neuroticism (see Online Methods, Supplementary Note, and Supplementary Tables 8–12 for details on cohorts, phenotype measures, genotyping, association models, and quality-control filters). For depressive symptoms (N = 180,866), we meta-analyzed publicly available results from a study performed by the Psychiatric Genomics Consortium (PGC)12 together with new results from analyses of the initial release of the UK Biobank data (UKB)13 and the Resource for Genetic Epidemiology Research on Aging (GERA) Cohort14. In UKB (N = 105,739), we constructed a continuous phenotype measure by combining responses to two questions, which ask about the frequency in the past two weeks with which the respondent experienced feelings of unenthusiasm/disinterest and depression/hopelessness. The other cohorts had ascertained case-control data on major depressive disorder (GERA: Ncases = 7,231, Ncontrols = 49,316; PGC: Ncases = 9,240, Ncontrols = 9,519).
For neuroticism (N = 170,910), we pooled summary statistics from a published study by the Genetics of Personality Consortium (GPC)4 with results from a new analysis of UKB data. The GPC (N = 63,661) harmonized different neuroticism batteries. In UKB (N = 107,245), our measure was the respondent’s score on a 12-item version of the Eysenck Personality Inventory Neuroticism scale15.
In both the depressive symptoms and neuroticism GWAS, the heterogeneous phenotypic measures are highly genetically correlated (Supplementary Table 1). As in our subjective well-being analyses, there is substantial inflation of the median test statistics (λGC = 1.168 for depressive symptoms, λGC = 1.317 for neuroticism), but the estimated LD Score intercepts (1.008 and 0.998, respectively) suggest that bias accounts for little or none of the inflation.
For depressive symptoms, we identified two lead SNPs, indicated in the Manhattan plot (Fig. 1b). For neuroticism, our meta-analysis yielded 16 loci that are independent according to our locus definition (Fig. 1c). However, 6 of these reside within a well-known inversion polymorphism16 on chromosome 8. We established that all genome-wide significant signals in the inversion region are attributable to the inversion, and we confirmed that the inversion is associated with neuroticism in both of our neuroticism datasets, the GPC and the UKB (Online Methods and Supplementary Note). In our list of lead SNPs (Table 1), we only retain the most strongly associated SNP from these 6 loci to tag the chromosome 8 inversion.
Another lead SNP associated with neuroticism, rs193236081, is located within a well-known inversion polymorphism on chromosome 17. We established that this association is attributable to the inversion polymorphism (Online Methods and Supplementary Note). Because this inversion yields only one significant locus and is genetically complex17, we hereafter simply use its lead SNP as its proxy. Our neuroticism GWAS therefore identified 11 lead SNPs, two of which tag inversion polymorphisms. A concurrent neuroticism GWAS using a subset of our sample reports similar findings18.
As shown in Table 1, the estimated effects of all lead SNPs associated with depressive symptoms and neuroticism are in the range 0.020 to 0.031 SDs per allele (R2 0.02% to 0.04%). In the UKB cohort we estimated the effect of an additional allele of the chromosome 8 inversion polymorphism itself on neuroticism to be 0.035 SDs (Supplementary Table 13). The inversion explains 0.06% of the variance in neuroticism (roughly the same as the total variance explained jointly by the 6 SNPs in the inversion region).
Genetic overlap across subjective well-being, depressive symptoms, and neuroticism
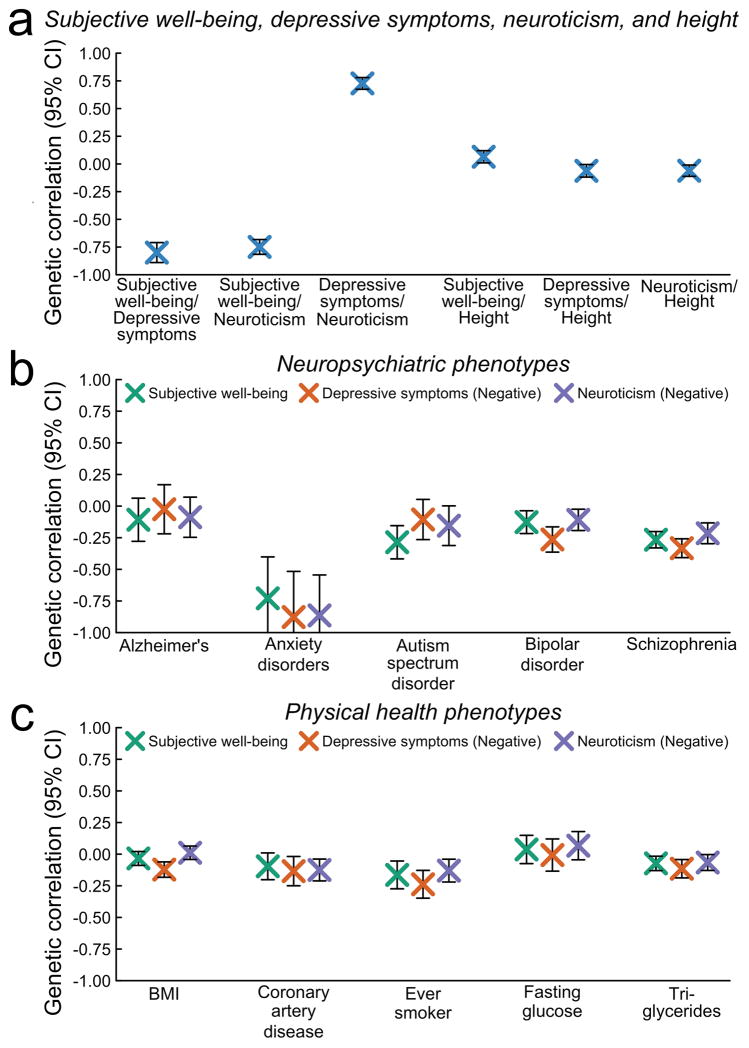
Figure 2a shows that the three pairwise genetic correlations between our phenotypes, estimated using bivariate LD Score regression10, are substantial: −0.81 (SE = 0.046) between subjective well-being and depressive symptoms, −0.75 (SE = 0.034) between subjective well-being and neuroticism, and 0.75 (SE = 0.027) between depressive symptoms and neuroticism. Using height as a negative control, we also examined pairwise genetic correlations between each of our phenotypes and height and, as expected, found all three to be modest, e.g., 0.07 with subjective well-being (Supplementary Table 1). The high genetic correlations between subjective well-being, depressive symptoms, and neuroticism may suggest that the genetic influences on these phenotypes are predominantly related to processes common across the phenotypes, such as mood, rather than being phenotype-specific.
Fig. 2. Genetic correlations with bars representing 95% confidence intervals.
The correlations are estimated using bivariate LD Score (LDSC) regression. (a) Genetic correlations between subjective well-being, depressive symptoms, and neuroticism (“our three phenotypes”), as well as between our three phenotypes and height. (b) Genetic correlations between our three phenotypes and selected neuropsychiatric phenotypes. (c) Genetic correlations between our three phenotypes and selected physical health phenotypes. In (b) and (c), we report the negative of the estimated correlation with depressive symptoms and neuroticism (but not subjective well-being).
Quasi-replication and Bayesian credibility analyses
We assessed the credibility of our findings using a standard Bayesian framework19,20 in which a positive fraction of SNPs have null effects and a positive fraction have non-null effects (Online Methods). For each phenotype, the non-null effect sizes are assumed to be drawn from a normal distribution whose variance is estimated from the GWAS summary statistics. As a first analysis, for each lead SNP’s association with its phenotype, we calculated the posterior probability of null association after having observed the GWAS results. We found that, for any assumption about the fraction of non-null SNPs in the range 1% to 99%, the probability of true association always exceeds 95% for all 16 loci (and always exceeds 98% for 14 of them).
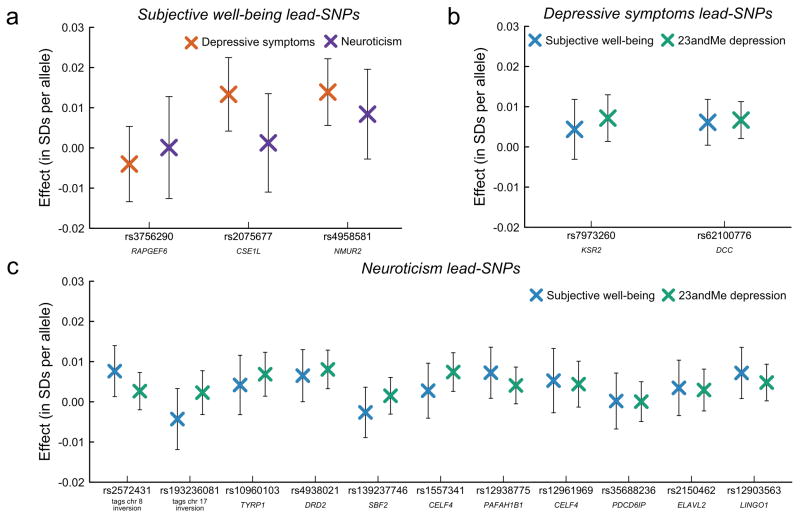
To further probe the credibility of the findings, we performed “quasi-replication” exercises (Online Methods) in which we tested the subjective well-being lead-SNPs for association with depressive symptoms and neuroticism. We similarly tested the depressive symptoms lead-SNPs and the neuroticism lead-SNPs for association with subjective well-being. Below, we refer to the phenotype for which the lead SNP was identified as the first-stage phenotype and the phenotype used for the quasi-replication as the second-stage phenotype. To avoid sample overlap, for each quasi-replication analysis we omitted any cohorts that contributed to the GWAS of the first-stage phenotype.
Results of the quasi-replication of the three subjective well-being lead-SNPs are shown in Figure 3a. For ease of interpretation, the reference allele for each association in the figure is chosen such that the predicted sign of the second-stage estimate is positive. We find that two out of the three subjective well-being lead-SNPs are significantly associated with depressive symptoms (p = 0.004 and p = 0.001) in the predicted direction. For neuroticism, where the second-stage sample size (N = 68,201) is about half as large, the subjective well-being-increasing allele has the predicted sign for all three SNPs, but none reach significance.
Fig. 3. Quasi-replication and lookup of lead SNPs.
In quasi-replication analyses, we examined whether (a) lead SNPs identified in the subjective well-being meta-analyses are associated with depressive symptoms or neuroticism, (b) lead SNPs identified in the analyses of depressive symptoms are associated with subjective well-being, and (c) lead SNPs identified in the analyses of neuroticism are associated with subjective well-being. The quasi-replication sample is always restricted to non-overlapping cohorts. In a separate lookup exercise, we examined whether lead SNPs for depressive symptoms and neuroticism are associated with depression in an independent sample of 23andMe customers (N = 368,890). The results from this lookup are depicted as green crosses in (b) and (c). Bars represent 95% CIs (not adjusted for multiple testing). For interpretational ease, we choose the reference allele so that positive coefficients imply that the estimated effect is in the predicted direction. Listed below each lead SNP is the nearest gene.
Figures 3b and 3c show the results for the depressive symptoms and neuroticism lead-SNPs, respectively. In each panel, the blue crosses depict results from the quasi-replications where subjective well-being is the second-stage phenotype. We find that the two depressive symptoms lead-SNPs have the predicted sign for subjective well-being, and one is nominally significant (p = 0.04). Finally, of the eleven neuroticism lead-SNPs, nine have the predicted sign for subjective well-being. Four of the eleven are nominally significantly associated with subjective well-being, all with the predicted sign. One of the four is the SNP tagging the inversion on chromosome 816. That SNP’s association with neuroticism (and likely with subjective well-being) is driven by its correlation with the inversion (Supplementary Fig. 2).
To evaluate what these quasi-replication results imply about the credibility of the 16 GWAS associations, we compared the observed quasi-replication record to the quasi-replication record expected given our statistical power. We calculated statistical power using our Bayesian framework, under the hypothesis that each lead SNP has a non-null effect on both the first- and second-stage phenotypes. Our calculations take into account both the imperfect genetic correlation between the first- and second-stage phenotypes and inflation of the first-stage estimates due to the well-known problem of winner’s curse (Online Methods). Of the 19 quasi-replication tests, our calculations imply that 16.7 would be expected to yield the anticipated sign and 6.9 would be significant at the 5% level. The observed numbers are 16 and 7. Our quasi-replication results are thus consistent with the hypothesis that none of the 16 genome-wide significant associations are chance findings, and in fact strengthen the credibility of our GWAS results (Supplementary Table 14).
Lookup of depressive symptoms and neuroticism lead-SNPs
Investigators of an ongoing large-scale GWAS of major depressive disorder (N = 368,890) in the 23andMe cohort shared association results for the loci identified in our depressive symptoms and neuroticism analyses (Online Methods and Supplementary Table 15)21. Because the depression sample overlaps with our subjective well-being sample, we did not request a lookup of the subjective well-being-associated SNPs.
In Figures 3b and 3c, the results are depicted as green crosses. For interpretational ease, we chose the reference allele so that positive coefficients imply that the estimated effect is in the predicted direction. All 13 associations have the predicted sign. Of the 11 neuroticism polymorphisms, four are significantly associated with depression at the 5% level. Both of the depressive symptoms lead-SNPs replicate (p = 0.004 and p = 0.015), with effect sizes (0.007 and −0.007 SDs per allele), close to those predicted by our Bayesian framework (0.008 and −0.006) (Supplementary Table 14 and Supplementary Table 15).
Panel A of Table 1 summarizes the results for the 16 lead SNPs identified across our separate GWA analyses of the three phenotypes. The right-most column summarizes the statistical significance of the quasi-replication and depression lookup analyses of each SNP.
Proxy-phenotype analyses
To identify additional SNPs associated with depressive symptoms, we conducted a two-stage “proxy phenotype” analysis (Online Methods). In the first stage, we ran a new GWAS of subjective well-being to identify a set of candidate SNPs. Specifically, from each locus exhibiting suggestive evidence of association (p < 10−4) with subjective well-being, we retained the SNP with the lowest p-value as a candidate. In the second stage, we tested these candidates for association with depressive symptoms at the 5% significance threshold, Bonferroni-adjusted for the number of candidates. We used an analogous two-stage procedure to identify additional SNPs associated with neuroticism. The first-stage subjective well-being sample differs across the two proxy-phenotype analyses (and from the primary subjective well-being GWAS sample) because we assigned cohorts across the first and second stages so as to maximize statistical power for the overall procedure.
For depressive symptoms, there are 163 candidate SNPs. 115 of them (71%) have the predicted direction of effect on depressive symptoms, 20 are significantly associated at the 5% significance level (19 in the predicted direction), and two remain significant after Bonferroni adjustment. For neuroticism, there are 170 candidate SNPs. 129 of them (76%) have the predicted direction of effect, all 28 SNPs significant at the 5% level have the predicted sign, and four of these remain significant after Bonferroni adjustment (Supplementary Fig. 3 and Supplementary Tables 16 and 17). Two of the four are the SNPs identified in the proxy-phenotype analysis for depressive symptoms.
Table 1 lists the four SNPs in total identified by the proxy-phenotype analyses.
Biological analyses
To shed some light on possible biological mechanisms underlying our findings, we conducted several analyses.
We began by using bivariate LD Score regression10 to quantify the amount of genetic overlap between each of our three phenotypes and ten neuropsychiatric and physical health phenotypes. Figures 2b and c display the estimates for subjective well-being and the negative of the estimates for depressive symptoms and neuroticism (since subjective well-being is negatively genetically correlated with depressive symptoms and neuroticism). Subjective well-being, depressive symptoms, and neuroticism have strikingly similar patterns of pairwise genetic correlation with the other phenotypes.
Figure 2b shows the results for the five neuropsychiatric phenotypes we examined: Alzheimer’s disease, anxiety disorders, autism spectrum disorder, bipolar disorder, and schizophrenia. For four of these phenotypes, genetic correlations with depression (but not neuroticism or subjective well-being) were reported in Bulik-Sullivan et al.10. For schizophrenia and bipolar disorder, our estimated correlations with depressive symptoms, 0.33 and 0.26, are substantially lower than Bulik-Sullivan et al.’s point estimates but contained within their 95% confidence intervals. By far the largest genetic correlations we estimate are with anxiety disorders: −0.73 with subjective well-being, 0.88 with depressive symptoms, and 0.86 with neuroticism. Genetic correlations estimated from GWAS data have not been previously reported for anxiety disorders.
Figure 2c shows the results for five physical health phenotypes that are known or believed to be risk factors for various adverse health outcomes: body mass index (BMI), ever-smoker status, coronary artery disease, fasting glucose, and triglycerides. The estimated genetic correlations are all small in magnitude, consistent with earlier work, although the greater precision of our estimates allows us to reject null effects in most cases. The signs are generally consistent with those of the phenotypic correlations reported in earlier work between our phenotypes and outcomes such as obesity22, smoking23,24, and cardiovascular health25.
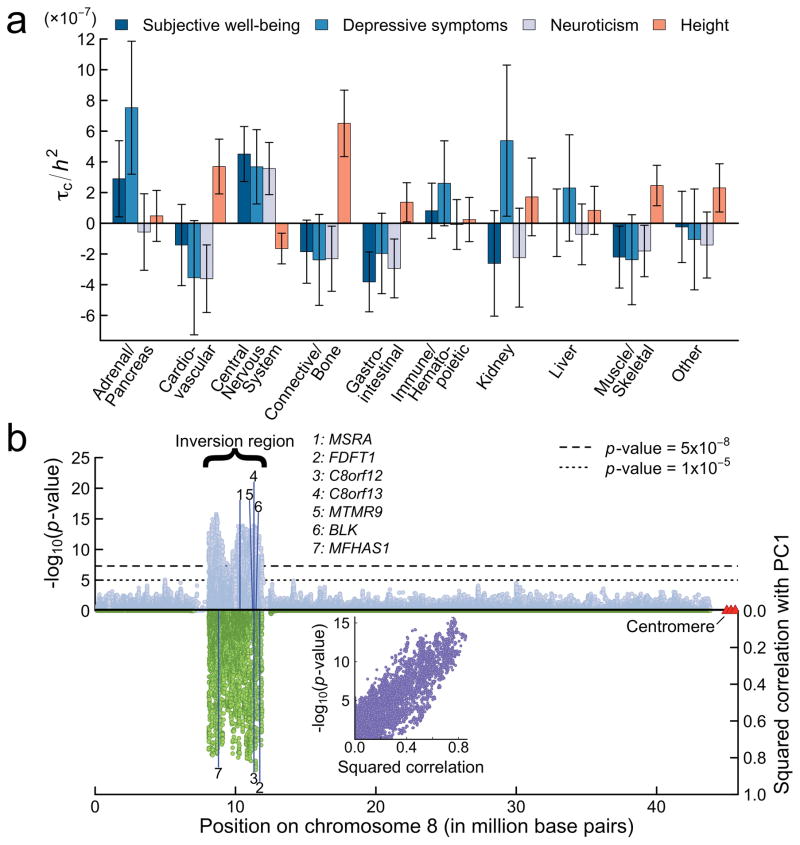
Next, to investigate whether our GWAS results are enriched in particular functional categories, we applied stratified LD Score regression26 to our meta-analysis results. In our first analysis, we report estimates for all 53 functional categories included in the “baseline model”; the results for subjective well-being, depressive symptoms, and neuroticism are broadly similar (Supplementary Tables 18–20) and are in line with what has been found for other phenotypes26. In our second analysis, the categories are groupings of SNPs likely to regulate gene expression in cells of a specific tissue. The estimates for subjective well-being, depressive symptoms, and neuroticism are shown in Figure 4a, alongside height, which is again included as a benchmark27 (see also Supplementary Table 21).
Fig. 4. Results from selected biological analyses.
(a) Estimates of the expected increase in the phenotypic variance accounted for by a SNP due to the SNP’s being in a given category (τc), divided by the LD Score heritability of the phenotype (h2). Each estimate of τc comes from a separate stratified LD Score regression, controlling for the 52 functional annotation categories in the “baseline model.” The bars represent 95% CIs (not adjusted for multiple testing). To benchmark the estimates, we compare them to those obtained from a recent study of height27. (b) Inversion polymorphism on chromosome 8 and the 7 genes for which the inversion is a significant cis-eQTL at FDR < 0.05. The upper half of the figure shows the Manhattan plot for neuroticism for the inversion and surrounding regions. The bottom half shows the squared correlation between the SNPs and the principal component that captures the inversion. The inlay plots the relationship, for each SNP in the inversion region, between the SNP’s significance and its squared correlation with the principal component that captures the inversion.
We found significant enrichment of CENTRAL NERVOUS SYSTEM for all three phenotypes and, perhaps more surprisingly, enrichment of ADRENAL/PANCREAS for subjective well-being and depressive symptoms. The cause of the ADRENAL/PANCREAS enrichment is unclear, but we note that the adrenal glands produce several hormones, including cortisol, epinephrine, and norepinephrine, known to play important roles in the bodily regulation of mood and stress. It has been robustly found that blood serum levels of cortisol in patients afflicted by depression are elevated relative to controls28.
While the above analyses utilize the genome-wide data, we also conducted three analyses (Online Methods) restricted to the 16 GWAS and four proxy-phenotype SNPs in Table 1. In brief, we ascertained whether each SNP (or a variant in strong linkage disequilibrium (LD) with it) falls into any of the following three classes: (i) resides in a locus for which genome-wide significant associations with other phenotypes have been reported (Supplementary Table 22), (ii) is nonsynonymous (Supplementary Table 23), and (iii) is an eQTL in blood or in one of 14 other tissues (although the non-blood analyses are based on smaller samples) (Supplementary Table 24). Here we highlight a few particularly interesting results.
We found that five of the 20 SNPs are in loci in which genome-wide significant associations have previously been reported. Two of these five are schizophrenia loci. Interestingly, one of them harbors the gene DRD2, which encodes the D2 subtype of the dopamine receptor, a target for antipsychotic drugs29 that is also known to play a key role in neural reward pathways30. Motivated by these findings, as well as by the modest genetic correlations with schizophrenia reported in Figure 2b, we examined whether the SNPs identified in a recent study of schizophrenia31 are enriched for association with neuroticism in our non-overlapping UKB sample (N = 107,245). We conducted several tests and found strong evidence of such enrichment (Supplementary Note). For example, we found that the p-values of the schizophrenia SNPs tend to be much lower than the p-values of a randomly selected set of SNPs matched on allele frequency (p = 6.50×10−71).
Perhaps the most notable pattern that emerges from our biological analyses is that the inversions on chromosomes 8 and 17 are implicated consistently across all analyses. The inversion-tagging SNP on chromosome 8 is in LD with SNPs that have previously been found to be associated with BMI32 and triglycerides33 (Supplementary Table 22). We also conducted eQTL analyses in blood for the inversion itself and found that it is a significant cis-eQTL for 7 genes (Supplementary Table 24). As shown in Figure 4b, all 7 genes are positioned in close proximity to the inversion breakpoints, suggesting that the molecular mechanism underlying the inversion’s effect on neuroticism could involve the relocation of regulatory sequences. Two of the genes (MSRA, MTMR9) are known to be highly expressed in tissues and cell types that belong to the nervous system, and two (BLK, MFHAS1) in the immune system. In the tissue-specific analyses, we found that the SNP tagging the inversion is a significant eQTL for two genes, AF131215.9 (in tibial nerve and thyroid tissue analyses) and NEIL2 (tibial nerve tissue), both of which are also located near the inversion breakpoint.
The SNP tagging the chromosome 17 inversion is a significant cis-eQTL for five genes in blood and is an eQTL in all 14 other tissues (Supplementary Table 24). It alone accounts for 151 out of the 169 significant associations identified in the 14 tissue-specific analyses. Additionally, the SNP is in near-perfect LD (R2 > 0.97) with 11 missense variants (Supplementary Table 23) in three different genes, one of which is MAPT. MAPT, which is also implicated in both the blood and the other tissue-specific analyses, encodes a protein important in the stabilization of microtubules in neurons. Associations have been previously reported between SNPs in MAPT (all of which are in strong LD with our inversion-tagging SNP) and neurodegenerative disorders, including Parkinson’s disease34 and progressive supranuclear palsy35, a rare disease whose symptoms include depression and apathy.
DISCUSSION
The discovery of genetic loci associated with subjective well-being, depression, and neuroticism has proven elusive. Our study identified several credible associations for two main reasons. First, our analyses had greater statistical power than prior studies because ours were conducted in larger samples. Our GWAS findings—three loci associated with subjective well-being, two with depressive symptoms, and eleven with neuroticism—support the view that GWAS can successfully identify genetic associations with highly polygenic phenotypes in sufficiently large samples5,36. A striking finding is that two of our identified associations are with inversion polymorphisms.
Second, our proxy-phenotype analyses further boosted power by exploiting the strong genetic overlap between our three phenotypes. These analyses identified two additional loci associated with neuroticism and two with both depressive symptoms and neuroticism. Through our quasi-replication tests, we also demonstrated how studying genetically overlapping phenotypes in concert can provide evidence on the credibility of GWAS findings. Our direct replication of the two genome-wide significant associations with depressive symptoms in an independent depression sample provides further confirmation of those findings (Fig. 2b and Supplementary Table 15).
We were able to assemble much larger samples than prior work in part because we combined data across heterogeneous phenotype measures. Our results reinforce the conclusions from our theoretical analysis that doing so increased our statistical power, but our strategy also has drawbacks. One is that mixing different measures may make any discovered associations more difficult to interpret. Research studying higher quality measures of the various facets of subjective well-being, depressive symptoms, and neuroticism is a critical next step. Our results can help facilitate such work because if the variants we identify are used as candidates, studies conducted in the smaller samples in which more fine-grained phenotype measures are available can be well powered.
Another limitation of mixing different measures is that doing so may reduce the heritability of the resulting phenotype, if the measures are influenced by different genetic factors. Indeed, our estimates of SNP-based heritability10 for our three phenotypes are quite low: 0.040 (SE = 0.002) for subjective well-being, 0.047 (SE = 0.004) for depressive symptoms, and 0.091 (SE = 0.007) for neuroticism. We correspondingly find that polygenic scores constructed from all measured SNPs explain a low fraction of variance in independent samples: ~0.9% for subjective well-being, ~0.5% for depressive symptoms, and ~0.7% for neuroticism (Online Methods). The low heritabilities imply that even when polygenic scores can be estimated using much larger samples than ours, they are unlikely to attain enough predictive power to be clinically useful.
According to our Bayesian calculations, the true explanatory power (corrected for winner’s curse) of the SNP with the largest posterior R2 is 0.003% for subjective well-being, 0.002% for depressive symptoms, and 0.011% for neuroticism (Supplementary Table 14). These effect sizes imply that in order to account for even a moderate share of the heritability, hundreds or (more likely) thousands of variants will be required. They also imply that our study’s power to detect variants of these effect sizes was not high—for example, our statistical power to detect the lead SNP with largest posterior R2 was only ~13%—which in turn means it is likely that there exist many variants with effect sizes comparable to our identified SNPs that evaded detection. These estimates suggest that many more loci will be found in studies with sample sizes realistically attainable in the near future. Consistent with this projection, when we meta-analyze the 54 SNPs reaching p < 10−5 in our analyses of depressive symptoms together with the 23andMe replication sample for depression, the number of genome-wide significant associations rises from 2 to 5 (Supplementary Table 15).
ONLINE METHODS
This article is accompanied by a Supplementary Note with further details.
GWAS of subjective well-being
Genome-wide association analyses were performed at the cohort level according to a pre-specified analysis plan. Genotyping was performed using a range of common, commercially available genotyping arrays. The analysis plan instructed cohorts to upload results imputed using the HapMap2 CEU (r22.b36) reference sample37. We meta-analyzed summary association statistics from 59 contributing cohorts with a combined sample size of 298,420 individuals. Before meta-analysis, a uniform set of quality-control (QC) procedures were applied to the cohort-level summary statistics, including but not limited to the EasyQC38 protocol. All analyses were restricted to European-ancestry individuals.
We performed a sample-size-weighted meta-analysis of the cohort-level summary statistics. To adjust standard errors for non-independence, we inflated them using the square root of the estimated intercept from a LD Score regression10. We also performed secondary, separate meta-analyses of positive affect (N = 180,281) and life satisfaction (N = 166,205) and a post hoc genome-wide analysis of subjective well-being in cohorts with 1000G-imputed data (N = 229,883); see Supplementary Figures 4–6.
Detailed cohort descriptions, information about cohort-level genotyping and imputation procedures, cohort-level measures, and quality-control filters are shown in Supplementary Tables 2–6. Supplementary Table 7 reports association results from the following four meta-analyses: the primary subjective well-being analysis, the life satisfaction analysis, the positive affect analysis, and the post hoc subjective well-being analysis. For each phenotype, we provide association results for the set of approximately independent SNPs that attained a p-value smaller than 10−5. We identify these SNP using the same clumping algorithm as for the lead SNPs, but with the p-value threshold set at 10−5 instead of genome-wide significance.
GWAS of depressive symptoms and neuroticism
Our auxiliary genome-wide association studies of DS and neuroticism were conducted in 1000G-imputed data, combining new genome-wide association analyses with publicly available summary statistics from previously published studies. We applied a similar QC protocol to that used in our primary subjective well-being analysis. In the DS meta-analysis (N = 180,866), we weighted the UKB analysis by sample size and the two case-control studies by effective sample size. In the neuroticism meta-analysis, we performed a sample-size-weighted fixed-effects meta-analysis of the UKB data and the publicly available summary statistics from a previous GWAS of neuroticism.
Detailed cohort descriptions, information about cohort-level genotyping and imputation procedures and quality-control filters are provided in Supplementary Tables 8–12. See Supplementary Figure 7 for quantile-quantile plots of the neuroticism and DS meta-analysis results. Association results for the set of approximately independent set of SNPs that attained a p-value smaller than 10−5 are supplied in Supplementary Table 25.
Population stratification
To quantify the fraction of the observed inflation of the mean test statistic that is due to bias, we used LD Score regression10. The estimated LD Score regression intercepts were all close to 1, suggesting no appreciable inflation of the test statistics attributable to population stratification in any of our subjective well-being, depressive symptoms, or neuroticism meta-analyses (Supplementary Fig. 8). For all three phenotypes, our estimates suggest that less than 2% of the observed inflation of the mean test statistic was accounted for by bias.
In our primary GWAS of subjective well-being, we also used two family-based analyses to test for and quantify stratification biases. These analyses used within-family (WF) estimates, the coefficients from regressing the difference in phenotype across siblings on the difference in siblings’ genotype (and controls). These WF estimates are not biased by population stratification because siblings share their ancestry entirely, and therefore differences in siblings’ genotypes cannot be due to the siblings being from different population groups. We meta-analyzed association statistics from WF analyses conducted in four cohorts.
In the first analysis, we estimated the fraction of SNPs for which the signs of the WF estimates were concordant with the signs of the estimates obtained from a GWAS identical to our primary subjective well-being GWAS except with the four family cohorts excluded. For the 112,884 approximately independent SNPs considered, we found a sign concordance of 50.83%, which is significantly greater than 50% (p = 1.04 × 10−8). Under the null hypothesis of no population stratification, the observed sign concordance matches the expected rate after winner’s curse adjustment nearly perfectly, 50.83% (Supplementary Fig 9).
The second analysis utilized the WF regression coefficient estimates (i.e., not only their signs) to estimate the amount of stratification bias. For each SNP j, let βj denote the GWAS estimate, and let βWF,j denote the WF estimate. Under the assumption that the causal effect of each SNP is the same within families as in the population, we can decompose the estimates as:
where βj is the true underlying GWAS parameter for SNP j, sj is the bias due to stratification (defined to be orthogonal to βj and Uj), and Uj and Vj are the sampling variances of the estimates with E(Uj)=E(Vj)=0. Whenever sj ≠ 0, the GWAS estimate of β̂j is biased away from the population parameter βj. The proportion of variance in the GWAS coefficients accounted for by true genetic signals can be written as:
In Supplementary Note, we show that with estimates β̂j and β̂WF,j (and their standard errors) from independent samples, it is possible to consistently estimate the above ratio. The 95% confidence interval for the ratio implies that between 72% and 100% of the signal in the GWAS estimates is a result of true genetic effects on subjective well-being rather than stratification.
Analyses of inversion polymorphisms
Two genome-wide significant SNPs for the neuroticism analysis are located within well-known inversion polymorphisms, on chromosomes 8 and 17. Using the genotypic data available for UKB participants, we called the inversion genotypes for UKB participants using a PCA-mixture method. For both inversions, the method clearly distinguishes 3 clusters of genotypes, corresponding to inversion genotypes (Supplementary Fig. 10). We validated the PCA-mixture procedure using existing methods designed to call inversion genotypes39 (Supplementary Table 26).
For both inversions, we established that the inversion-tagging SNPs were always located in close proximity of the inversion region (Fig. 3b and Supplementary Figs. 10–11). Supplementary Tables 27–28 list the twenty variants that most strongly correlate with the PCs that capture the inversion polymorphisms on chromosome 8 and 17, respectively. In additional analyses, we confirmed that the inversion is associated with neuroticism and subjective well-being in independent cohorts (Supplementary Tables 29–30 and Supplementary Fig. 12–13).
Proxy-phenotype analyses
In these analyses, we used a two-stage approach that has been successfully applied in other contexts6. In the first stage, we conducted a meta-analysis of our first-stage “proxy phenotype” and used our clumping procedure to identify the set of approximately independent SNPs at the p-value threshold of 10−4. In the second stage, we tested SNPs identified in stage 1 (or high-LD proxies for them) for association with a second-stage phenotype in an independent (non-overlapping) sample. In our analyses, we used our primary phenotype of subjective well-being as the proxy-phenotype. We conducted one analysis with depressive symptoms as the second-stage phenotype, and one analysis with neuroticism as the second-stage phenotype. In the analyses, we omit cohorts from the first-stage or second-stage as needed to ensure that the samples in the two stages are non-overlapping. Supplementary Table 31 lists the cohort restrictions imposed. These cohort restrictions, as well as the p-value threshold of 10−4, were chosen before the data were analyzed on the basis of statistical power calculations.
To test for cross-phenotype enrichment, we used a non-parametric procedure that tests whether the lead SNPs are more strongly associated with the second-stage phenotype than randomly chosen sets of SNPs with a similar distribution of allele frequencies (Supplementary Note).
To test the individual lead SNPs for experiment-wide significance, we examined whether any of the lead SNPs (or their high-LD proxies) are significantly associated with the second-stage phenotype at the Bonferroni-adjusted significance level of 0.05/Y.
Genetic correlations
We used bivariate LD Score regression10 to quantify the amount of genetic heterogeneity among the phenotypic measures pooled in each of our three separate meta-analyses. For subjective well-being, we estimated a pairwise correlation of 0.981 (SE = 0.065) between life satisfaction and positive affect, 0.897 (SE = 0.017) between “wellbeing” (our measure that combines life satisfaction and positive affect) and life satisfaction, and 1.031 (SE = 0.019) between positive affect and wellbeing. For depressive symptoms, we estimated a genetic correlation of 0.588 (SE = 0.242) between GERA and PGC, 0.972 (SE = 0.216) between GERA and UKB, and 0.797 (SE = 0.108) between UKB and PGC. Finally, we estimated a genetic correlation of 1.11 (SE = 0.14) between the measures of neuroticism in the UKB analyses and the summary statistics from a previously published meta-analysis4.
Bayesian credibility analyses
To evaluate the credibility of our findings, we use a standard Bayesian framework19 in which our prior distribution for any SNP’s effect is:
Here, π is the fraction of non-null SNPs, and is the variance of the non-null SNPs for trait j ∈ {subjective well-being, depressive symptoms, neuroticism}. In this framework, credibility is defined as the probability that a given SNP is non-null.
We begin with univariate analyses of the GWAS results that do not incorporate the additional information from the quasi-replication analyses of the 16 lead SNPs reported in Table 1. We use the three subjective well-being-associated SNPs to illustrate our approach, but we use analogous procedures when analyzing depressive symptoms and neuroticism. We calculate credibility for each value π ∈ 0.01,0.02,…,0.99. For each assumed value of π, we estimate by maximum likelihood (Supplementary Note). For each SNP, we use Bayes’ rule to obtain a posterior estimate of credibility for each of the assumed values of π. Supplementary Figure 14 shows that for all considered values of π and all three SNPs, the posterior probability that the SNP is null is below 1%. Similar analyses of the depressive symptoms and neuroticism SNPs show that the posterior probability never exceeds 5%.
In our joint analyses, we consider two phenotypes with genetic correlation rg. We make the simplifying assumption that the set of null SNPs is the same for both phenotypes. The joint distribution of a SNP’s effect on the two phenotypes is then given by
With coefficient estimates, β̂1 and β̂2, obtained from non-overlapping samples, the variance-covariance matrix of the estimation error will be diagonal. We denote the diagonal entries of this matrix, which represent the variances of the estimation error in the two samples, by and . This gives us the joint prior distribution
To select parameter values for the prior, we use the estimates of rg reported in Supplementary Table 1, and we estimate the parameters π, , and from GWAS summary statistics using a maximum likelihood procedure. For this procedure, we make the standard assumption10,40 that the variance of a SNP’s effect size is inversely proportional to the variance of its genotype, 2×MAF×(1−MAF).
The credibility estimates follow from applying Bayes’ Rule to calculate either the probability that the SNP is non-null (an event denoted C) given only the first-stage estimate, P(C|β̂1), or the probability that the SNP is non-null conditional on the results of both the first-stage GWAS and the quasi-replication analysis, P(C|β̂1, β̂2). Credibility estimates for our lead SNPs are in Supplementary Table 14.
To calculate the expected record of a replication or quasi-replication study, we assume that the SNP is non-null for both phenotypes. (This is analogous to a standard power calculation for a single phenotype, in which the SNP is assumed to be non-null.) Under this assumption, β̂1 and β̂2 are jointly normally distributed, implying that the conditional distribution of β̂2 given β̂1 is
Using this equation, we can calculate the probability that the GWAS estimates will have concordant signs across the two phenotypes, or that the GWAS estimate of the second-stage phenotype will reach some level of significance. These probabilities can be summed over the set of lead SNPs to generate the expected number of SNPs meeting the criterion.
To obtain effect-size estimates for a SNP that are adjusted for the winner’s curse (Supplementary Table 32), we use the mean of the posterior distribution of the SNP’s effect, conditional on the quasi-replication result and the SNP being non-null. We derive the posterior distribution and expected R2 in the Supplementary Note.
Lookup of depressive symptoms and neuroticism-associated SNPs in an independent depression study
We partnered with the investigators of an ongoing large-scale GWAS of major depressive symptoms (N = 368,890) to follow up on the associations identified in the depressive symptoms and neuroticism analyses. The participants of the study were all European-ancestry customers of 23andMe, a personal genomics company, who responded to online survey questions about mental health. We did not request results for the SNPs identified in the subjective well-being or proxy-phenotype analyses, since these were both conducted in samples that overlap with 23andMe’s depression sample. For details on association models, quality-control filters, and the ascertainment of depression status, we refer to the companion study21. The p-values we report are based on standard errors that have been inflated by the square by the intercept from an LD score regression10.
Polygenic prediction
To evaluate the predictive power of a polygenic score derived from the subjective well-being meta-analysis results, we used two independent hold-out cohorts: the Health and Retirement Study (HRS41) and the Netherlands Twin Register (NTR42,43). To generate the weights for the polygenic score, we performed meta-analyses of the pooled subjective well-being phenotype excluding each of the holdout cohorts, applying a minimum-sample-size filter of 100,000 individuals (Supplementary Note). The results from these analyses are reported in Supplementary Table 33 and depicted in Supplementary Figure 15.
Biological annotation
For the biological annotation of the 20 SNPs in Table 1, we generated a list of LD partners for each of the original SNPs. A SNP was considered an LD partner for the original SNP if (i) its pairwise LD with the original SNP exceeded R2 = 0.6 and (ii) it was located within 250kb of the original SNP. We also generated a list of genes residing within loci tagged by our lead SNPs (Supplementary Table 34).
We used the NHGRI GWAS catalog44 to determine which of our 20 SNPs (and their LD partners) were in LD with SNPs for which genome-wide significant associations have been previously reported. Since the GWAS catalog does not always include the most recent GWAS results available, we included additional recent GWAS studies. We used the tool HaploReg45 to identify nonsynonymous variants in LD with any of the 20 SNPs or their LD partners.
We examined whether the 20 polymorphisms in Table 1 were associated with gene expression levels (Supplementary Table 24 and Supplementary Note). The cis-eQTL associations were performed in 4,896 peripheral-blood gene expression and genome-wide SNP samples from two Dutch cohorts measured on the Affymetrix U219 platform42,43,46. We also performed eQTL lookups of our 20 SNPs in the Genotype-Tissue Expression Portal47,48. We restricted the search to the following trait-relevant tissues: hippocampus, hypothalamus, anterior cingulate cortex (BA24), putamen (basal ganglia), frontal cortex (BA9), nucleus accumbens (basal ganglia), caudate (basal ganglia), cortex, cerebellar hemisphere, cerebellum, tibial nerve, thyroid, adrenal gland, and pituitary.
Finally, using a gene co-expression database49, we explored the predicted functions of genes co-locating with the 20 SNPs in Table 1 (Supplementary Table 35).
Supplementary Material
Acknowledgments
This research was carried out under the auspices of the Social Science Genetic Association Consortium (SSGAC). The SSGAC seeks to facilitate studies that investigate the influence of genes on human behavior, well-being, and social-scientific outcomes using large genome-wide association study meta-analyses. The SSGAC also provides opportunities for replication and promotes the collection of accurately measured, harmonized phenotypes across cohorts. The SSGAC operates as a working group within the CHARGE consortium. This research has also been conducted using the UK Biobank Resource. The study was supported by funding from the U.S. National Science Foundation (EAGER: “Workshop for the Formation of a Social Science Genetic Association Consortium”), a supplementary grant from the National Institute of Health Office of Behavioral and Social Science Research, the Ragnar Söderberg Foundation (E9/11), the Swedish Research Council (421-2013-1061), The Jan Wallander and Tom Hedelius Foundation, an ERC Consolidator Grant (647648 EdGe), the Pershing Square Fund of the Foundations of Human Behavior, and the NIA/NIH through grants P01-AG005842, P01-AG005842-20S2, P30-AG012810, and T32-AG000186-23 to NBER and R01-AG042568-02 to the University of Southern California. We are grateful to Peter M. Visscher for advice, support, and feedback. We thank Samantha Cunningham and Nishanth Galla for research assistance. A full list of acknowledgments is provided in the Supplementary Note.
Footnotes
URLs:
Genotype-Tissue Expression Portal www.GTExportal.org
Social Science Genetic Association Consortium (SSGAC) website: http://www.thessgac.org/#!data/kuzq8.
ACCESSION CODES: For neuroticism and depressive symptoms, we provide meta-analysis results from the combined analyses for all variants. For subjective well-being, meta-analysis results for all variants are provided for the full sample excluding 23andMe, for which only up to 10,000 SNPs can be reported. Therefore, for the subjective well-being meta-analysis, we provide results for 10,000 SNPs. Meta-analysis results can be downloaded from the SSGAC website.
AUTHOR CONTRIBUTIONS: M.B., D.J.B., D.C., J.E.D.N., P.D.K., and R.F.K. designed and oversaw the study. A.O. and B.B. were responsible for quality control and meta-analyses. Bioinformatics analyses were carried out by J.B., T.E., M.A.F., J.R.G., J.L., S.F.W.M., M.N., and H.J.W. Other follow-up analyses were conducted by M.A.F., J.B., P.T., A.O., B.B., and R.K.L. Especially major contributions to the writing and editing were made by M.B., D.J.B., J.B., D.C., J.E.D.N., P.K., A.O., and P.T. All authors contributed to and critically reviewed the manuscript.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
References
- 1.Kendler KS, Myers J. The genetic and environmental relationship between major depression and the five-factor model of personality. Psychol Med. 2009;40:801. doi: 10.1017/S0033291709991140. [DOI] [PubMed] [Google Scholar]
- 2.Weiss A, Bates TC, Luciano M. Happiness is a personal(ity) thing: The genetics of personality and well-being in a representative sample. Psychol Sci. 2008;19:205–210. doi: 10.1111/j.1467-9280.2008.02068.x. [DOI] [PubMed] [Google Scholar]
- 3.Bartels M, Cacioppo JT, van Beijsterveldt TCEM, Boomsma DI. Exploring the association between well-being and psychopathology in adolescents. Behav Genet. 2013;43:177–190. doi: 10.1007/s10519-013-9589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Moor MHM, et al. Meta-analysis of genome-wide association studies for neuroticism, and the polygenic association with Major Depressive Disorder. JAMA Psychiatry. 2015;72:642–650. doi: 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyman S. Mental health: Depression needs large human-genetics studies. Nature. 2014;515:189–191. doi: 10.1038/515189a. [DOI] [PubMed] [Google Scholar]
- 6.Rietveld CA, et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc Natl Acad Sci U S A. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kahneman D, Deaton A. High income improves evaluation of life but not emotional well-being. Proc Natl Acad Sci U S A. 2010;107:16489–16493. doi: 10.1073/pnas.1011492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kahneman D, Riis J. In: The Science of Well-Being. Uppter F, Baylis N, Keverne B, editors. Oxford University Press; 2005. pp. 285–301. [Google Scholar]
- 9.Bartels M, Boomsma DI. Born to be happy? The etiology of subjective well-being Behav Genet. 2009;39:605–615. doi: 10.1007/s10519-009-9294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudlow C, et al. UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.dbGaP. Resource for Genetic Epidemiology Research on Adult Health and Aging (GERA) 2015 at < http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000674.v1.p1>.
- 15.Eysenck HJ, Eysenck SBG. Manual of the Eysenck Personality Questionnaire. Hodder and Stroughton; 1975. [Google Scholar]
- 16.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genet. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg KM, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet. 2012;44:872–80. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DJ, et al. Genome-wide analysis of over 106,000 individuals identifies 9 neuroticism-associated loci. 2015 doi: 10.1101/032417. bioRxiv 032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilhjálmsson BJ, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyde CL, et al. Common genetic variants associated with major depressive disorder among individuals of European descent. Nat Genet [Google Scholar]
- 22.Roberts RE, Kaplan Ga, Shema SJ, Strawbridge WJ. Are the obese at greater risk for depression? Am J Epidemiol. 2000;152:163–170. doi: 10.1093/aje/152.2.163. [DOI] [PubMed] [Google Scholar]
- 23.Glassman AJ, et al. Smoking, smoking cessation, and major depression. J Am Med Assoc. 1990;264:1546–1549. [PubMed] [Google Scholar]
- 24.Shahab L, West R. Differences in happiness between smokers, ex-smokers and never smokers: Cross-sectional findings from a national household survey. Drug Alcohol Depend. 2012;121:38–44. doi: 10.1016/j.drugalcdep.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Rugulies R. Depression as a predictor for coronary heart disease: A review and meta-analysis. Am J Prev Med. 2002;23:51–61. doi: 10.1016/s0749-3797(02)00439-7. [DOI] [PubMed] [Google Scholar]
- 26.Finucane HK, et al. Partitioning heritability by functional category using GWAS summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stetler C, Miller GE. Depression and hypothalamic-pituitary-adrenal activation: a quantitative summary of four decades of research. Psychosom Med. 2011;73:114–126. doi: 10.1097/PSY.0b013e31820ad12b. [DOI] [PubMed] [Google Scholar]
- 29.Seeman P. Dopamine D2 receptors as treatment targets in schizophrenia. Clin Schizophr Relat Psychoses. 2010;4:56–73. [PubMed] [Google Scholar]
- 30.Vallone D, Picetti R, Borrelli E. Structure and function of dopamine receptors. Neurosci Biobehav Rev. 2000;24:125–132. doi: 10.1016/s0149-7634(99)00063-9. [DOI] [PubMed] [Google Scholar]
- 31.Ripke S, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer CCA, et al. Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20:345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Höglinger GU, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan PF. Don’t give up on GWAS. Mol Psychiatry. 2012;17:2–3. doi: 10.1038/mp.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium TIH. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler TW, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cáceres A, González JR. Following the footprints of polymorphic inversions on SNP data: from detection to association tests. Nucleic Acids Res. 2015;43:e53. doi: 10.1093/nar/gkv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonnega A, et al. Cohort Profile: the Health and Retirement Study (HRS) Int J Epidemiol. 2014;43:576–585. doi: 10.1093/ije/dyu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willemsen G, et al. The Adult Netherlands Twin Register: Twenty-Five Years of Survey and Biological Data Collection. Twin Res Hum Genet. 2013;16:271–281. doi: 10.1017/thg.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Beijsterveldt CEM, et al. The Young Netherlands Twin Register (YNTR): Longitudinal Twin and Family Studies in Over 70,000 Children. Twin Res Hum Genet. 2013;16:252–267. doi: 10.1017/thg.2012.118. [DOI] [PubMed] [Google Scholar]
- 44.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penninx BWJH, et al. The Netherlands study of depression and anxiety (NESDA): Rationale, objectives and methods. Int J Methods Psychiatr Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardlie KG, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fehrmann RSN, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.