Abstract
Background: Various methods are currently used for the early detection of West Nile virus (WNV) but their outputs are not quantitative and/or do not take into account all available information. Our study aimed to test a multivariate syndromic surveillance system to evaluate if the sensitivity and the specificity of detection of WNV could be improved.
Methods: Weekly time series data on nervous syndromes in horses and mortality in both horses and wild birds were used. Baselines were fitted to the three time series and used to simulate 100 years of surveillance data. WNV outbreaks were simulated and inserted into the baselines based on historical data and expert opinion. Univariate and multivariate syndromic surveillance systems were tested to gauge how well they detected the outbreaks; detection was based on an empirical Bayesian approach. The systems' performances were compared using measures of sensitivity, specificity, and area under receiver operating characteristic curve (AUC).
Results: When data sources were considered separately (i.e., univariate systems), the best detection performance was obtained using the data set of nervous symptoms in horses compared to those of bird and horse mortality (AUCs equal to 0.80, 0.75, and 0.50, respectively). A multivariate outbreak detection system that used nervous symptoms in horses and bird mortality generated the best performance (AUC = 0.87).
Conclusions: The proposed approach is suitable for performing multivariate syndromic surveillance of WNV outbreaks. This is particularly relevant, given that a multivariate surveillance system performed better than a univariate approach. Such a surveillance system could be especially useful in serving as an alert for the possibility of human viral infections. This approach can be also used for other diseases for which multiple sources of evidence are available.
Key Words: : Bayes, Horses, Multivariate detection, Syndromic surveillance, West Nile
Introduction
West Nile virus (WNV) is a zoonotic mosquito-borne arbovirus, mainly transmitted by mosquitoes from the genus Culex (family Culicidae). Main reservoir hosts are birds, but the virus also affects various nonavian species, including horses and humans, with dramatic consequences for public health and for the equine industry, that is, potentially fatal encephalitis in humans and horses (Campbell et al. 2002, Castillo-Olivares and Wood 2004). In Europe, and more specifically in France, WNV lineage I emerged in the 1960s and several outbreaks have been documented since that time (Calistri et al. 2010). Even if this lineage is now considered endemic in a large part of Europe, the number of reported outbreaks is presently increasing in Southern and Eastern Europe, particularly, in Italy, Greece, and Bulgaria (Di Sabatino et al. 2014).WNV lineage II has been introduced in Europe in 2004 and spread in several parts of Europe. This lineage induces more cases and more severe symptoms than lineage I in humans, horses, and birds (Bakonyi et al. 2006, Calzolari et al. 2013, Hernández-Triana et al. 2014). As an example, in Greece, 197 neuroinvasive human cases and 35 deaths were reported in 2010 with lineage II (Danis et al. 2011). All these elements contribute to make WNV a growing concern in Europe. Currently, in France and in most countries, the surveillance of WNV is mainly passive, that is, based on the vigilance of owners and veterinary practitioners who declare the cases. To improve early detection of WNV outbreaks, the major challenge is to develop more integrated and quantitative approaches (Beck et al. 2013, Bellini et al. 2014a).
Syndromic surveillance is defined as “the (near) real-time collection, analysis, interpretation and dissemination of health-related data to enable the early identification of the impact—or absence of impact—of potential threats. Syndromic surveillance is based not on the laboratory-confirmed diagnosis of a disease but on non-specific health indicators including clinical signs, symptoms as well as proxy measures” (Triple S Project 2011). In Europe, the surveillance of nervous syndromes in horses is considered as one of the most cost-effective surveillance systems in the European context (Chevalier et al. 2011) and has been shown to detect an outbreak of WNV 3 weeks before laboratory identification in the South of France (Leblond et al. 2007a; Saegerman et al. 2016). In the United States, instead, increased mortality in wild birds is one of the most timely indicators of virus activity (Brown 2012). Mortality in wild birds had rarely been reported in Europe until the recent explosive spread of lineage II in 2008–2009 in Hungary and Austria, which suggests that this parameter could be also incorporated into future monitoring systems in Europe (Bakonyi et al. 2013). This is consistent with recent experimental infections of European wild birds with various WNV strains, which generated an average mortality rate of 43% (Sotelo et al. 2011, Dridi et al. 2013, Ziegler et al. 2013, Del Amo et al. 2014a, 2014b). Apart from mortality in wild birds and nervous symptoms in horses, WNV is also associated with mortality in horses, which could constitute another signal of a WNV outbreak. Considering that horses and birds should be affected by WNV before humans (Kulasekera et al. 2001, Leblond et al. 2007a), a surveillance system based on the analysis of these data could be especially useful in serving as early warning for possible human viral infections.
Multivariate syndromic surveillance combines different syndromic data sources available (Sonesson and Frisén 2005, Frisén et al. 2010) and should give better results for outbreak detection in terms of specificity and sensitivity than univariate methods alone. However, at the time of writing, multivariate syndromic surveillance has never been implemented for the detection of WNV outbreaks. The aim of our study was to evaluate the performance of a multivariate syndromic surveillance system in detecting WNV using three data sets: nervous syndromes in horses, mortality in horses, and mortality in wild birds. Mortality will be considered in our study as a syndrome. We focused on the French Mediterranean coast, which is a particularly high-risk area for WNV outbreaks. Indeed, in France, WNV has only ever been identified in this area according to the last outbreaks occurring in 2000, 2004, 2006, and 2015 (Murgue et al. 2001, Autorino et al. 2002, Anonymous 2007, Leblond et al. 2007a, Kutasi et al. 2011, Lecollinet et al. 2016). This French region is especially at risk because mammalian and avian hosts, bridging vectors, and large protected wetlands with numerous migratory birds are all present.
Materials and Methods
Data sources
Nervous syndromes in horses
Data on nervous syndromes in horses are collected through the passive surveillance system “Réseau d'Epidémio-Surveillance en Pathologie Equine (RESPE).” This French network for the surveillance of equine diseases (www.respe.net/) collects standardized declarations from veterinary practitioners registered as sentinels. In the RESPE database, nervous symptoms in horses are defined as any signs of impairment of the central nervous system, that is, ataxia, paresis, paralysis and/or recumbency, and/or behavioral disorder. Nervous disorders with evidence of traumatic or congenital origins are excluded. All the samples sent for laboratory diagnosis are systematically tested for two diseases, WNV and equine herpes virus-1, and the results are registered in the RESPE database. Currently, the collected data are mainly used to produce alerts when cases with positive laboratory diagnoses are identified. To obtain an outbreak-free baseline data set, we used data from 2006 to 2013 that included only the 44 declarations without positive laboratory test results from the region of the French Mediterranean coast. The time series of nervous syndromes in horses is designated NervSy in subsequent sections.
Mortality in horses
Data on mortality in horses have been centralized since 2011 in the “EDI-SPAN” database, managed by all the French fallen stock companies and the French Ministry of Agriculture (Perrin et al. 2012). As WNV does not produce perinatal mortality, we only considered the 8742 dead adult horses collected around the French Mediterranean coast between 2011 and 2014. The time series of mortality in adult horses is designated DeadHorse in subsequent sections.
Mortality in wild birds
Data on mortality in wild birds are collected through the event-based surveillance system “SAGIR,” the national French surveillance network of diseases in wild birds and mammals, which collects declarations from field workers (e.g., hunters, technicians from departmental hunting federations, and environmental inspectors from the French National Hunting and Wildlife Agency [ONCFS]). Surveillance relies on diagnosis at a local veterinary laboratory (Decors et al. 2014). Between 2007 and 2013, 292 dead wild birds were collected and necropsied around the French Mediterranean coast. The time series of the number of necropsied wild birds is designated DeadBird in subsequent sections.
Data modeling and simulation
Baselines modeling
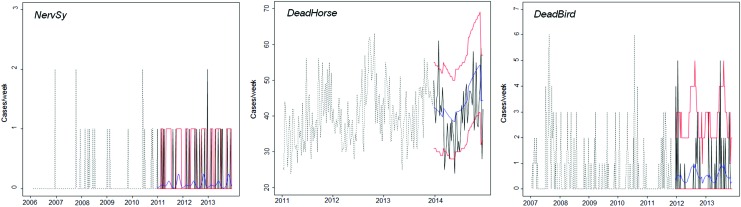
All time series were aggregated weekly. Using visual examination, abnormal peaks were observed only in DeadBird due to health troubles occurring in the wild bird population (i.e., intoxication). These extreme values were removed based on a method adapted from Tsui et al. (Tsui et al. 2001): the entire data set was first fitted to a negative binomial (NB) distribution (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/vbz), and then, values above the 95% confidence interval were deleted and replaced with the average value of the four previous weeks.
To calibrate the models, we used NervSy data from 2006 to 2010, DeadHorse data from 2011 to 2013, and DeadBird data from 2007 to 2011. Instead, to validate the quality of predictions, we used NervSy data from 2011 to 2013, DeadHorse data from 2014, and DeadBird data from 2012 to 2013. To define the background noise of the time series without outbreaks, we fitted alternative regression models based on Poisson and NB distributions (Supplementary Table S2). Models were implemented in R x64 version 3.0.2. Dynamic regression was performed with the functions glm (package {stats}) and glm.nb (package {MASS}). The expected number of counts at time t was estimated with the predict functions of the respective packages.
Models were evaluated using the Akaike information criterion (Bozdogan 1987), and the adjusted deviance (deviance/degree of freedom) was used as a measure of goodness-of-fit (GOF). The agreement between predicted and observed values was assessed according to the root-mean-squared error (RMSE) (Chai and Draxler 2014). The criterion was assessed within the calibration period (RMSEc) and within the validation period (RMSEv). In either case, the lower the value, the better the predictive performance of the model.
Baseline simulation
For each time series, the best regression model was used to predict the expected value of each week of the next simulated year. Distribution of cases for each week was defined as a Poisson distribution with lambda equals to equaling the predicted value for the same week. Weekly samples from 100 fictive years were generated by random sampling from the previous distributions as proposed by Dórea et al. (2013).
WNV outbreaks modeling
The weekly counts of cases of five real European WNV outbreaks (Murgue et al. 2001, Autorino et al. 2002, Anonymous 2007, Leblond et al. 2007a; Kutasi et al. 2011) were fitted to the NB distribution, and the resulting distribution of the additional number of nervous cases due to WNV during an outbreak was NB (mu = 3.12, theta = 1.150). The mortality among horses clinically affected by WNV was fitted to a normal distribution (mean = 0.384, standard deviation = 0.128) based on the studies by Murgue et al. 2001, Autorino et al. 2002, Ward et al. 2006, and Leblond et al. 2007a. The NervSy data set did not provide the real number of clinically affected horses, so we assumed that only 50% of horses with nervous symptoms were declared to Réseau d'Epidémio-Surveillance en Pathologie Equine (RESPE). To estimate the real number of clinically affected horses, we simulated RESPE declarations of nervous symptoms associated with 100 WNV outbreaks and doubled the counts of horses obtained. The related weekly count of dead adult horses was then deduced and fitted to the NB distribution NB (mu = 3, theta = 2.005). The distribution of the weekly number of dead birds was estimated by expert's opinions to be NB (mean = 2.23, theta = 3.34). Experts were European diplomates in equine internal medicine and persons involved in SAGIR network, RESPE network, and reference laboratories. They based their estimation on data available in the literature (Bakonyi et al. 2013); (Sotelo et al. 2011, Dridi et al. 2013, Ziegler et al. 2013, Del Amo et al. 2014a, 2014b), their personal knowledge acquired during the observation of real WNV outbreaks in Hungary, France, Italy, and Spain during the last decade, and their knowledge of equine and wild birds diseases in general.
WNV outbreaks simulation
Data on real WNV outbreaks are scarce, so we used simulated outbreaks to evaluate our detection system. For each syndrome, the distribution of the number of cases during an outbreak was estimated with the fitdist function of the package {fitdistrplus}. Time series for each syndrome during 100 fictive outbreaks of 8 weeks was simulated by randomly sampling the corresponding distribution. All the weeks within an epidemic time period have thus the same probability to have a high (or low) number of cases.
Simulated WNV outbreaks insertion in simulated baselines
One simulated outbreak was inserted in each year of simulated baseline. The outbreaks related to nervous cases in horses were randomly inserted, followed by the corresponding outbreaks related to wild bird mortality, such that the time lag between the first dead bird and the first nervous case in horses due to WNV was 0, 1, or 2 weeks according to Kulasekera et al. 2001. The corresponding horse mortality outbreaks were inserted such that half of the affected horses died the week of onset of clinical signs and half died the week after (Cantile et al. 2000, Trock et al. 2001, Bunning et al. 2002, Ward et al. 2006). A summary of time lag between nervous symptoms in horses, horse mortality, and wild bird mortality is available in Supplementary Figure S1.
Outbreak detection
Bayesian framework
Bayesian hypothesis testing is based on two mutually exclusive hypotheses, which can be expressed in the syndromic surveillance context as H1, “there is an ongoing outbreak of WNV (or another event with similar symptoms),” and H0, “there is no ongoing outbreak (Andersson et al. 2014). The relative probability of the two hypotheses can be expressed as a ratio (Opri) that represents our a priori belief about the disease status:
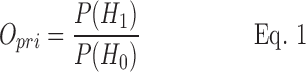 |
When evidence in favor (or not) of each hypothesis is observed, we can build the a posteriori belief about the disease status (Opost):
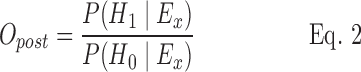 |
where P(H1|Ex) is the probability of H1 given the evidence E observed in time series x, and P(H0 |Ex) is the probability of H0 given the evidence E observed in time series x in a particular week.
Using this general framework with the application of Bayes' theorem, Opost can be calculated as follows:
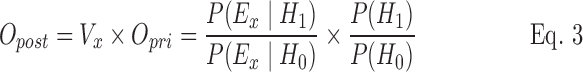 |
where Vx is the value of evidence, P(Ex|H1) is the probability of observing the number of reported cases of syndrome x in a particular week, given that H1 is true, and P(Ex|H0) is the probability of observing the number of reported cases of syndrome x in a particular week given that H0 is true.
To estimate P(Ex|H1) and P(Ex|H0), information on the probability distribution for the number of reported cases in non-outbreak and outbreak situations is used. The probability of Ex (observation of n cases in time series x) during an outbreak is calculated as follows:
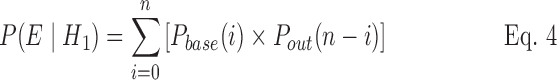 |
where Pbase(i) is the probability of drawing i cases from the baseline distribution in time series x, and Pout(i) is the probability of drawing i cases from the outbreak distribution in time series x based on the shape of the outbreak, as previously simulated.
To detect outbreaks, several values for Opost were tested to serve as alarm thresholds.
Combining time series
When the three time series were combined, Vtot incorporated evidence from NervSy, DeadHorse, and DeadBird, respectively, denoted as ENervSy, EDeadHorse, and EDeadBird. Assuming that the three sources of evidence were conditionally independent, given outbreak status and seasonality of baselines, Vtot was calculated as
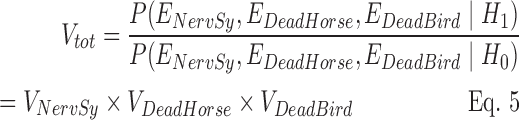 |
and Opost_tot was calculated as
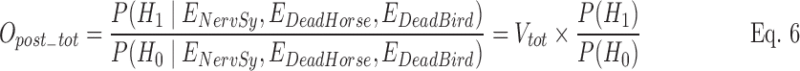 |
Performance assessment
Sensitivity (Se) and specificity (Sp) were calculated as follows:
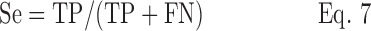 |
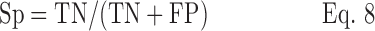 |
where TP is the number of true positive alarms, TN the number of true negative alarms, FP the number of false positive alarms, and FN the number of false negative alarms.
The receiver operating characteristic (ROC) curve was generated in R by testing various alarm thresholds, and the area under ROC curve (AUC) was calculated with the auc function of the package {flux}. A larger AUC represented a better detection performance.
Results
Modeling time series and simulating data
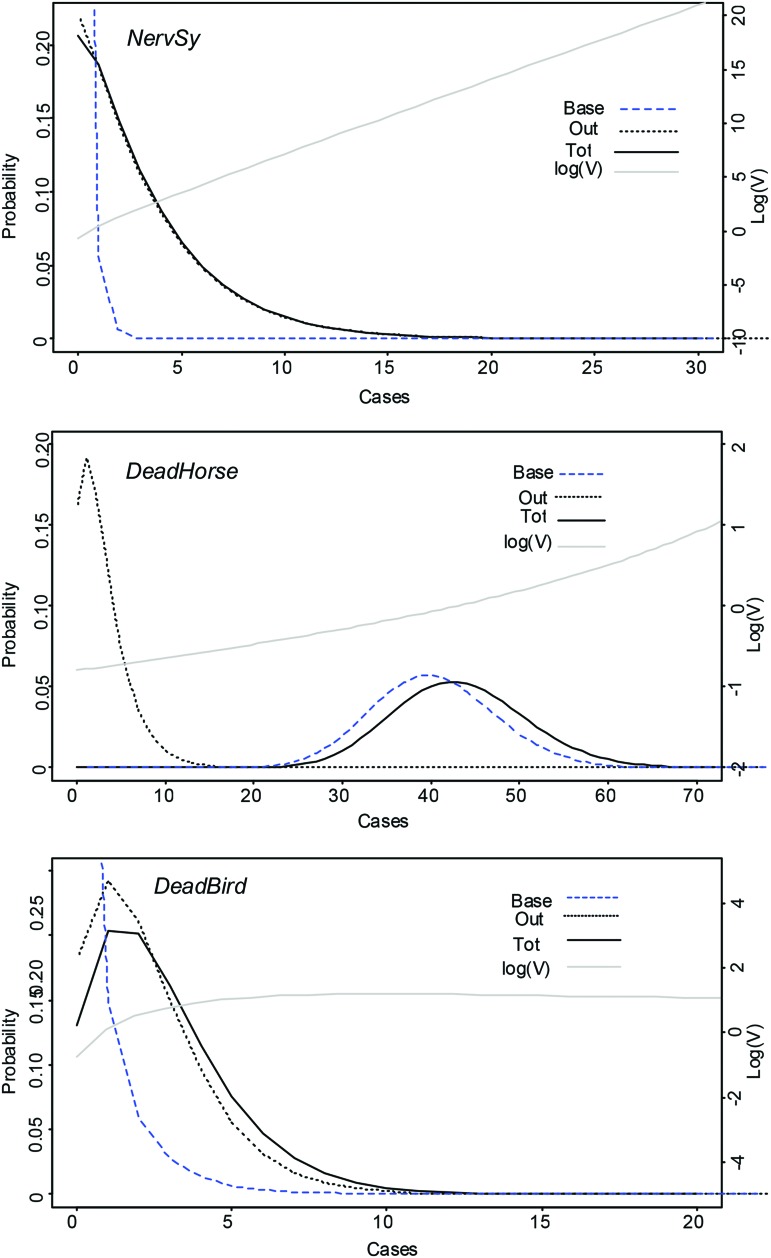
For all time series, the best fits were obtained for NB distributions. The resulting model parameters are summarized in Table 1, and corresponding baselines and predictions are shown in Figure 1. The probabilities of observing n cases and the resulting value of V (P(E|H1)/P(E|H0)) during a non-outbreak (P(E|H0)) and an outbreak (P(E|H1)) situation for each time series are summarized in Figure 2.
Table 1.
Models and Model Parameters Obtained for the Three Time Series
| Negative binomial distribution | ||||||
|---|---|---|---|---|---|---|
| Formulae | Theta | Mean | AIC | GOF | RMSEc | RMSEv |
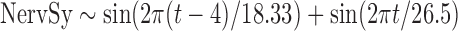 |
0.413 | 0.077 | 143 | 0.279 | 0.30 | 0.39 |
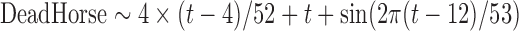 |
176 | 40.3 | 1063 | 1.016 | 7.06 | 8.57 |
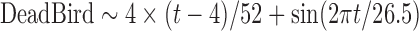 |
0.373 | 0.520 | 497 | 0.675 | 1.03 | 1.05 |
Theta is the dispersion parameter as defined in the function glm.nb (package {MASS}) in R x64 version 3.0.2.
AIC, Akaike information criterion; GOF, goodness-of-fit; RMSE, root-mean-squared error.
FIG. 1.
Three time series considered. NervSy: number of declaration of nervous syndrome in horses without a positive laboratory result. DeadHorse: number of dead adult horses collected by French fallen stock companies. DeadBird: number of dead wild birds autopsied with values above the 95% confidence interval deleted. Dotted lines, training data; solid black lines, test data; solid blue lines, predicted value; solid red lines, 95% confidence interval. Color images available online at www.liebertpub.com/vbz
FIG. 2.
Value of evidence and probabilities of observing n cases during a non-outbreak (Base) and an outbreak (Out) situation. Base, distribution of cases distribution into the baseline; Out, distribution of cases related to a WNV outbreak; Tot, distribution of cases during an outbreak (Base+Out); Log(V) = log10(P(n|outbreak)/P(n|baseline)). Out was estimated with fitdistr function of the package {fitdistrplus} and was based for NervSy on NB (mu = 3.12, theta = 1.150), for DeadHorse on NB (mu = 3, theta = 2.005), and for DeadBird on NB (mean = 2.23, theta = 3.34). NB, negative binomial; WNV, West Nile virus. Color images available online at www.liebertpub.com/vbz
Outbreak detection
We estimated the respective performance of each univariate system (NervSy, DeadHorse, and DeadBird) in detecting WNV outbreaks without considering any a priori values for disease status (Opri = 1). Examples of simulated baselines with inserted outbreaks and associated variations in log10(V) are presented in Supplementary Figure S2.
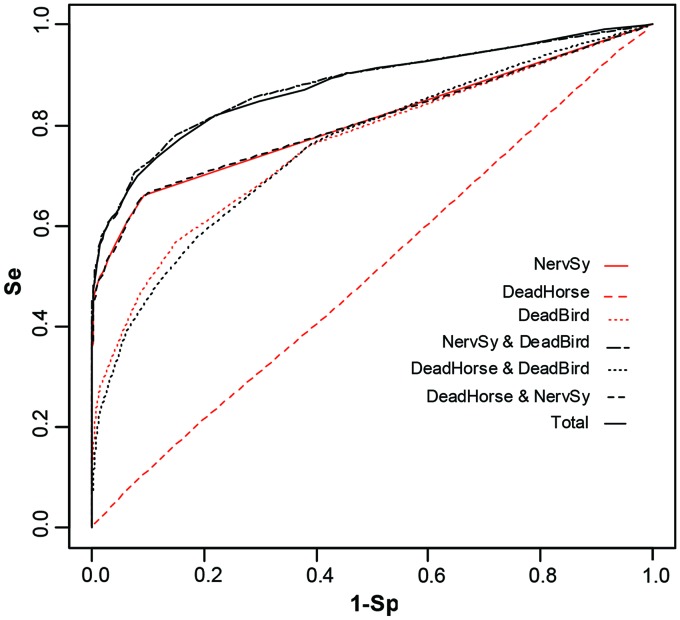
The best results for univariate outbreak detection were obtained for NervSy, which outperformed analyses using DeadHorse and DeadBird (Fig. 3 and Table 2). DeadBird models yielded intermediary detection performances, whereas models using DeadHorse were not able to discriminate between outbreak and non-outbreak situations (AUC ≈0.50).
FIG. 3.
Receiver operating characteristic curves for univariate and multivariate outbreak detection using NervSy, DeadHorse, and DeadBird. Color images available online at www.liebertpub.com/vbz
Table 2.
Area Under the ROC Curve and Standard Error for Univariate and Multivariate Outbreak Detection Using NervSy, DeadHorse, and DeadBird
| NervSy | DeadHorse | DeadBird | NervSy & DeadBird | NervSy & DeadHorse | DeadHorse & DeadBird | Total | |
|---|---|---|---|---|---|---|---|
| AUC | 0.80 | 0.50 | 0.75 | 0.87 | 0.80 | 0.75 | 0.87 |
| Standard error | 0.0082 | 0.0097 | 0.0089 | 0.0068 | 0.0081 | 0.0089 | 0.0068 |
AUC, area under ROC curve; ROC, receiver operating characteristic.
The best results for multivariate outbreak detection were obtained for analyses that combined NervSy with DeadBird data, which gave similar results to a combination of the three time series (Fig. 3 and Table 2). The results of using NervSy combined with DeadBird were also better than those obtained with each time series alone. For example, for a specificity set at 0.80, the sensitivity of the detection reached 0.80 with the combined NervSy and DeadBird series, whereas it was 0.67 with NervSy and 0.60 with DeadBird alone.
Discussion
This is the first time that a real assessment of sensitivity and specificity has been implemented for WNV syndromic surveillance. Previous early warning systems developed for WNV only identified risk factors of WNV outbreaks, but did not evaluate the detection performances of those systems (Gosselin et al. 2005, Shuai et al. 2006, El Adlouni et al. 2007, Brown 2012, Chaskopoulou et al. 2013, Bellini et al. 2014b; Rosà et al. 2014, Valiakos et al. 2014). Only two attempts to assess the sensitivity and specificity of surveillance have been made (Leblond et al. 2007a, Andersson et al. 2014), but the parameters of interest were only evaluated based on a limited number of outbreaks, which did not allow any conclusions to be drawn regarding the overall system performance. Timeliness has occasionally been evaluated, but only based on a limited number of real WNV outbreaks, and has not been associated with a further evaluation of system performance (Eidson et al. 2001, Mostashari et al. 2003, Johnson et al. 2006, Veksler et al. 2009, Calzolari et al. 2013, Chaintoutis et al. 2014). In our study, we have refrained from assessing timeliness as there is currently little or no data to support assumptions on the temporal course on WNV outbreak in Europe, especially, in wild birds. Indeed, we are currently only able to estimate the number of cases expected during an epidemic time period, but not the difference between the number of expected cases at the start of an outbreak and later on. All the weeks within an epidemic time period are thus independent and have the same probability to have a high (or low) number of cases. In this situation, assessing which one is detected first would be not informative about the timeliness of our detection. However, further studies should be conducted on that point to rule on the efficiency of such surveillance in serving as an early warning system for possible human viral infections.
Our results indicated that when using a univariate detection method, NervSy was the best indicator of WNV outbreaks. This is consistent with the number of expected cases during an outbreak compared to the baseline of each time series considered (i.e., high number of cases for NervSy, moderate number of cases for DeadBird, and low number of cases for DeadHorse). Indeed, models based only on the DeadHorse data resulted in poor detection performance at the regional level, because mortality in horses is mainly due to causes other than WNV. To implement such a surveillance system on the field, it would be necessary to assess the cost-effectiveness of the system to define, in close collaboration with decision-makers, the best balance between sensitivity and specificity. In addition, the real representativeness of data sets is still unknown and should be assessed as they might have a great impact on system performances. However, it is hoped that our promising results will promote the timely collection and analysis of relevant data and the implementation of such studies.
The best detection performance was obtained using multivariate syndromic surveillance based on reports of nervous symptoms in horses (NervSy) and wild bird mortality (DeadBird). It is complicated to know how different data sets complement one another. However, we can expect that dead birds would be mainly used to signal the start of an outbreak and that horses confirm the occurrence. To our knowledge, this is the first time that multivariate syndromic surveillance has been implemented for WNV detection. Our results not only offer a wide range of opportunities but also raise questions regarding practical implementation on the field of such multivariate system. In the model, the value of evidence compares the probability of observing syndromes under baseline conditions and during a WNV outbreak, and the calculation of specificity refers to false alarms from random aberrations. Consequently, peaks in the syndromic data streams due to other causes, such as equine herpes virus-1 for NervSy or Avian Influenza or intoxication for DeadBird, will be presented as evidence in favor of WNV. The Bayesian framework offers the possibility to include differential diagnoses and specifies the prior probability and expected impact on the distribution of counts in each data stream. Doing so would enable us to estimate the posterior probability and evidence in favor of a WNV outbreak. However, such a model would be very complicated and hard to support with data. Instead, we explicitly define our hypothesis of interest. When the model triggers an alarm, the distinction between WNV and other diagnoses will be made using field investigations.
The Bayesian framework is a comprehensive and logical way to combine syndromic data from several data streams and it seems well adapted for multivariate WNV detection using three indicators for WNV outbreak detection. This framework provides a means of weighting the results from syndromic surveillance, and thus, additional information can be easily added. Then, a next step in the early detection of WNV outbreaks should be to test the efficiency of the method with other data, such as the predicted abundance of mosquitoes (Calistri et al. 2014, Rosà et al. 2014), environmental risk factors (Tran et al. 2014), and risk of introduction (Brown et al. 2012, Bessell et al. 2014). In addition, the Bayesian approach could be easily adapted to spatiotemporal analysis. Such approach could be especially relevant for WNV surveillance as there are strong links between environment and WNV outbreaks, and as we expect local clusters of cases (Mostashari et al. 2003, Leblond et al. 2007b). Without integrating a spatiotemporal approach, the usefulness of a multivariate syndromic approach could be limited, especially for vector-borne disease surveillance, and thus, the next step would be to develop and test a spatiotemporal model. However, the quality of geographical information of reported cases used in our study is currently insufficient to implement spatiotemporal analysis. In future studies, it would be interesting to improve data quality to test if spatiotemporal analysis could also improve WNV detection and to rule on the usefulness of DeadHorse time series. Indeed, using another spatiotemporal scale, local clusters of deaths in horses might be used as a signal of a WNV outbreak.
Conclusion
The proposed approach gives promising results for improving surveillance of WNV in France and maybe also more generally in Europe. It offers a comprehensive and logical way to combine syndromic data from several data streams, which can be relevant to improve the surveillance of many other diseases (e.g., Bluetongue virus combining data from milk yield and stillbirths, or Japanese encephalitis combining data on nervous symptoms in horses, and reproductive losses in swine). However, questions remain on the practical implementation on the field of such multivariate system, especially regarding interpretation of combined signals, and on the detection's timeliness to serve as an early signal for possible human WNV infections in Europe. It is hoped that our results will support the implementation of further studies to solve these questions and that they will contribute to develop more collaborative work between existing surveillance networks.
Supplementary Material
Acknowledgments
Data on mortality in horses were provided by the French Ministry of Agriculture. Data on mortality in wild birds were provided by the event-based surveillance system “SAGIR,” the French surveillance network for wild birds and mammals. Data on nervous symptoms in horses were provided by “RESPE” (Réseau d'Epidémio-Surveillance en Pathologie Equine). The authors thank all persons involved in these networks (hunters, technicians of FDC, environmental inspectors of ONCFS, local veterinary laboratories, veterinarians, and fallen stock companies) for collecting these data and all these institutes for providing them. This research was carried out as part of the VICE project funded under the FP7 EMIDA ERA-NET initiative. National funding within ERA-Net was provided by the Dutch Ministry of Economic Affairs (project no. BO-20-009-009) and by the Swedish Research Council Formas. This work was also partially financially supported by IFCE (Institut Français du Cheval et de l'Equitation) and Vetagro Sup (National Veterinary School of Lyon).
Author Disclosure Statement
No competing financial interests exist.
References
- Andersson MG, Faverjon C, Via F, Legrand L, et al. Using bayes' rule to define the value of evidence from syndromic surveillance. PLoS One 2014; 9:e111335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous. Programme de surveillance vétérinaire de la fièvre West-Nile. Direction générale de l'alimentation; 2007. Report No.: DGAL/SDSPA/N2007-8136. Available at http://agriculture.gouv.fr/IMG/pdf/dgaln20078136z.pdf (in French)
- Autorino GL, Battisti A, Deubel V, Ferrari G, et al. West Nile virus epidemic in horses, Tuscany Region, Italy. Emerg Infect Dis 2002; 8:1372–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakonyi T, Ivanics É, Erdélyi K, Ursu K, et al. Lineage 1 and 2 strains of encephalitic West Nile Virus, Central Europe. Emerg Infect Dis 2006; 12:618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakonyi T, Ferenczi E, Erdélyi K, Kutasi O, et al. Explosive spread of a neuroinvasive lineage 2 West Nile virus in Central Europe, 2008/2009. Vet Microbiol 2013; 165:61–70 [DOI] [PubMed] [Google Scholar]
- Beck C, Jimenez-Clavero M, Leblond A, Durand B, et al. Flaviviruses in Europe: complex circulation patterns and their consequences for the diagnosis and control of West Nile disease. Int J Environ Res Public Health 2013; 10:6049–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini R, Calzolari M, Mattivi A, Tamba M, et al. The experience of West Nile virus integrated surveillance system in the Emilia-Romagna region: five years of implementation, Italy, 2009 to 2013. Euro Surveill 2014a; 19:pii: [DOI] [PubMed] [Google Scholar]
- Bellini R, Zeller H, Bortel WV. A review of the vector management methods to prevent and control outbreaks of West Nile virus infection and the challenge for Europe. Parasit Vectors 2014b; 7:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessell PR, Robinson RA, Golding N, Searle KR, et al. Quantifying the risk of introduction of West Nile virus into Great Britain by migrating passerine birds. Transbound Emerg Dis 2014. [Epub ahead of print]; DOI: 10.1111/tbed.12310 [DOI] [PubMed]
- Bozdogan H. Model selection and Akaike's information criterion (AIC): the general theory and its analytical extensions. Psychometrika 1987; 52:345–370 [Google Scholar]
- Brown G. California Mosquito-borne Virus Surveillance & Response Plan. Mosquito & Vector Control Association of California, University of California at Davis, 2012:57 Available at www.cdph.ca.gov/HealthInfo/discond/Documents/CAResponsePlanMay2012.pdf [Google Scholar]
- Brown EBE, Adkin A, Fooks AR, Stephenson B, et al. Assessing the risks of West Nile virus-infected mosquitoes from transatlantic aircraft: implications for disease emergence in the United Kingdom. Vector Borne Zoonotic Dis 2012; 12:310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunning ML, Bowen RA, Cropp CB, Sullivan KG, et al. Experimental infection of horses with West Nile virus. Emerg Infect Dis 2002; 8:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri P, Giovannini A, Hubalek Z, Ionescu A, et al. Epidemiology of West Nile in Europe and in the Mediterranean Basin. Open Virol J 2010; 4:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri P, Savini L, Candeloro L, Di Sabatino D, et al. A transitional model for the evaluation of West Nile virus transmission in Italy. Transbound Emerg Dis 2014. [Epub ahead of print]; DOI: 10.1111/tbed.12290 [DOI] [PubMed] [Google Scholar]
- Calzolari M, Monaco F, Montarsi F, Bonilauri P, et al. New incursions of West Nile virus lineage 2 in Italy in 2013: the value of the entomological surveillance as early warning system. Vet Ital 2013; 49:315–319 [DOI] [PubMed] [Google Scholar]
- Campbell GL, Marfin AA, Lanciotti RS, Gubler DJ. West Nile virus. Lancet Infect Dis 2002; 2:519–529 [DOI] [PubMed] [Google Scholar]
- Cantile C, Di Guardo G, Eleni C, Arispici M. Clinical and neuropathological features of West Nile virus equine encephalomyelitis in Italy. Equine Vet J 2000; 32:31–35 [DOI] [PubMed] [Google Scholar]
- Castillo-Olivares J, Wood J. West Nile virus infection of horses. Vet Res 2004; 35:467–483 [DOI] [PubMed] [Google Scholar]
- Chai T, Draxler RR. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci Model Dev 2014; 7:1247–1250 [Google Scholar]
- Chaintoutis SC, Dovas CI, Papanastassopoulou M, Gewehr S, et al. Evaluation of a West Nile virus surveillance and early warning system in Greece, based on domestic pigeons. Comp Immunol Microbiol Infect Dis 2014; 37:131–141 [DOI] [PubMed] [Google Scholar]
- Chaskopoulou A, Dovas CI, Chaintoutis SC, Kashefi J, et al. Detection and early warning of West Nile virus circulation in Central Macedonia, Greece, using sentinel chickens and mosquitoes. Vector Borne Zoonotic Dis 2013; 13:723–732 [DOI] [PubMed] [Google Scholar]
- Chevalier V, Lecollinet S, Durand B. West Nile virus in Europe: a comparison of surveillance system designs in a changing epidemiological context. Vector Borne Zoonotic Dis 2011; 11:1085–1091 [DOI] [PubMed] [Google Scholar]
- Danis K, Papa A, Theocharopoulos G, Dougas G, et al. Outbreak of West Nile virus infection in Greece, 2010. Emerg Infect Dis 2011; 17:1868–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decors A, Hars J, Faure E, Quintaine T, et al. Le réseau Sagir: un outil de vigilance vis-à-vis des agents pathogènes exotiques. Bull Épidémiologique Santé Anim Aliment 2014; 66:35–39 [Google Scholar]
- Del Amo J, Llorente F, Figuerola J, Soriguer RC, et al. Experimental infection of house sparrows (Passer domesticus) with West Nile virus isolates of Euro-Mediterranean and North American origins. Vet Res 2014a; 45:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Amo J, Llorente F, Pérez-Ramirez E, Soriguer RC, et al. Experimental infection of house sparrows (Passer domesticus) with West Nile virus strains of lineages 1 and 2. Vet Microbiol 2014b; 172:542–547 [DOI] [PubMed] [Google Scholar]
- Di Sabatino D, Bruno R, Sauro F, Danzetta ML, et al. Epidemiology of West Nile disease in Europe and in the Mediterranean Basin from 2009 to 2013. BioMed Res Int 2014; 2014:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dórea FC, McEwen BJ, McNab WB, Revie CW, et al. Syndromic surveillance using veterinary laboratory data: data pre-processing and algorithm performance evaluation. J R Soc Interface 2013; 10:20130114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi M, Vangeluwe D, Lecollinet S, van den Berg T, et al. Experimental infection of Carrion crows (Corvus corone) with two European West Nile virus (WNV) strains. Vet Microbiol 2013; 165:160–166 [DOI] [PubMed] [Google Scholar]
- Eidson M, Kramer L, Stone W, Hagiwara Y, et al. Dead bird surveillance as an early warning system for West Nile virus. Emerg Infect Dis 2001; 7:631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Adlouni S, Beaulieu C, Ouarda T, Gosselin P, et al. Effects of climate on West Nile Virus transmission risk used for public health decision-making in Quebec. Int J Health Geogr 2007; 6:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisén M, Andersson E, Schiöler L. Evaluation of multivariate surveillance. J Appl Stat 2010; 37:2089–2100 [Google Scholar]
- Gosselin P, Lebel G, Rivest S, Douville-Fradet M. The Integrated System for Public Health Monitoring of West Nile Virus (ISPHM-WNV): a real-time GIS for surveillance and decision-making. Int J Health Geogr 2005; 4:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Triana LM, Jeffries CL, Mansfield KL, Carnell G, et al. Emergence of West Nile virus lineage 2 in Europe: a review on the introduction and spread of a mosquito-borne disease. Front Public Health 2014; 2:271 Available at www.ncbi.nlm.nih.gov/pmc/articles/PMC4258884/ [DOI] [PMC free article] [PubMed]
- Johnson GD, Eidson M, Schmit K, Ellis A, et al. Geographic prediction of human onset of West Nile virus using dead crow clusters: an evaluation of year 2002 data in New York State. Am J Epidemiol 2006; 163:171–180 [DOI] [PubMed] [Google Scholar]
- Kulasekera VL, Kramer L, Nasci RS, Mostashari F, et al. West Nile virus infection in mosquitoes, birds, horses, and humans, Staten Island, New York, 2000. Emerg Infect Dis 2001; 7:722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutasi O, Bakonyi T, Lecollinet S, Biksi I, et al. Equine encephalomyelitis outbreak caused by a genetic lineage 2 West Nile virus in Hungary: Lineage 2 West Nile virus encephalomyelitis. J Vet Intern Med 2011; 25:586–591 [DOI] [PubMed] [Google Scholar]
- Leblond A, Hendrikx P, Sabatier P. West Nile virus outbreak detection using syndromic monitoring in horses. Vector Borne Zoonotic Dis 2007a; 7:403–410 [DOI] [PubMed] [Google Scholar]
- Leblond A, Sandoz A, Lefebvre G, Zeller H, et al. Remote sensing based identification of environmental risk factors associated with West Nile disease in horses in Camargue, France. Prev Vet Med 2007b; 79:20–31 [DOI] [PubMed] [Google Scholar]
- Lecollinet S, Beck C, Lebond A, Marcillaud-Pitel C, et al. Re-emerging West Nile virus in horses from South Eastern France, 2015. J Equine Vet Sci 2016; 39:S26eS32 [Google Scholar]
- Mostashari F, Kulldorff M, Hartman JJ, Miller JR, et al. Dead bird clusters as an early warning system for West Nile virus activity. Emerg Infect Dis 2003; 9:641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgue B, Murri S, Zientara S, Durand B, et al. West Nile outbreak in horses in southern France, 2000: the return after 35 years. Emerg Infect Dis 2001; 7:692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin J-B, Ducrot C, Vinard J-L, Morignat E, et al. Assessment of the utility of routinely collected cattle census and disposal data for syndromic surveillance. Prev Vet Med 2012; 105:244–252 [DOI] [PubMed] [Google Scholar]
- Rosà R, Marini G, Bolzoni L, Neteler M, et al. Early warning of West Nile virus mosquito vector: climate and land use models successfully explain phenology and abundance of Culex pipiens mosquitoes in north-western Italy. Parasit Vectors 2014; 7:269 Available at www.biomedcentral.com/content/pdf/1756-3305-7-269.pdf [DOI] [PMC free article] [PubMed]
- Saegerman C, Alba-Casals A, García-Bocanegra I, Dal Pozzo F, et al. Clinical sentinel surveillance of equine West Nile fever, Spain. Transbound Emerg Dis 2016; 63:184–193. Available at http://doi.wiley.com/10.1111/tbed.12243 [DOI] [PubMed]
- Shuai J, Buck P, Sockett P, Aramini J, et al. A GIS-driven integrated real-time surveillance pilot system for national West Nile virus dead bird surveillance in Canada. Int J Health Geogr 2006; 5:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonesson C, Frisén M. Multivariate surveillance. In: Lawson AB, Kleinman K, eds. Spatial and Syndromic Surveillance for Public Health. John Wiley & Sons, Ltd., Chichester, UK, 2005:153–166 [Google Scholar]
- Sotelo E, Gutierrez-Guzmán A, del Amo J, Llorente F, et al. Pathogenicity of two recent Western Mediterranean West Nile virus isolates in a wild bird species indigenous to Southern Europe: the red-legged partridge. Vet Res 2011; 42:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran A, Sudre B, Paz S, Rossi M, et al. Environmental predictors of West Nile fever risk in Europe. Int J Health Geogr 2014; 13:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triple S Project. Guideline for designing and implementing a syndromic surveillance system 2011. Available at www.syndromicsurveillance.eu/Triple-S_guidelines.pdf
- Trock SC, Meade BJ, Glaser AL, Ostlund EN, et al. West Nile virus outbreak among horses in New York State, 1999 and 2000. Emerg Infect Dis 2001; 7:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui FC, Wagner MM, Dato V, Chang CC. Value of ICD-9 coded chief complaints for detection of epidemics. Proc AMIA Symp 2001; 711–715 [PMC free article] [PubMed] [Google Scholar]
- Valiakos G, Papaspyropoulos K, Giannakopoulos A, Birtsas P, et al. Use of Wild Bird Surveillance, Human Case Data and GIS Spatial Analysis for Predicting Spatial Distributions of West Nile Virus in Greece. PLoS One 2014; 9:e96935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veksler A, Eidson M, Zurbenko I. Assessment of methods for prediction of human West Nile virus (WNV) disease from WNV-infected dead birds. Emerg Themes Epidemiol 2009; 6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Schuermann JA, Highfield LD, Murray KO. Characteristics of an outbreak of West Nile virus encephalomyelitis in a previously uninfected population of horses. Vet Microbiol 2006; 118:255–259 [DOI] [PubMed] [Google Scholar]
- Ziegler U, Angenvoort J, Fischer D, Fast C, et al. Pathogenesis of West Nile virus lineage 1 and 2 in experimentally infected large falcons. Vet Microbiol 2013; 161:263–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.