Abstract
RNA synthesis in eukaryotes is divided among three RNA polymerases (RNAP). RNAP III transcribes hundreds of tRNA genes and fewer additional short RNA genes. We survey recent work on transcription by RNAP III including an atomic structure, mechanisms of action, interactions with chromatin and retroposons, and a conserved link between its activity and a tRNA modification that enhances mRNA decoding. Other new work suggests important mechanistic connections to oxidative stress, autoimmunity and cancer, embryonic stem cell pluripotency, and tissue-specific developmental effects. We consider that, for some of its complex functions, variation in RNAP III activity levels lead to nonuniform changes in tRNA pools that can shift the translation profiles of key codon-biased mRNAs with resultant phenotypes or disease states.
RNA polymerase III empowers tRNA genes as individual genetic units
Unlike bacteria and archaea, eukaryotes divide RNA synthesis among three RNA polymerases (RNAPs). RNAP I synthesizes large ribosomal RNA (rRNA). RNAP II synthesizes thousands of mRNAs and numerous noncoding RNAs (ncRNA). RNAP III uniquely synthesizes only short RNAs, most of which are tRNAs, but also include 5S rRNA, U6 snRNA, the short ncRNA component of RN'ases P, the mitochondrial RNA processing (MRP) RNA, the signal recognition particle SRP RNA [1], and in higher eukaryotes a number of micro and other small RNAs [2]. Plants also contain RNAPs IV & V, which are specialized homologs of RNAP II [3]. In contrast to bacteria, which have polycistronic genes that contain multiple tRNAs together with rRNA, eukaryotic tRNA genes are monocistronic (rare tRNA-tRNA dicistronics exist) and exclusive synthesis by RNAP III affords their independent transcriptional control. However, while tRNA gene occupancy by transcription factors in growing yeast is more or less uniform and their synthetic output is thought to be uniformly high, animals exert some tRNA gene-specific expression that is poorly understood and likely reflects local chromatin effects and cell type-specificity [4].
RNAP III activity is linked to posttranscriptional modification of specific tRNAs, which activates them for codon-specific mRNA decoding [5]. Because of wobble decoding most eukaryote genomes require tRNA genes for only 42 or so isoacceptors, although the gene copy number for each of these can vary widely in any given species [6]. Some yeasts and other single cell eukaryotes exhibit good correlation between tRNA gene copy number and cellular tRNA abundance, which is harmonized with translation efficiency and fidelity for their mRNAs [7]. However, data from higher eukaryotes suggest that different subsets of tRNA genes are active in different cell types [4].
Altering the relative amounts of different tRNAs can affect translation generally as well as the hierarchical translation efficiency of individual mRNAs that bear different codon biases. For such mRNAs, the relative abundances of tRNAs can determine ribosome pausing and protein folding apart from general translation efficiency [7]. The control of tRNAs as independent units of expression holds potential for differential mRNA translation. tRNAs have a unique type and range of influence on the expression of genetic information. Because the gene copy numbers for different tRNA isoacceptors are highly variable among and even within species [8–10], they should be considered as dynamic genetic elements that influence phenotype.
That tRNA genes are independent units not only provides genomes with the potential to adapt to novel translational demands, but also with a stable transcription machinery that has chromatin-organizing and replication-associated genome integrity activities [11–13]. A theme of this survey is that the RNAP III system provides potential for differential control that could theoretically coordinate tRNA supply with codon demand during growth, stress and differentiation.
Upstream and internal promoters differentially control class III genes
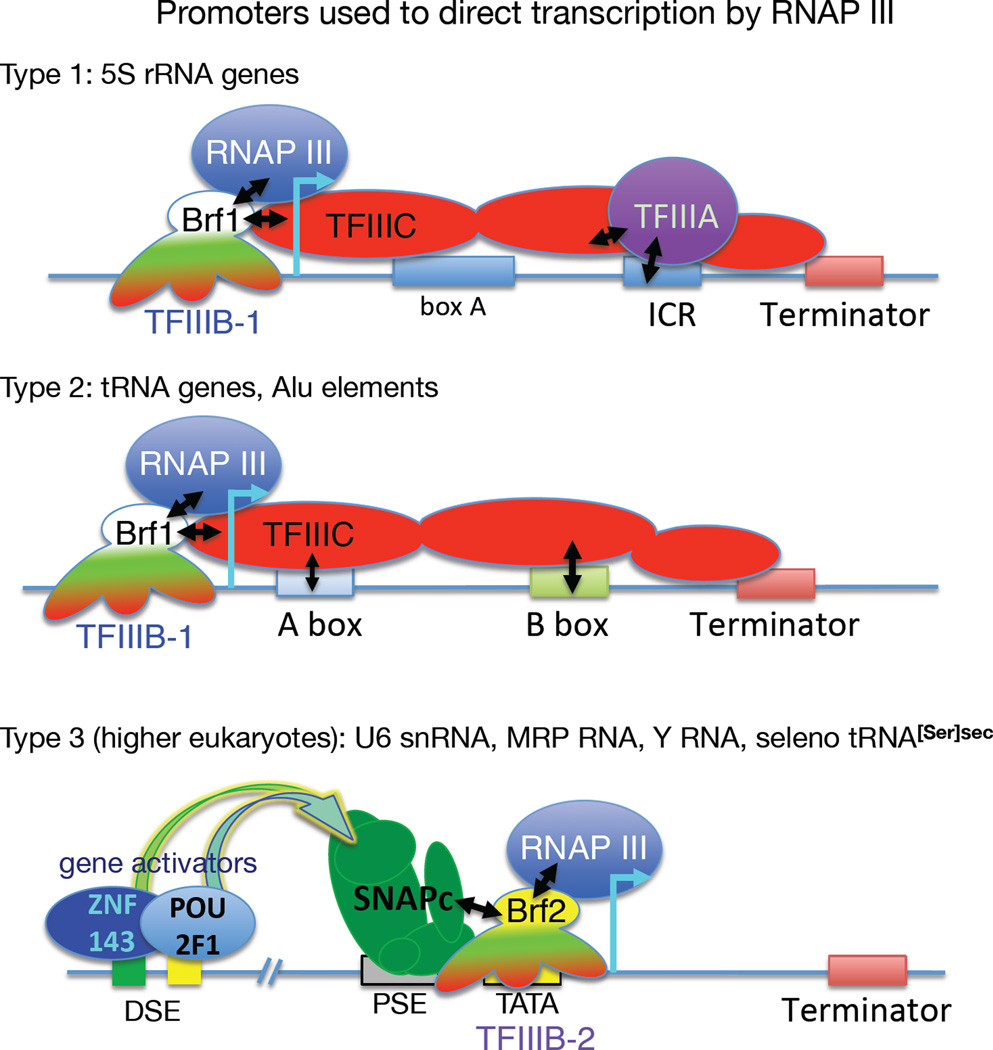
For most RNAP III transcribed genes, which are referred to as class III genes, a three-subunit TFIIIB complex containing TATA binding protein (TBP), TFIIB-related factor-1 (Brf1), and B" (B double prime, Bdp1) recruits RNAP III and directs its initiation. Depending on the class III gene type, 1, 2 or 3, other factors, TFIIIA, TFIIIC or SNAPc, bind sequence-specific promoter elements, and guide TFIIIB onto upstream DNA which directs initiation [14] (Fig. 1). TFIIIC is a 6-subunit complex that recognizes the internal promoter elements of tRNA genes, the A and B boxes (Fig. 1). In chromatin, TFIIIC exhibits plasticity as it accommodates a range of distances between the A and B box, and the B box and terminator [15]. High resolution structures reveal how the TFC4 subunit of TFIIIC interacts with Brf1 and Bdp1 to recruit TFIIIB [16].
Figure 1. Class III gene promoter types and initiation assembly factors used for RNAP III.
All class III genes use TFIIIB-1 or TFIIIB-2 as the transcription initiation factor for RNAP III. TFIIIB-1 and -2 are three-subunit complexes comprised of TBP (TATA-binding protein), BDP (B" or B double prime) and Brf1 or Brf2 respectively (see text). The type 1 (internal) promoter is used by 5S rRNA genes. It requires the 5S gene-specific transcription factor, TFIIIA, as well as Brf1-containing TFIIIB-1 and the six-subunit TFIIIC complex represented here as three subdomains (red ovals). The type 2 (internal) promoter is used by tRNA genes and requires TFIIIB-1 and TFIIIC, the latter of which recognizes the A and B box internal promoter elements (distinct from box A of 5S gene) which are downstream of the start site of transcription (cyan bent arrow). The type 3 promoters of higher eukaryotes are external, upstream of the start site of transcription and require the multisubunit transcription factor SNAPc, which recognizes the proximal sequence element (PSE) and a TFIIIB variant containing Brf2 rather than Brf1 designated TFIIIB-2 that binds the TATA box element. The selenocysteine tRNA[Ser]secUCA gene, as well as the U6 snRNA, MRP RNA, RNase P snRNA, and the hY RNA genes, use type 3 promoters. An important feature of the type 3 promoter is a distal sequence element (DSE) which is multipartite and recognized by activators ZNF143 and POU2F1 (see text), as schematized here for the tRNA[Ser]sec gene [17]. The terminator elements are also indicated.
Higher eukaryotes have further distinguished the tRNA and 5S rRNA genes from the type 3 promoter genes such as those encoding RNAs U6, 7SK, RNAse P, MRP and others [14]. A related feature is that while the tRNA genes are self-contained and potentially susceptible to repression by chromatin, type 3 genes include extensive upstream control regions, including a proximal sequence element (PSE) occupied by the multisubunit SNAPc, and a distal sequence element (DSE) occupied by transcriptional activators ZNF143 and POU2F1 (also known as Oct-1 and SBF) [17] (Fig 1). This wiring and activator involvement may render type 3 genes more refractory to tissue-specific and chromatin-mediated repression than some tRNA genes. In addition, Brf2 replaces Brf1 in TFIIIB for these type 3 promoters [17] (Fig. 1). Both Brf1 and Brf2 make contact with initiation-specific subunits of RNAP III. Moreover, Brf2 is a critical factor in an oxidative stress response that is mediated by a special tRNA[Ser]Sec that inserts the amino acid selenocysteine (sec) in a small set of mRNAs that share a reprogrammed UGA codon. The resulting selenoproteins are involved in oxidative stress and oxidative chemistry [18].
High resolution structure of RNAP III clarifies function
A major advance in understanding RNAP III came from three recent structures [19]. Two of these are different conformers of the 17-subunit enzyme at 4.6 and 4.7 Å that differ in the relation of the stalk, which is formed by initiation-important subunits, C17 and C25, to the trimeric initiation subcomplex proper, formed by C82, C34 and C31. The two conformers likely reflect different states of the initiation process [19]. The third structure, at 3.9 Å, is in elongation mode with a RNA:DNA hybrid in the active center and downstream duplex DNA in the cleft. This shows how the C82/34/31 subcomplex packs on the C1 clamphead to aid duplex DNA binding and shows interconnectivity between the subunits C53/C37 and C11, which form the termination-reinitiation subcomplex, with each other and other motifs [19].
More remarkably, the complex revealed unusually tight binding of the downstream DNA and unusually loose binding of the RNA:DNA hybrid in the active center. The authors noted that these differences reflect on the specialization of RNAP III to acutely respond to the weak oligo(rU:dA) hybrid that is formed prior to termination [19]. The enlightening structures are supported by existing biochemical and genetic data [20], and suggest that RNAP III termination is more sensitive to active center mechanisms than are other RNAPs. They are also supported by functional data indicating elongation rate as a major determinant of RNAP III termination and transcription output [21]. Other termination features are detailed below.
Tracking RNAP III termination
In order to supply cells with ten times more tRNAs than ribosomes, RNAP III must initiate and terminate with high efficiency [22], and this is enabled by proficient recycling [23]. That RNAP III can terminate efficiently at a simple tract of oligo-T (on the nontemplate DNA strand) while homologous RNAPs require additional cis-acting elements and/or ancillary factors, had been enigmatic [24]. Two recent studies provided new insights into this. Biochemical analysis revealed that the oligo-T terminator contains more information than expected [25], and the new RNAP III structures indicate a relatively loose RNA:DNA hybrid binding in the active center [19].
The unusually weak thermodynamic stability of oligo(rU:dA) that forms upon transcription of the template strand of the terminator element is destabilizing to RNAP complexes [26]. Yet, this alone is insufficient for RNAPs that bypass simple oligo-T tracts [27]. Recent data suggest that the unpaired oligo-T nontemplate strand carries sequence-specific information that directs a novel termination pathway [25]. Moreover, the sequence of the nontemplate DNA appears to work primarily via the Rpc37 subunit of RNAP III, explaining in part the elongation-slowing effect of this subunit during termination [25]. Thus, the RNAP III oligo-T termination signal contains separate information in each strand that combine to execute termination [25].
High resolution TFIIIB structure predicts new function
Around the same time that the RNAP III structure appeared, so did one for a human TFIIIB-Brf2 complex which led to new insights into an oxidative stress response via Brf2, tRNA[Ser]Sec, and cancer. It was known that Brf1 and Brf2 are differentially expressed in cancers [28] and that Brf2 appears to be oncogenic in lung, where its increased levels indicate poor prognosis [29, 30].
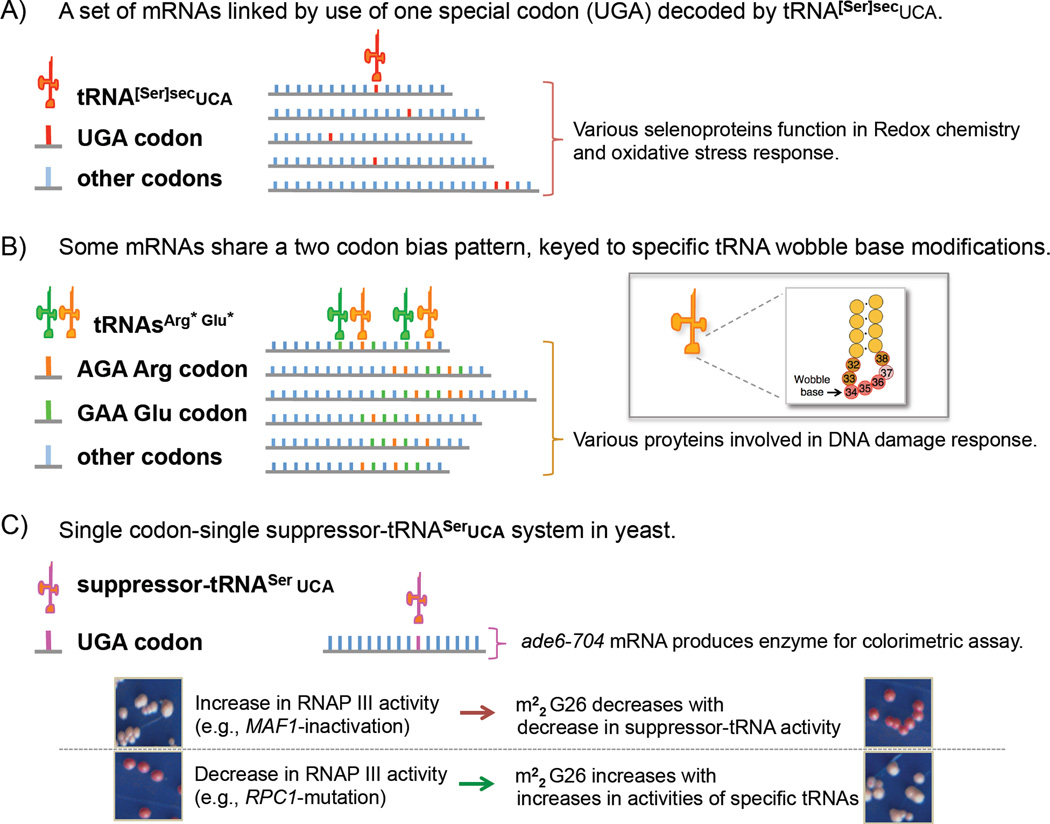
In higher eukaryotes, tRNA[Ser]Sec is the only tRNA gene known to have a type 3 Brf2-dependent promoter [14, 18]. Selenocysteine is encoded by a UGA codon in particular mRNAs involved in oxidative stress and metabolism [31]. Translation of these mRNAs share dependence on the specialized tRNA[Ser]Sec (Fig 2A), providing a unique transcriptional control mechanism.
Figure 2. RNAP III supplies tRNA, organized by transcriptional and/or posttranscriptional mechanisms, to fill codon demand for mRNA translation programs.
A) The type 3 promoter-driven, Brf2-dependent, mammalian-essential selenocysteine tRNA[Ser]secUCA is required for the decoding of reprogrammed UGA codons present in a small subset of mRNAs (about two dozen in human) that produce selenoproteins involved in oxidative chemistry and oxidative stress response [18]. The cartoon represents an example of keying a tRNA of a particular specificity to a subset of mRNAs that share the same pattern of codon bias. B) Some mRNAs share a two codon-bias. A subset of yeast mRNAs with biased content of AGA and GAA codons are depicted whose efficient translation rely on a wobble base modification mediated by Trm9 methylase [32, 33]. The inset shows the anticodon stem loop region of tRNA with the wobble base indicated. C) Schematic of suppressor-tRNASerUCA used to study single codon-specific colorimetric reporter mRNA ade6-704 in yeast that revealed the evolutionarily conserved link between RNAP III activity rate, tRNA m22G26 modification, and tRNA activity [5, 103]. The lower panels show yeast colony color under the RNAP III activity conditions indicated.
Cysteine is an oxidizable amino acid that can act as a redox sensor [18 and refs therein]. The recent structures of Brf2-DNA revealed that its cysteine-361 interfaces with the type 3 promoter DNA [18]. Oxidation of C361 inhibits formation of the TFIIIB-DNA complex, acting as a redox switch [18]. Such down-regulation of tRNA[Ser]Sec leads to decreased selenoprotein production. As noted by Vannini and coworkers, Brf2-oxidation-mediated regulation of tRNA[Ser]Sec has important implications for cancer [18].
This link between a specific tRNA and a set of mRNAs serves as an example for how translation of a group of mRNAs can be titrated by the expression levels of particular cognate tRNAs that distinguish specific codons [7]. As another example, work from Begley identified a set of mRNAs that share biased use of two codons whose decoding is mediated by and coupled to the wobble base modification differences in two cognate tRNAs [32, 33] (Fig 2B).
Maf1 regulates RNAP III: metabolic economy and tRNA activity
Maf1 is a repressor of RNAP III whose effects are analogous to decreasing the available amount of active RNAP III [34]. Thus, Maf1 regulation affects the ability of RNAP III to actively engage pre-existing transcription initiation complexes.
Growing cells use significant energy, in the form of NTPs, to make tRNAs [22]. Growth support by RNAP III is critical to cancer [35] as oncogenes such as MYC activate, while tumor suppressors Rb and p53 repress, RNAP III [36, 37]. Under certain starvation conditions, Maf1 repression of RNAP III curtails tRNA synthesis [34, 38, 39]. Maf1 expression can decrease growth and tumor progression in some settings [40]. Maf1 is under control of the target of rapamycin (TOR) kinase [22, 34], which integrates various stimuli to maintain growth and homeostasis [41]. Upon deletion of Maf1 in otherwise normal mice, the increased RNAP III activity produces excessive pre-tRNAs but mature tRNA levels do not increase. The maf1-null mice fail to gain weight apparently due to general metabolic inefficiency, suggesting that the purpose of the tRNA synthesis-repressive function of Maf1 is to support metabolic economy [39].
Though Maf1 should repress tRNA transcription and effect expression of mature tRNAs globally, initial observations in maf1-mutant yeast suggested a more complex situation as revealed by a phenotype known as paradoxical 'antisuppression' [42, 43]. In this case the strain contained a suppressor-tRNA gene whose expression suppresses a premature stop codon in a mRNA whose translation into a synthetic enzyme is followed by a simple colorimetric assay. However, upon mutation of MAF1 and global increase in tRNA synthesis, the suppression phenotype was paradoxically decreased. Nonuniform differences in tRNA gene transcripts were indeed detected, pre-tRNAs with introns accumulated more than other pre-tRNAs in maf1-deleted cells due to limiting capacity of tRNA splicing and associated machinery [44, 45].
RNAP III activity rate is linked to a functional tRNA modification
The 'maf1-antisuppression paradox' was explained by a posttranscriptional tRNA modification that is inversely linked to overall RNAP III activity and that enhances the specific activity of tRNA for mRNA decoding [5] (Fig. 2C). Similar to exceeding the capacity of tRNA splicing machinery, synthesis of the tRNA modification, dimethyl-guanosine-26 (m22G26), becomes limiting as tRNA transcription increases. The study used a novel method, tRNA-HydroSeq, to monitor tRNAs and their modifications. While the levels of the suppressor-tRNA were elevated in maf1-mutant cells, its specific activity was low due to hypomodification of m22G26, with consequent codon-specific effects on mRNA translation [5]. Trm1, the G26-methyltransferrase, is limiting in cells and the increase in tRNA transcription in maf1 mutants overwhelms its modification capacity [5] (Fig 3A–B, tRNA asterisks). Accordingly, over-expression of Trm1 reversed the antisuppression phenotype. This was shown in divergent yeasts with different tRNA isoacceptors and each had its codon-specific effects on mRNA translation. Interestingly, several of the other tRNA substrates of Trm1 also revealed changes in their m22G26 modification content, including in varying growth conditions [5]. Thus, increases and decreases in overall RNAP III activity leads to nonuniform alterations of relative tRNA activities with consequent effects on translation (Fig 3B–D).
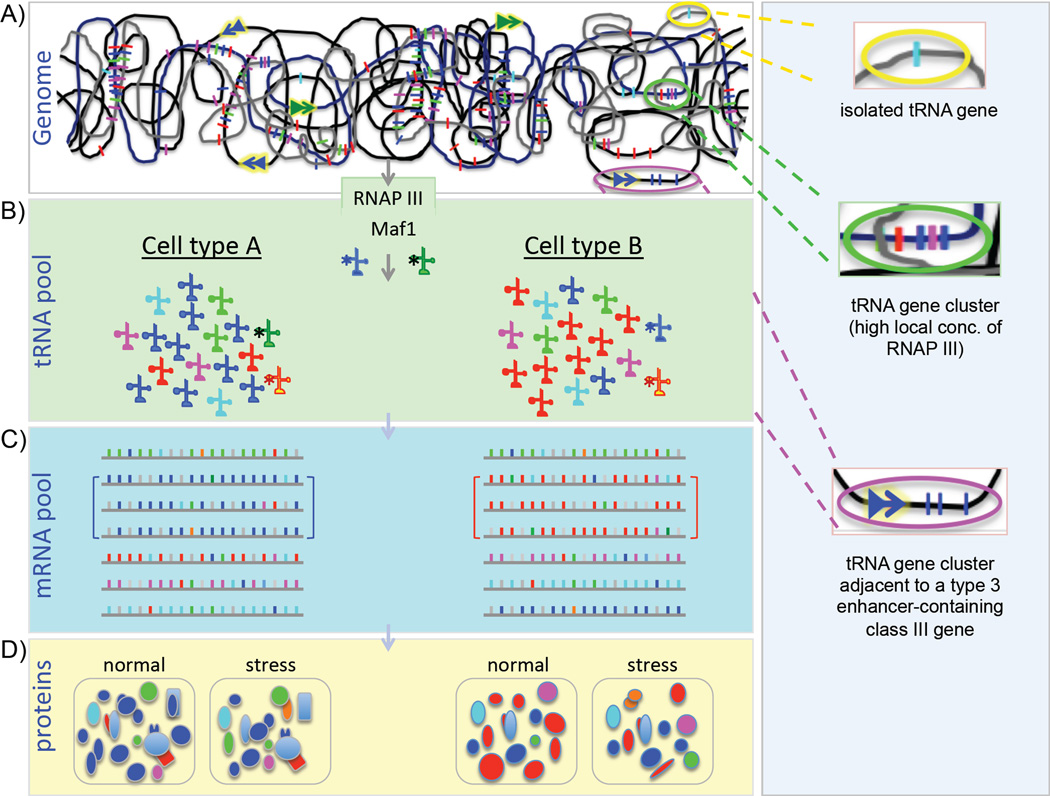
Figure 3. Model of tRNA-specific gene and modification effects of RNAP III.
A) Highly schematized genomic DNA containing interspersed tRNA genes (individual colored bars) in isolation or in clusters as found on mammalian chromosomes, in open dispersed or concentrated regions, representing different chromatin states. The double arrows represent type 3 promoters with their gene enhancers and activators. Three ovals of yellow, green and magenta are expanded from top to bottom, in the vertical panel to the right. These depict an isolated tRNA gene (yellow oval), a tRNA gene cluster which might benefit from a relatively high local concentration of recycling RNAP III as compared to an isolated tRNA gene (green oval), and a tRNA gene cluster adjacent to a type 3 gene with its enhancer (magental oval, see Fig. 1). B) Different hypothetical cell types, derived from the same genome as above but with different complements of tRNAs depicted in five colors to match the codon use bias in the mRNA pools below (C) in the same cells. Note in this hypothetical model, each cell type contains a set of mRNAs that share a codon use bias, indicated by brackets, that match the predominant tRNA in the tRNA pool. These mRNAs would have translational advantage over other mRNAs and produce more proteins as reflected in the lower panels (D). Under conditions when general RNAP III activity is decreased, for example due to Maf1-mediated repression, a subset of tRNAs become hypermodified at G26 (m22G26), as indicated by asterisks in (B). Under these stress conditions, the relative activities for mRNA decoding changes and the resulting protein compositions change. D) For each cell type is shown two protein pools, one under normal conditions (left) and the other after Maf1-mediated stress-induced repression of general RNAP III activity and resultant changes in codon-specific tRNA decoding activities due to m22G26 (right) [5].
This link between RNAP III activity and nonuniform changes in tRNA activities was shown to have been conserved through eukaryotic evolution [5]. Other conditions that alter RNAP III activity, including stimulation, repression and/or mutation, may have similar effects.
RNAP III mutations cause tissue- and disease-specific disorders
As noted earlier, chromatin modification as well as RNAP III and factor occupancy studies suggest cell-type specific transcription of tRNA genes in isolated cells from higher eukaryotes [4, 46–48]. In animals, there are relatively few but well documented cases of tissue-specific control of type 2 promoters. A most recent example is the n-Tr20 gene which produces a tRNAArgUCU species only in brain and other parts of the central nervous system (CNS) [49] although the mechanism involved in its selective expression is unknown. Most of the other cases come from specialized silk glands of silk worms and frog oocytes [50–52]. For these, the differential expression patterns appear to reflect unique binding sensitivities of the tissue-specific class III genes to TFIIIB or TFIIIC [50, 52]. Because the more recent studies suggest that different subsets of tRNA genes are active in different isolated cell types [4] that are not expected to differ significantly in RNAP III, TFIIIB or TFIIIC levels, we should also suspect that differential chromatin organization may be a determinant (Fig. 3A).
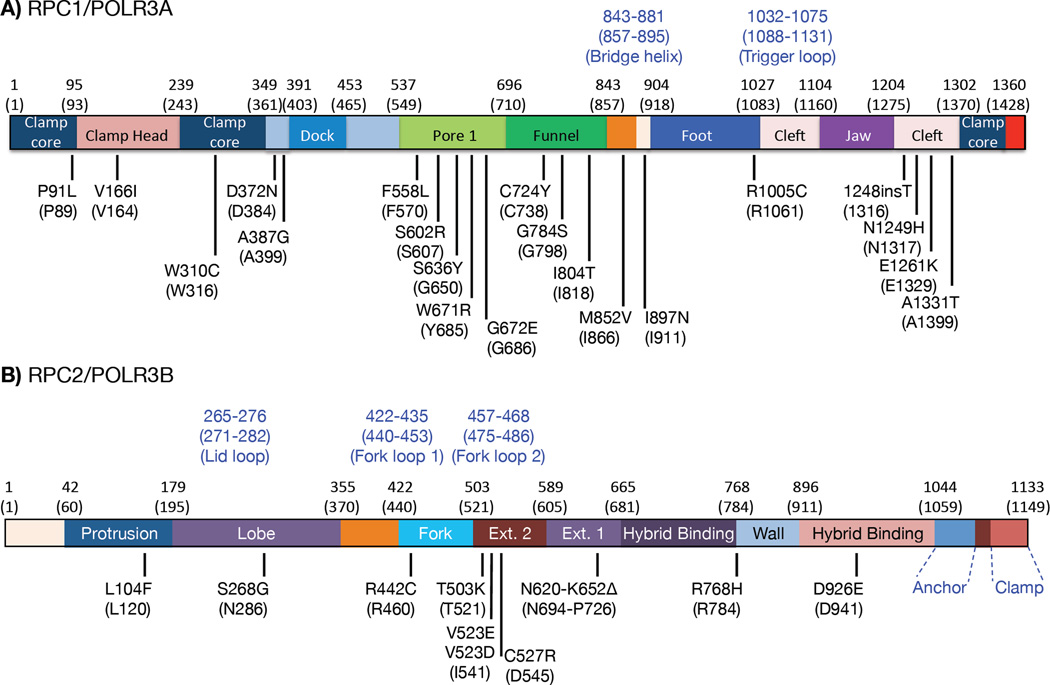
Mutations in RNAP III subunits are linked to tissue-specific defects. In human, a relatively large number of variant alleles encoding the two largest RNAP III subunits, Rpc1 and Rpc2, contain 'mutations' causative of CNS hypomyelinating leukodystrophy (HLD) [53–56] (Fig 4). Similarly in Zebrafish, mutations in Rpc2 cause a digestive and neurodevelopmental disorder [57]. In addition, mutations in the TFIIIB-1 subunits BDP1 and BRF1 reportedly cause hearing and neurodevelopmental disorders, respectively [58, 59] (BDP1 is also a subunit of TFIIIB-2).
Figure 4. Distribution of HLD (hypomyelinating leukodystrophy) mutations in the human RPC1 and RPC2 subunits of RNAP III.
Reported mutations in HLD patients were mapped to the linear representation models of RPC1/POLR3A (A) and RPC2/POLR3B (B) with some of the major structural regions indicated in different colored rectangles as defined by the cryo-EM structure of budding yeast (S. cerevisiae) RNAP III [19]. Numbers in parenthesis above the rectangles indicate the S. cerevisiae amino acid positions of the boundaries of these regions with the homologous human position numbers given above. Numbers in blue font indicate selected structural elements. Amino acid substitutions caused by patient mutations are indicated below the rectangles with the S. cerevisiae amino acid identity and position numbers in parentheses; as can be seen by comparing identities at each of the mutated positions, the vast majority are identical in yeast and human. The human mutations were as reported: [54–56, 104–108].
Although RNAP III mutations that reduce RNAP III activity may be most manifested in tissues with the most proliferative cells [60] this may not explain the observed sensitivity of neural tissue, which after differentiation is considered to be post-mitotic. Several other observations also suggest a link between neurosensitivity and defective tRNA biogenesis specifically, suggesting that the deficiency in RNAP IIIopathies is tRNA(s). Mutations in genes encoding a tRNA splicing factor or a nuclear tRNA gene n-Tr20 cause neurodevelopmental disorder [49, 61–63]. The tRNA splicing-associated pathology involves aberrant tRNA fragments that sensitize motor neurons to an oxidative stress pathway [64, 65]. Characterization of a mouse ataxia phenotype uncovered n-Tr20 a CNS-specific tRNAArgUCU whose T-loop stem mutation leads to motor neuron degeneration in a specific genetic variant background [49]. Theoretically, other tissue- and cell-specific class III gene transcripts may be essential in their respective cell types. Also, depending on the exact deficiency in the RNAP III component, different class III genes may be differentially affected, a precedent for which is the unique sensitivity of the yeast RPR1 (RNase P RNA) gene promoter to mutations in BDP1 [66]. The human genes encoding RNAse P RNA and MRP RNA rely on type 3 promoters, dependent on Brf2, [1]. Notable is the plethora of variant alleles in humans with MRP RNA promoter SNPs associated with the disorder cartilage-hair hypoplasia [67].
Another model is more tRNA- and codon-centric. RNAP III deficiency would lead to tissue-specific changes in tRNA activities due to transcriptional and posttranscriptional mechanisms (Fig 3A–D). Accordingly, the translation profiles of cell type-specific mRNAs would be altered hierarchically, and uniquely dependent on codon use, and the most sensitive would determine phenotype. This disease model accounts for pleiotropy and phenotypic diversity and is expanded upon in the next section. This persective also fits with a conserved but plastic way for the RNAP III system and the variable tRNAome contents of different species to maintain match between tRNA supply and codon demand [7, 68]. This model is extended to a specific disease involving RNAP III in the next section.
Some HLD mutations decrease RNAP III activity and nonuniformly alter tRNA
Most HLD cases carry mutations located throughout the Rpc1 or Rpc2 subunits (Fig 4). These two largest subunits together form an extended active center in RNAP III and also provide surface contact points for other subunits. HLD mutations localize throughout RPC 1 and 2 and likely affect different functional aspects of the enzyme complex (Fig 4) including assembly, stability, initiation, elongation and/or termination [53, 54]. Two such mutations decreased tRNA transcription in Schizosachharomyces pombe [5]. The link between RNAP III activity and tRNA m22G26 was also seen for these mutants, as revealed by a codon-specific mRNA translation phenotype due to alteration of m22G26-dependent tRNA specific activity [5]. A similar alteration in codon-specific translation may occur in human cells bearing these mutations.
mRNA codon use and tRNA supply are thought to match in healthy tissue. Pathological alteration of tRNA pools due to HLD mutations may lead not only to altered translation efficiency profiles of key mRNAs, but could also alter translational fidelity. Imbalances in the normal tRNA supply may induce ribosome pausing on certain mRNAs and even amino acid-tRNA misincorporation, both of which could cause polypeptide misfolding-related proteopathy [69]. Codon use by brain-specific genes has been remarkably conserved through mammalian evolution [70], suggesting that the CNS may be especially vulnerable to disturbance of the tRNA supply/codon demand balance [7], possibly reflecting particular sensitivity to protein misfolding.
Histone methylation of Alu short interspersed elements represses RNAP III
RNAP III is the driving force behind the most successful of all known mobile retroposons, the Alu short interspersed elements (SINEs), about a million of which reside in human DNA. These and other retroelements are interspersed throughout the genome via reverse transcribed RNA. Alu SINEs use the same type 2 promoter to direct synthesis of their transcripts as do tRNA genes. Similar SINEs are found in primate and rodent genomes (B1-Alu), and other SINEs exhibit extensive homology to specific tRNA sequences (e.g., rodent ID SINEs). Because the promoter resides downstream of the transcription start site, the RNA that 'retroposes' back into the genome creates a near fully equipped potential transcription unit. Many Alu type repeats can form active RNAP III transcription complexes in naked genomic DNA but are prevented from doing so in chromatin [71]. Although Alu DNA methylation was thought to be responsible, new data indicate that histone methylation at Alu loci is the main repressive determinant [72].
Alu and related SINEs derived from RNAP III transcrips are not autonomous retroposons but are effective at commandeering the retrotransposition machinery of another type of mobile genetic element, the long interspersed element (LINE) which encode reverse transcriptase and integrase to propogate themselves [73]. Thus, there is an intimate relationship between class III genes and long retrotransposons.
Long retrotransposons target TFIIIB-RNAP III gene complexes as integration sites in yeast
The S. cerevisiae retrotransposon Ty3 inserts precisely upstream of class III genes [74 and refs therein], which is facilitated by TFIIIB (but not TFIIIC) [75, 76] and La, the latter being likely involved in Ty3 RNA processing or stability but possibly also initiation site targeting [77]. Thus, strong evidence has indicated that Ty3 requires TFIIIB for integration and that interactions with TFIIIB at its binding site defines the site at which Ty3 integration occurs in S. cerevisiae. Ty1 is another S. cerevisiae retrotransposon related to Ty3 but with its own identity and distinct integration characteristics that also targets tRNA genes. The RNAP III associated factor, Bdp1 can recruit the chromatin remodeler Isw2p and influence Ty1 integration [78]. More recently, a molecular target and primary determinant of Ty1 integration site specificity was uncovered. Focusing on RNAP III itself, Bridier-Nahmias et al. found that its AC40 subunit interacts with and is the molecular target of the Ty1 integrase [79]. Independent studies found that the Ty1-Integrase interacts with RNAP III-specific subunits to promote integration upstream of class III genes [80]. It would appear that RNAP III subunits mediate direct functional interactions with Ty1 integrase and this is a basis of integration upstream of class III transcription complexes in S. cerevisiae. Thus, although Ty3 and Ty1 are distinct retrotransposons, exhibit different target preference characteristics and are influenced by different host factors, they target different components of RNAP III class III gene complexes.
Intranuclear organization: activity, processing and nearby silencing
Hundreds to thousands of tRNA genes [6] are interspersed throughout eukaryotic genomes at copy numbers that vary widely among species and significantly among individuals [8–10]. Also, as tRNA genes host stable transcription complexes that challenge the progression of DNA replication forks, these genes have been integrated into a system of genome stability [11, 12] and also punctuate other intergenic activities [81]. Thus, tRNA genes are multifaceted dynamic elements that can provide several types of spatio-functional organization. In this section, we describe how active tRNA genes can behave as barriers to the spread of repressive heterochromatin [82], and how some TFIIIC sites tether to the nuclear periphery and have chromatin organizer activity [83].
In budding yeast, active tRNA genes cluster at the nucleolus [84, 85] via TFIIIC binding to condensin [86]. In addition, they can repress nearby RNAP II-transcribed genes by tRNA gene mediated silencing (tgm), which is distinct from other silencing mechanisms [87, 88]. Nucleolar clustering and tgm can be distinguished by factors that are required for one but not the other; nucleolar localization is necessary for tgm but not sufficient [88 and refs therein]. An intriguing factor is Mod5, a tRNA modification enzyme that confers tgm by associating with tRNA genes and their nascent transcripts even if they are not substrates for modification [88].
Similar silencing was uncovered in human cells but involving Ago2 [89]. In this case, Ago2 found at tRNA and 5S rRNA genes (but not type 3 promoters lacking TFIIIC) is involved in silencing nearby RNAP II-transcribed genes [89]. Ago2 mediates this function without its usual RNAi-mediated silencing partner, DICER, and instead interacts with the nascent tRNA and 5S rRNA transcripts in chromatin and represses nearby RNAP II-transcribed genes [89].
Other data indicate that association of tRNA genes and nuclear pore proteins enhance tRNA synthesis and nuclear export in yeast [90, 91]. Efficient docking of tRNA genes to the pores requires the chromosome movement protein, cohesin, and the tRNA nuclear export factor, Los1 [91]. Nuclear pore proteins are also associated with and influence tRNA gene expression in nematodes. A subset of class III snoRNA and tRNA genes in C. elegans associate with nuclear pores, which also promotes processing of their transcripts [92, 93]. Thus, increasing data from varied sources suggest that chromatin-associated nascent transcripts of tRNA and other TFIIIC-bearing genes contribute to higher order genomic activities.
Two RPC32 isoforms of vertebrate RNAP III and a link to pluripotency
Earlier work employing transformed cells identified two isoforms of RNAP III, Pol IIIα and IIIβ. Gene duplication in an ancient vertebrate created a specialized homolog of the RPC32 subunit of RNAP III [94]. RPC32 is part of a trimeric subcomplex (C62/39/32) in human RNAP III [95] that is homologous to yeast subcomplex C82/34/31, discussed earlier. Screening for genes upregulated in undifferentiated embryonic stem (ES) cells identified the gene POLR3G, which encodes RPC32 [96]. A novel RPC32 paralog, RPC32β was identified from a distinct gene, offering a molecular basis for the distinct RNAP III isoforms [97]. RPC32(α) defines Pol IIIα, is mostly restricted to ES and transformed cells, and is dispensable, whereas RPC32β (defining Pol IIIβ) is ubiquitous and essential [97]. Expression of RPC32α but not β increases with transformation, and increasing or decreasing its expression induces or inhibits invasive growth [97]. RPC32α appears to contribute to maintenance of pluripotency in ES cells, consistent with its control by two key pluripotency regulators, NANOG and OCT4 [98].
Genome-wide analysis found that RPC32α and β occupied all class III genes in the cell type examined [94]. However, the transcription factor MYC occupies the RPC32α promoter, as it does for all other RNAP III subunit genes except the RPC32β promoter, suggesting independent regulation [94]. A recent structure of the human RPC62-RPC32 protein dimer suggests that the paralog-specific functions of RPC32 α and β reside in their terminal extensions that would appear to lie at the surface of RNAP III [99]. This arrangement supports the idea of paralog-specific responses of Pol III α and β to effectors such as Maf1 as proposed [94], consistent with data from earlier studies of the yeast counterparts, as recently noted in a report of the human RPC62-RPC32 structure [99 and refs therein]. It also raises the possibility that RPC32α and RPC32β may contribute to differential tRNA and other class III gene expression in different cell types and during vertebrate development.
A new link from RNAP III to cancer and development of autoimmunity
Antibodies against the largest RNAP III subunit, RPC1 (encoded by POLR3A), and few other specific proteins are found in scleroderma patients. In those with anti-RNAP III antibodies, there is an association between the onset of scleroderma and cancer [100, 101]. Sequencing of POLR3A uncovered distinct somatic missense mutations in three patients, and loss of heterozygosity at POLR3A specifically in the cancer tissue of three other of the eight patients examined [102]. Two of the three mutations, E1072Q and I104T, are in highly conserved RPC1 regions. E1072 is in the trigger loop and when mutated was found to activate RNAP III in S. pombe [21]. Further studies are required to understand if the altered RNAP III activity plays a functional role in the disease.
CONCLUDING REMARKS
Various recent advances have increased our understanding of RNAP III in three major areas, 1) integrating basic mechanisms and high resolution structure; 2) unexpected consequences of MAF1 mutation or inactivation on energy homeostasis, body weight management and tRNA modification; and 3) tissue-specific and disease-specific disorders associated with and/or caused by RNAP III mutations.
The first near-atomic resolution structures of elongating RNAP III and the accompanying insight into transcription termination sets the stage for examining how an RNAP enzyme complex can undergo rapid transcription reinitiation, hopefully uncovering cancer susceptibilities. The discovery that Brf2 uniquely controls of tRNA[Ser]Sec and other type 3 gene loci in higher eukaryotes in response to oxidative stress, separate from the hundreds of other tRNAs, raises several important questions and opportunities for promising links to cancer.
Understanding the mechanism and extent of tissue-specific tRNA gene transcription in relation to the chromatin context is a critical focus for the near future. By contrast, deciphering the mechanisms of RNAP III-associated disease is on the horizon. While deficient transcription of any non-tRNA class III gene may cause a specific phenotype, we should consider the potential effects of nonuniform changes in the tRNAs that accompany global alteration of RNAP III activity. Thus, a more distant goal is to understand how tRNA activities change in a tissue-specific manner in response to regulation of or mutation in RNAP III to alter mRNA translation profiles resulting in a phenotype. This includes the somatic mutations in RPC1 that are associated with cancer. In any case, it would be fruitful to focus on the HLD mutations as a model RNAP III pathology.
Outstanding Questions Box.
Since tRNA genes share similar promoters and tissue-specific class III transcription factors are unknown, what is a mechanistic basis of the apparent cell type-specific tRNA gene activity? To what extent does local chromatin context influence cell type-specific tRNA gene activity in higher eukaryotes, and what are the determinants?
Now that a high-resolution structure for RNAP III has been solved, can structures for intermediates of transcription initiation and termination be isolated? Which residues contribute to the active center, are involved in elongation, or in termination, and how do they affect RNA synthesis rate?
Since each strand of the RNAP III terminator contains separate functional information for termination, to what extent does nucleotide identity at key positions affect the template or nontemplate strand function? Can such information improve understanding of how noncanonical terminators work?
To what extent does Brf2 residence at different type 3 promoters change during oxidative stress? How is RNAP III recruited to type 3 promoters?
How do alterations in RNAP III activity, change the mRNA translational profile, the proteome, of cells?
Do the different vertebrate RNAP III isoforms α and β, containing RPC32α or RPC32β respectively, respond differently to effectors such as Maf1? Do RPC32α and RPC32β play different roles during vertebrate development or in different somatic cell types?
Do the newly uncovered autoimmune and cancer-associated mutations in RPC1 represent a new mechanism by which cancers activate RNAP III for increased tRNA synthesis?
How does deficiency of a housekeeping enzyme, RNAP III, lead to tissue-specific defects, presumably as is the case for HLD? Is it due to deficient synthesis of a tissue-specific transcript, or tRNAs in general? How do mRNA translational profiles change?
Trends Box.
The first atomic structures of RNAP III bring novel insight into the mechanisms that promote efficient synthesis of short RNAs. A loose active center, tight DNA binding cleft and other features will drive experiments to target RNA synthesis in cancer.
A Crystal structure of Brf2 with type 3 promoter DNA uncovered a novel switch mechanism for selenocysteine-tRNA synthesis and control of the oxidative stress response.
The dimethyl-G26 modification found on some tRNAs is linked to RNAP III and regulated by Maf1, which is conserved from yeast to human. Because dimethyl-G26 increases tRNA activity for decoding, it alters cellular mRNA translational profiles.
Mutations throughout the two largest RNAP III subunits cause the developmental neurological disorder hypomyelenating leukodystrophy (HLD) These mutations likely decrease tRNA synthesis, affect dimethyl-G26, and alter mRNA translation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome research. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of noncoding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 4.White RJ. Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet. 2011;12:459–463. doi: 10.1038/nrg3001. [DOI] [PubMed] [Google Scholar]
- 5.Arimbasseri AG, Blewett1 NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ. RNA Polymerase III Output is Functionally Linked to tRNA Dimethyl-G26 Modification. PLoS Genetics. 2015 doi: 10.1371/journal.pgen.1005671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2015 Dec 15; doi: 10.1093/nar/gkv1309. pii: gkv1309. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maraia RJ, Iben JR. Different types of secondary information in the genetic code. RNA. 2014;20:977–984. doi: 10.1261/rna.044115.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iben JR, Maraia RJ. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA. 2012;18:1358–1372. doi: 10.1261/rna.032151.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iben JR, Maraia RJ. tRNA gene copy number variation in humans. GENE. 2014;536:376–884. doi: 10.1016/j.gene.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parisien M, Wang X, Pan T. Diversity of human tRNA genes from the 1000-genomes project. RNA biology. 2013;10:1853–1867. doi: 10.4161/rna.27361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clelland BW, Schultz MC. Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription. 2010;1:115–125. doi: 10.4161/trns.1.3.13735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen VC, Clelland BW, Hockman DJ, Kujat-Choy SL, Mewhort HE, Schultz MC. Replication stress checkpoint signaling controls tRNA gene transcription. Nat Struct Mol Biol. 2010;17:976–981. doi: 10.1038/nsmb.1857. [DOI] [PubMed] [Google Scholar]
- 13.Yona AH, Bloom-Ackermann Z, Frumkin I, Hanson-Smith V, Charpak-Amikam Y, Feng Q, Pilpel Y. tRNA genes rapidly change in evolution to meet novel translational demands. Elife. 2013;2:e01339. doi: 10.7554/eLife.01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 15.Nagarajavel V, Iben JR, Howard BH, Maraia RJ, Clark DJ. Global 'bootprinting' reveals the elastic architecture of the yeast TFIIIB-TFIIIC transcription complex in vivo. Nucleic Acids Res. 2013;41:8135–8143. doi: 10.1093/nar/gkt611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Male G, von Appen A, Glatt S, Taylor NM, Cristovao M, Groetsch H, Muller CW. Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly. Nat Commun. 2015;6:7387. doi: 10.1038/ncomms8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS genetics. 2012;8:e1003028-e. doi: 10.1371/journal.pgen.1003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouge J, Satia K, Guthertz N, Widya M, Thompson AJ, Cousin P, Vannini A. Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2. Cell. 2015;163:1375–1387. doi: 10.1016/j.cell.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann N, Jakobi A, Moreno-Morcillo M, Glatt S, Kosinski J, Hagen W, Muller C. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015 doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraia RJ, Rijal K. Structural biology: A transcriptional specialist resolved. Nature. 2015 doi: 10.1038/nature16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rijal K, Maraia RJ. An active site-controlled mechanism of RNA polymerase III termination and its link to a limiting step in transcription. Submitted. 2016 [Google Scholar]
- 22.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta. 2013;1829:361–375. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2014;5(1):e27639. doi: 10.4161/trns.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arimbasseri AG, Rijal K, Maraia RJ. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbagrm.2012.10.006. S1874-9399(12)00177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arimbasseri AG, Maraia RJ. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell. 2015;58:1124–1132. doi: 10.1016/j.molcel.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin FH, Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arimbasseri AG, Maraia RJ. A high density of cis-information terminates RNA Polymerase III on a two-rail track. RNA biology. 2015 doi: 10.1080/15476286.2015.1116677. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabarcas S, Jacob J, Veras I, Schramm L. Differential expression of the TFIIIB subunits Brf1 and Brf2 in cancer cells. BMC molecular biology. 2008;9:74-. doi: 10.1186/1471-2199-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Tian H, Yue W, Li L, Li S, Qi L, Si L. TFIIB-related factor 2 over expression is a prognosis marker for early-stage non-small cell lung cancer correlated with tumor angiogenesis. PLoS One. 2014;9:e88032. doi: 10.1371/journal.pone.0088032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian Y, Lu M, Yue W, Li L, Li S, Gao C, Tian H. TFIIB-related factor 2 is associated with poor prognosis of nonsmall cell lung cancer patients through promoting tumor epithelial-mesenchymal transition. Biomed Res Int. 2014;2014:530786. doi: 10.1155/2014/530786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Commans S, Bock A. Selenocysteine inserting tRNAs: an overview. FEMS Microbiol Rev. 1999;23:335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 32.Begley U, Dyavaiah M, Patil A, Rooney JP, Direnzo D, Young CM, Begley TJ. Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015;11:e1005706. doi: 10.1371/journal.pgen.1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boguta M. Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim Biophys Acta. 2013;1829:376–384. doi: 10.1016/j.bbagrm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 37.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 38.Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- 39.Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Willis IM. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palian BM, Rohira AD, Johnson SA, He L, Zheng N, Dubeau L, Johnson DL. Maf1 Is a Novel Target of PTEN and PI3K Signaling That Negatively Regulates Oncogenesis and Lipid Metabolism. PLoS Genet. 2014;10:e1004789. doi: 10.1371/journal.pgen.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boguta M, Czerska K, Zoladek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185:291–296. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- 43.Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Boguta M. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–5040. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciesla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, Boguta M. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol. 2007;27:7693–7702. doi: 10.1128/MCB.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karkusiewicz I, Turowski TW, Graczyk D, Towpik J, Dhungel N, Hopper AK, Boguta M. Maf1, repressor of RNA polymerase III, indirectly affects tRNA processing. J Biol Chem. 2011 doi: 10.1074/jbc.M111.253310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cairns BR. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kutter C, Brown GD, Goncalves A, Wilson MD, Watt S, Brazma A, Odom DT. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet. 2011;43:948–955. doi: 10.1038/ng.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Ackerman SL. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan HS, Young LS, White CN, Sprague KU. Silk gland-specific tRNA(Ala) genes interact more weakly than constitutive tRNA(Ala) genes with silkworm TFIIIB and polymerase III fractions. Mol Cell Biol. 1994;14:1806–1814. doi: 10.1128/mcb.14.3.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stutz F, Gouilloud E, Clarkson SG. Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes Dev. 1989;3:1190–1198. doi: 10.1101/gad.3.8.1190. [DOI] [PubMed] [Google Scholar]
- 52.Reynolds WF, Johnson DL. Differential expression of oocyte-type class III genes with fraction TFIIIC from immature or mature oocytes. Mol Cell Biol. 1992;12:946–953. doi: 10.1128/mcb.12.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thiffault I, Wolf NI, Forget D, Guerrero K, Tran LT, Choquet K, Bernard G. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat Commun. 2015;6:7623. doi: 10.1038/ncomms8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernard G, Chouery E, Putorti ML, Tetreault M, Takanohashi A, Carosso G, Brais B. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89:415–423. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tetreault M, Choquet K, Orcesi S, Tonduti D, Balottin U, Teichmann M, Bernard G. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89:652–655. doi: 10.1016/j.ajhg.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitsu H, Osaka H, Sasaki M, Takanashi J, Hamada K, Yamashita A, Matsumoto N. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomalrecessive hypomyelinating leukoencephalopathy. Am J Hum Genet. 2011;89:644–651. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yee NS, Gong W, Huang Y, Lorent K, Dolan AC, Maraia RJ, Pack M. Mutation of RNA polymerase III subunit rpc2/polr3b leads to deficiency of the RNA cleavage subunit, Rpc11/Polr3k, and disrupts zebrafish digestive system development. PLoS Biol. 2007;5:2484–2492. doi: 10.1371/journal.pbio.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borck G, Hog F, Dentici ML, Tan PL, Sowada N, Medeira A, Kubisch C. BRF1 mutations alter RNA polymerase III-dependent transcription and cause neurodevelopmental anomalies. Genome research. 2015;25:609. [PMC free article] [PubMed] [Google Scholar]
- 59.Girotto G, Abdulhadi K, Buniello A, Vozzi D, Licastro D, d'Eustacchio A, Gasparini P. Linkage study and exome sequencing identify a BDP1 mutation associated with hereditary hearing loss. PLoS One. 2013;8:e80323. doi: 10.1371/journal.pone.0080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall L, Goodfellow SJ, White RJ. Diminished Activity of RNA Polymerase III Selectively Disrupts Tissues with the Most Actively Dividing Cells. PLoS Biol. 2007;5:e286. doi: 10.1371/journal.pbio.0050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, Lupski JR. Human CLP1 mutations alter tRNA biogenesis, affecting both peripheral and central nervous system function. Cell. 2014;157:636–650. doi: 10.1016/j.cell.2014.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaffer AE, Eggens VR, Caglayan AO, Reuter MS, Scott E, Coufal NG, Gleeson JG. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Penninger JM. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weitzer S, Hanada T, Penninger JM, Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley interdisciplinary reviews. 2015;6:47–63. doi: 10.1002/wrna.1255. [DOI] [PubMed] [Google Scholar]
- 65.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishiguro A, Kassavetis GA, Geiduschek EP. Essential roles of Bdp1, a subunit of RNA polymerase III initiation factor TFIIIB, in transcription and tRNA processing. Mol Cell Biol. 2002;22:3264–3275. doi: 10.1128/MCB.22.10.3264-3275.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattijssen S, Welting TJ, Pruijn GJ. RNase MRP and disease. Wiley interdisciplinary reviews. 2010;1:102–116. doi: 10.1002/wrna.9. [DOI] [PubMed] [Google Scholar]
- 68.Schmitt BM, Rudolph KL, Karagianni P, Fonseca NA, White RJ, Talianidis I, Kutter C. High-resolution mapping of transcriptional dynamics across tissue development reveals a stable mRNA-tRNA interface. Genome research. 2014;24:1797–1807. doi: 10.1101/gr.176784.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 70.Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russanova VR, Driscoll CT, Howard BH. Adenovirus 2 preferentially stimulates pol III transcription of Alu elements by relieving repression: a potential role for chromatin. Mol. Cell Biol. 1995;15:4282–4290. doi: 10.1128/mcb.15.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Varshney D, Vavrova-Anderson J, Oler AJ, Cowling VH, Cairns BR, White RJ. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat Commun. 2015;6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahl V, Keller H, Schmidt S, Weichenrieder O. Retrotransposition and Crystal Structure of an Alu RNP in the Ribosome-Stalling Conformation. Mol Cell. 2015;60:715–727. doi: 10.1016/j.molcel.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Qi X, Daily K, Nguyen K, Wang H, Mayhew D, Rigor P, Sandmeyer S. Retrotransposon profiling of RNA polymerase III initiation sites. Genome research. 2012;22:681–692. doi: 10.1101/gr.131219.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yieh L, Hatzis H, Kassavetis G, Sandmeyer SB. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J Biol Chem. 2002;277:25920–25928. doi: 10.1074/jbc.M202729200. [DOI] [PubMed] [Google Scholar]
- 76.Aye M, Dildine SL, Claypool JA, Jourdain S, Sandmeyer SB. A truncation mutant of the 95-kilodalton subunit of transcription factor IIIC reveals asymmetry in Ty3 integration. Mol Cell Biol. 2001;21:7839–7851. doi: 10.1128/MCB.21.22.7839-7851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aye M, Sandmeyer SB. Ty3 requires yeast La homologous protein for wild-type frequencies of transposition. Mol Microbiol. 2003;49:501–515. doi: 10.1046/j.1365-2958.2003.03568.x. [DOI] [PubMed] [Google Scholar]
- 78.Bachman N, Gelbart ME, Tsukiyama T, Boeke JD. TFIIIB subunit Bdp1p is required for periodic integration of the Ty1 retrotransposon and targeting of Isw2p to S. cerevisiae tDNAs. Genes Dev. 2005;19:955–964. doi: 10.1101/gad.1299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, Flores A, Lesage P. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science. 2015;348:585–588. doi: 10.1126/science.1259114. [DOI] [PubMed] [Google Scholar]
- 80.Cheung S, Ma L, Chan PH, Hu HL, Mayor T, Chen HT, Measday V. Ty1-Integrase interacts with RNA Polymerase III specific subcomplexes to promote insertion of Ty1 elements upstream of Pol III-transcribed genes. J Biol Chem. 2016 doi: 10.1074/jbc.M115.686840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korde A, Rosselot JM, Donze D. Intergenic transcriptional interference is blocked by RNA polymerase III transcription factor TFIIIB in Saccharomyces cerevisiae. Genetics. 2014;196:427–438. doi: 10.1534/genetics.113.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McFarlane RJ, Whitehall SK. tRNA genes in eukaryotic genome organization and reorganization. Cell Cycle. 2009;8:3102–3106. doi: 10.4161/cc.8.19.9625. [DOI] [PubMed] [Google Scholar]
- 83.Noma KI, Cam1 HP, Maraia RJ, Grewal SIS. A novel function for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 84.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haeusler RA, Engelke DR. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 2006;34:4826–4836. doi: 10.1093/nar/gkl656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang L, Haeusler RA, Good PD, Thompson M, Nagar S, Engelke DR. Silencing near tRNA genes requires nucleolar localization. J Biol Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, Engelke DR. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A. 2013;110:E3081–E3089. doi: 10.1073/pnas.1219946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woolnough JL, Atwood BL, Giles KE. Argonaute 2 Binds Directly to tRNA Genes and Promotes Gene Repression in cis. Mol Cell Biol. 2015;35:2278–2294. doi: 10.1128/MCB.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruben GJ, Kirkland JG, MacDonough T, Chen M, Dubey RN, Gartenberg MR, Kamakaka RT. Nucleoporin mediated nuclear positioning and silencing of HMR. PLoS One. 2011;6:e21923. doi: 10.1371/journal.pone.0021923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen M, Gartenberg MR. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 2014;28:959–970. doi: 10.1101/gad.236729.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell. 2013;51:840–849. doi: 10.1016/j.molcel.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maraia RJ, Arimbasseri AG. It's Sno'ing on Pol III at nuclear pores. Genome Biol. 2013;14:137. doi: 10.1186/gb4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Renaud M, Praz V, Vieu E, Florens L, Washburn MP, l'Hote P, Hernandez N. Gene duplication and neofunctionalization: POLR3G and POLR3GL. Genome research. 2014;24:37–51. doi: 10.1101/gr.161570.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes & Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 96.Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Andrews PW. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Human Molecular Genetics. 2005;14:3129–3140. doi: 10.1093/hmg/ddi345. [DOI] [PubMed] [Google Scholar]
- 97.Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, Teichmann M. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci U S A. 2010;107:4176–4181. doi: 10.1073/pnas.0914980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wong RC, Pollan S, Fong H, Ibrahim A, Smith EL, Ho M, Donovan PJ. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells. 2011;29:1517–1527. doi: 10.1002/stem.714. [DOI] [PubMed] [Google Scholar]
- 99.Boissier F, Dumay-Odelot H, Teichmann M, Fribourg S. Structural analysis of human RPC32beta-RPC62 complex. J Struct Biol. 2015;192:313–319. doi: 10.1016/j.jsb.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 100.Shah AA, Rosen A, Hummers L, Wigley F, Casciola-Rosen L. Close temporal relationship between onset of cancer and scleroderma in patients with RNA polymerase I/III antibodies. Arthritis Rheum. 2010;62:2787–2795. doi: 10.1002/art.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moinzadeh P, Fonseca C, Hellmich M, Shah AA, Chighizola C, Denton CP, Ong VH. Association of anti-RNA polymerase III autoantibodies and cancer in scleroderma. Arthritis Res Ther. 2014;16:R53. doi: 10.1186/ar4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Joseph CG, Darrah E, Shah AA, Skora AD, Casciola-Rosen LA, Wigley FM, Rosen A. Association of the autoimmune disease scleroderma with an immunologic response to cancer. Science. 2014;343:152–157. doi: 10.1126/science.1246886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rijal K, Maraia RJ, Arimbasseri AG. A methods review on use of nonsense suppression to study 3' end formation and other aspects of tRNA biogenesis. Gene. 2015;556:35–50. doi: 10.1016/j.gene.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Daoud H, Tetreault M, Gibson W, Guerrero K, Cohen A, Gburek-Augustat J, Bernard G. Mutations in POLR3A and POLR3B are a major cause of hypomyelinating leukodystrophies with or without dental abnormalities and/or hypogonadotropic hypogonadism. J Med Genet. 2013;50:194–197. doi: 10.1136/jmedgenet-2012-101357. [DOI] [PubMed] [Google Scholar]
- 105.Potic A, Brais B, Choquet K, Schiffmann R, Bernard G. 4H Syndrome With Late-Onset Growth Hormone Deficiency Caused by POLR3A Mutations. Arch Neurol. 2012;69:720–723. doi: 10.1001/archneurol.2011.1963. [DOI] [PubMed] [Google Scholar]
- 106.Shimojima K, Shimada S, Tamasaki A, Akaboshi S, Komoike Y, Saito A, Yamamoto T. Novel compound heterozygous mutations of POLR3A revealed by whole-exome sequencing in a patient with hypomyelination. Brain Dev. 2014;36:315–321. doi: 10.1016/j.braindev.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 107.107.Tamura A, Niwa A, Ii Y, Sasaki R, Tomimoto H, Saitsu H. A case of hypomyelinating leukodystrophy with new homozygous mutation in POLR3A. Rinsho Shinkeigaku. 2013;53:624–629. doi: 10.5692/clinicalneurol.53.624. [DOI] [PubMed] [Google Scholar]
- 108.Terao Y, Saitsu H, Segawa M, Kondo Y, Sakamoto K, Matsumoto N, Nomura Y. Diffuse central hypomyelination presenting as 4H syndrome caused by compound heterozygous mutations in POLR3A encoding the catalytic subunit of polymerase III. Journal of the neurological sciences. 2012;320:102–105. doi: 10.1016/j.jns.2012.07.005. [DOI] [PubMed] [Google Scholar]