Abstract
Introduction: Dengue is a significant arboviral infection that represents a major public health concern worldwide. The infection is endemic in most parts of South East Asia, sub-Saharan Africa, and Latin America. Among the four dengue virus (DENV) serotypes, DENV-2 has been reported to be the predominant serotype in Saudi Arabia since 1992. However, virological and epidemiological data of DENV-2 from Saudi Arabia are severely deficient and require further investigations.
Methods: Full genome sequencing of a recent DENV-2 isolate and phylogenetic analysis of all available DENV-2 sequences from Saudi Arabia.
Results: Based on full genome and envelope (E) gene sequence, we show that a recent isolate (DENV-2-Jeddah-2014) belongs to the Indian subcontinent lineage of the Cosmopolitan genotype with close similarity to recent strains from Pakistan. Interestingly, the E gene sequence of DENV-2-Jeddah-2014 isolate was slightly divergent from those previously identified in Saudi Arabia between 1992 and 2004 with three to nine amino acid (aa) substitutions. While our data show that the Cosmopolitan genotype is still circulating in Saudi Arabia, they highlight four distinct genetic groups suggesting at least four independent introductions into the Kingdom.
Conclusions: The close clustering of DENV-2 isolates reported from Saudi Arabia between 1992 and 2014 with strains from countries providing the highest numbers of pilgrims attending either Hajj or Umrah pilgrimages (Indonesia, Pakistan, India) clearly suggests a role for pilgrims or expatriates coming from DENV endemic countries in DENV-2 importation into Saudi Arabia. Accordingly, continuous monitoring of the circulation of DENVs in Saudi Arabia must be implemented to undertake effective control and management strategies in the Kingdom. Screening of the pilgrims coming to perform Hajj and Umrah might help prevent the introduction of new DENV strains, which is expected to increase the burden of the disease not only in Saudi Arabia but also in other countries.
Key Words: Dengue virus, Full genome, Jeddah, Phylogenetic analysis sequencing, Saudi Arabia
Introduction
Dengue virus (DENV) is a major health concern all over the world particularly in tropical and subtropical regions (Gubler 1998, World Health Organization 2012). It infects more than 50 million individuals in countries in the Middle East, Africa, Asia, Europe, and the United States (San Martín et al. 2010, Medlock et al. 2015, Pem-Novosel et al. 2015). Serious health conditions such as dengue with warning signs or severe dengue infection can occur during reinfection with heterologous serotypes (Anoop et al. 2010, Figueiredo et al. 2011).
The viral genome size of DENVs varies from 10.6 to 11 kb and contains a single open reading frame (ORF) encoding three structural and seven nonstructural proteins (Green and Rothman 2006). DENV virion consists of the capsid (C), membrane (M), and envelope (E) proteins, whereas the nonstructural proteins are composed of the NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 proteins. Although the four DENV serotypes (DENV-1 to 4) share 65–70% nucleotide (nt) sequence similarity (Green and Rothman 2006), they are antigenically and genetically distinct flaviviruses (Henchal and Putnak 1990). Based on the nt sequence of the E protein, all the four serotypes have been further divided into different genotypes (Wang et al. 2000, Weaver and Vasilakis 2009). Phylogenetic and epidemiological studies have shown an association between the increased air travel and the spread of DENV serotypes in diverse geographical areas as well as the potential association between certain genotypes and disease severity (Kyle and Harris 2008, Rico-Hesse 2010, Nunes et al. 2014).
In Saudi Arabia, millions of Muslims come to Makkah and Al-Madinah during certain time of the year (Hajj season) to perform Hajj and throughout the year for Umrah. Pilgrims arriving from dengue endemic areas can represent a major source for DENV importation into Saudi Arabia. This is evident by the spread of DENV infections in western part of Saudi Arabia particularly in cities like Jeddah, Makkah, and Al-Madinah (Fakeeh and Zaki 2001, 2003, Ayyub et al. 2006, Khan et al. 2008, Zaki et al. 2008, Ahmed 2010, El-Badry et al. 2014, Azhar et al. 2015). In these cities, DENV outbreaks were first reported to be caused by DENV-1 and DENV-2 serotypes in 1994 (Fakeeh and Zaki 2001), and subsequently by DENV-3 in 1997 (Fakeeh and Zaki 2001). These three serotypes seem to continue circulating in the western part of Saudi Arabia until now (Ayyub et al. 2006, Khan et al. 2008, Zaki et al. 2008, Ahmed 2010, El-Badry et al. 2014, Azhar et al. 2015). In addition, the presence and high prevalence of Aedes aegypti in Jeddah and the surrounding areas (Alikhan et al. 2014) increase the risk of autochthonous spread of DENV imported with pilgrims from endemic areas.
The availability of complete genome sequences from Saudi Arabia could provide critical information about the genetic relatedness of locally isolated DENVs to other circulating strains or genotypes in other regions. It could also shed some light on the geographical origin of these viruses. We recently reported the first full genome sequence of DENV-1 isolate from Jeddah and showed the continued spread of DENV-1 Asian genotype in this region (Azhar et al. 2015). However, data on other DENV serotypes are severely deficient in Saudi Arabia. The lack of such data and the need for reference information for future studies prompted us to extend our previous studies. In this report, the complete sequence of a DENV-2 strain isolated in the city of Jeddah in 2014 was used together with seven available partial sequences of DENV-2 viruses from Saudi Arabia to determine whether they were derived from a single ancestral strain or they represented multiple introductions.
Materials and Methods
Diagnosis, virus isolation, and genome sequencing
Serum and plasma samples were collected and stored at −80°C until testing from a patient suffering from dengue-like symptoms who was admitted to King Abdulaziz University Hospital in 2014. Ethical approval was obtained from the Unit of Biomedical Ethics in King Abdulaziz University hospital (Reference Nos. 19–14). The methods were carried out in accordance with the approved guidelines. The plasma and serum samples were screened for DENV IgG and IgM antibodies (Abs) using the Panbio Dengue IgM Capture Kit and Panbio Dengue IgG Kit (Panbio) according to the manufacturer's instructions. RNA was extracted from plasma or culture supernatant using the QIAamp Viral RNA Mini Kit (Qiagen) according to the manufacturer's instructions. Eluted RNA was screened for DENV RNA by real-time RT-PCR using primers and probes as previously described (Drosten et al. 2002). Mosquito Aedes albopictus C6/36 cells were inoculated with 100 μL of patient serum, maintained in a complete Dulbecco's modified Eagle's medium supplemented with 2% fetal calf serum and antibiotics, and incubated in a humidified atmosphere at 28°C in 5% CO2. Viral RNA was extracted from C6/36 cell supernatants and analyzed by real-time RT-PCR. DENV RNA-positive culture supernatants were used for whole viral genome sequencing. Viral RNA extracted from culture supernatant was subjected to RT-PCR amplification and cycle sequencing as described previously (Christenbury et al. 2010, Azhar et al. 2015) using primer pairs covering the whole length of the viral genome. The GenBank accession number given for the deposited DENV-2-Jeddah-2014 sequence is KJ830750.
Sequence and phylogenetic analysis
The complete nt sequence of DENV-2-Jeddah-2014 isolate was initially searched for similarity using the BLAST program (www.ncbi.nlm.nih.gov/BLAST/) (Altschul et al. 1997), and DENV-2 sequences that showed a higher score were selected for further analysis. Sequences were then multiply aligned using ClustalW (www.ebi.ac.uk/clustalw) and nt sequences identity matrix and amino acid (aa) substitutions were analyzed. The evolutionary history was inferred by using the maximum likelihood method based on the general time reversible model gamma distributed with invariant sites (Tamura et al. 2013). The tree with the highest log likelihood (−26343.5636) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach. A discrete gamma distribution was used to model evolutionary rate differences among sites (five categories [+G parameters = 0.4655]). The analysis involved 1065 nucleotide sequences. All positions with less than 95% site coverage were eliminated, so that there were a total of 1485 positions in the final data set. Evolutionary analysis was conducted in MEGA6 (Tamura et al. 2013). In the phylograms, identification of countries was based on the ISO Alpha-2 code (www.iso.org/obp/ui/#search). Analysis for putative recombinations was done using Simplot program (Lole et al. 1999).
Results
Diagnosis and virus isolation
Serological testing of the patient's sample showed positivity for anti-DENV IgM but not IgG Abs, suggesting an acute primary dengue infection. The patient's samples as well as the inoculated cell culture supernatant tested positive for DENV RNA by real-time RT-PCR confirming the acute infection status.
Complete genome sequencingand phylogenetic analysis
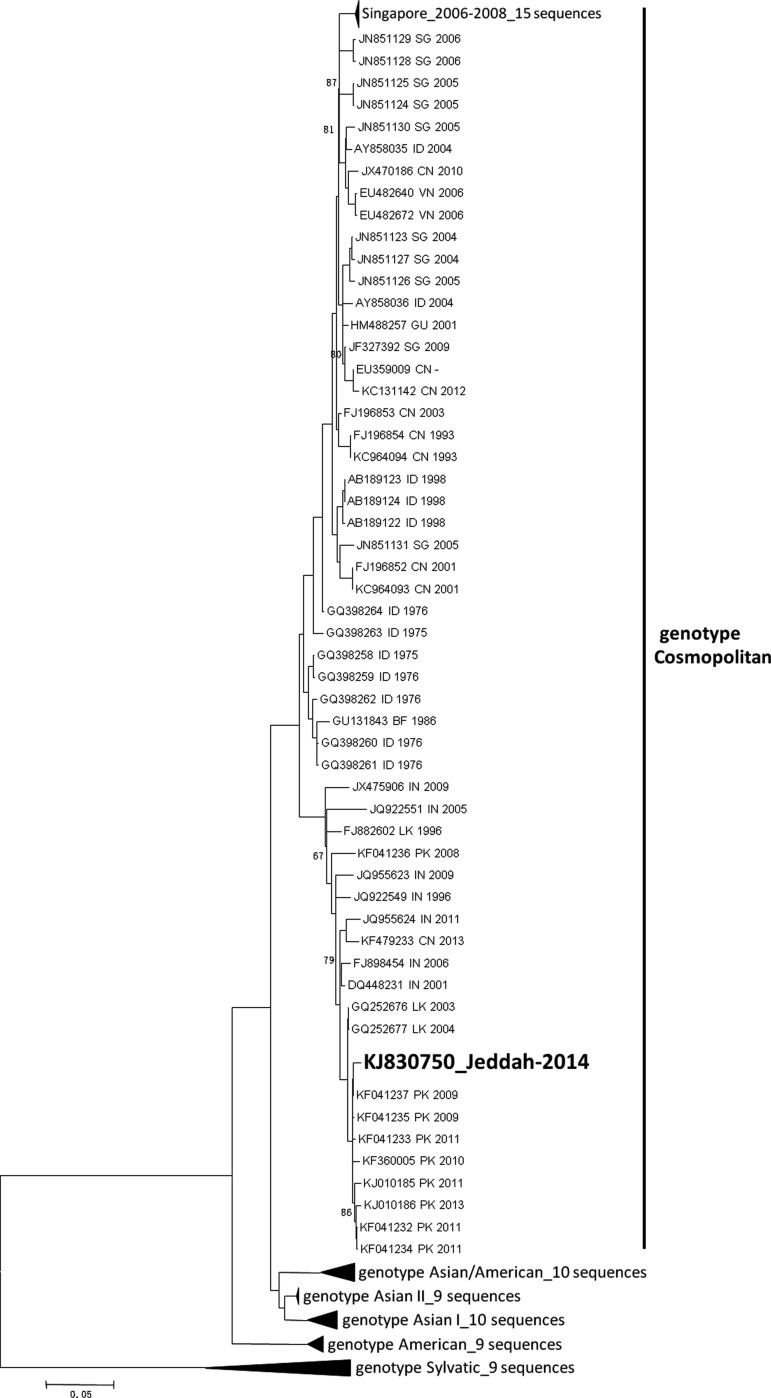
All the sequenced fragments were assembled into a single contig using the Geneious software (www.biomatters.com), and the complete genome of DENV-2-Jeddah-2014 isolate was found to contain 10,718 nt with a single ORF coding for 3391 aa. BLAST query using the nt sequence of DENV-2-Jeddah-2014 isolate against the NCBI GenBank database (www.ncbi.nlm.nih.gov) showed the highest nt identity (99.0%) with an isolate from Pakistan collected in 2009 (KF041237) and lowest nt identity (93.0%) with an isolate collected from Papua New Guinea in 1944 (KM204118). Identities of 99% or more were observed with several DENV-2 isolates collected between 2009 and 2013 from Pakistan (KF041237, KF041235, KF041233, KF041234, KF041232, KJ701507, KF360005, KJ010185, KJ010186) and isolates collected from Sri Lanka between 2003 and 2004 (GQ252676, GQ252677). Lower similarities ranging between 93% and 98% were observed with other strains isolated from India, Pakistan, China, Burkina Faso, Indonesia, Sri Lanka, Vietnam, Taiwan, Singapore, Brunei, Guam, and Australia. Phylogenetic analysis of the full genome sequences showed that the DENV-2-Jeddah-2014 isolate belongs to the Indian subcontinent lineage of the Cosmopolitan genotype of DENV-2 viruses (Fig. 1). Comparing the aa sequences of DENV-2-Jeddah-2014 isolate with its closest match from Pakistan (KF041237) showed eight aa substitutions only in both structural and nonstructural proteins (Table 1). Interestingly, most nt mutations observed in DENV-2-Jeddah-2014 compared to related strains that have at least 95% nt similarity were synonymous mutations with 51–58 aa changes even in comparison to oldest strains collected in 1975 (GQ398263) and 1976 (GQ398264) from Indonesia, and in 1983 (EU056810) and 1986 (GU131843) from Burkina Faso. Furthermore, testing for probable recombination events in the same data set of DENV-2 isolates did not show any recombination events with any of the tested strains (data not shown).
FIG. 1.
Phylogenetic tree of DENV-2-Jeddah-2014 with 118 selected sequences, including all the 71 Cosmopolitan sequences based on complete coding sequences. BF, Burkina Faso; CN, China; DENV, dengue virus; GU, Guam; ID, Indonesia; IN, India; LK, Sri Lanka; PK, Pakistan; SG, Singapore; VN, Vietnam.
Table 1.
Amino Acid Substitutions Observed in the DENV-2-Jeddah-2014 Isolate Compared to the Most Closely Related Isolate
| Protein | Position | KJ830750-Jeddah-2014 | KF041237-Pakistan-2009 |
|---|---|---|---|
| CP | 71 | I | T |
| 92 | M | L | |
| E | 640 | G | E |
| NS1 | 953 | S | F |
| NS2A | 1246 | S | T |
| NS2B | 1400 | K | R |
| NS4B | 2263 | I | T |
| 2393 | V | I |
DENV, dengue virus.
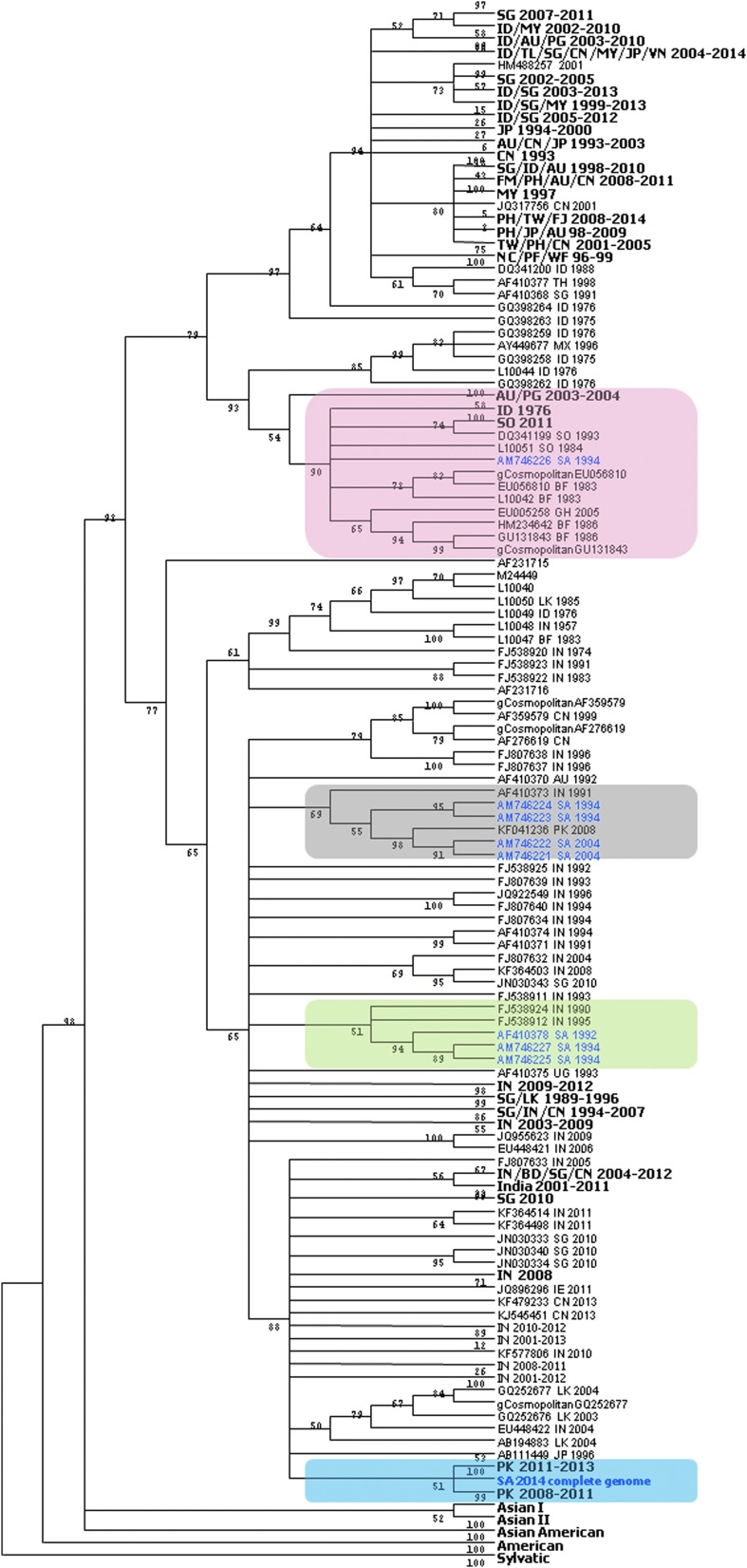
Since most of the DENV-2 sequences in the GenBank are from the E region compared to full genomes, we analyzed the E gene of DENV-2-Jeddah-2014 isolate and compared it to other DENV-2 E genes. Similar to full genome, phylogenetic analysis of the E protein gene of DENV-2-Jeddah-2014 strain showed clustering with isolates circulated in Pakistan from 2008 to 2013 (Fig. 2). Also, it showed that all reported DENV-2 isolates from Saudi Arabia between 1992 and 2014 belong to the Cosmopolitan genotype. Interestingly, the DENV-2-Jeddah-2014 strain E gene sequence was quite different from other previously reported DENV-2 strains isolated from Saudi Arabia between 1992 and 2004. Comparing aa sequence of the E protein between these strains showed three to nine aa differences (Table 2).
FIG. 2.
Phylogenetic tree of DENV-2-Jeddah-2014 based on E gene sequences with selected isolates. Taxa in boldface letters represent sequences that were collected from the same region at the same period of time. Pink box shows clustering of the Jeddah isolate from 1994. Gray box shows clustering of the Jeddah isolates from 1994 to 2004. Green box shows clustering of the Jeddah isolates from 1992 to 1994. Blue box shows clustering of the Jeddah isolate from 2014. Tree shows the closest relatives of Saudi DENV-2 sequences are therefore: SA, Saudi Arabia; AU, Australia; PG, Papua New Guinea; ID, Indonesia; SO, Somalia; BF, Burkina Faso; GH, Ghana; CN, China; IN, India; PK, Pakistan; UG, Uganda; SG, Singapore; LK, Sri Lanka.
Table 2.
Amino Acid Changes in the E Protein of DENV-2 Isolates from Saudi Arabia
| Position | KJ830750-SA-2014 | AF410378-SA-1992 | AM746226-SA-1994 | AM746225-SA-1994 | AM746224-SA-1994 | AM746223-SA-1994 | AM746221-SA-2004 | AM746222-SA-2004 |
|---|---|---|---|---|---|---|---|---|
| 46 | I | T | ||||||
| 93 | K | R | R | |||||
| 129 | I | V | ||||||
| 141 | V | I | I | I | ||||
| 164 | V | I | ||||||
| 171 | T | S | ||||||
| 202 | E | K | ||||||
| 226 | I | T | T | T | T | T | T | T |
| 322 | V | I | I | I | I | I | I | |
| 359 | T | A | M | |||||
| 360 | G | E | E | E | E | E | E | E |
| 390 | S | N | ||||||
| 461 | V | A | A |
Only aa changes are shown and empty cells indicate similar aa to KJ830750-SA-2014 isolate.
aa, Amino acid.
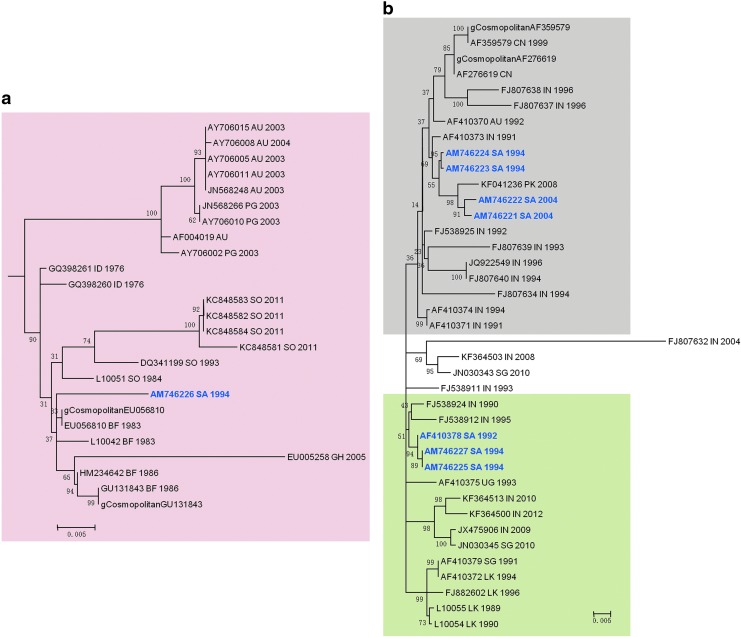
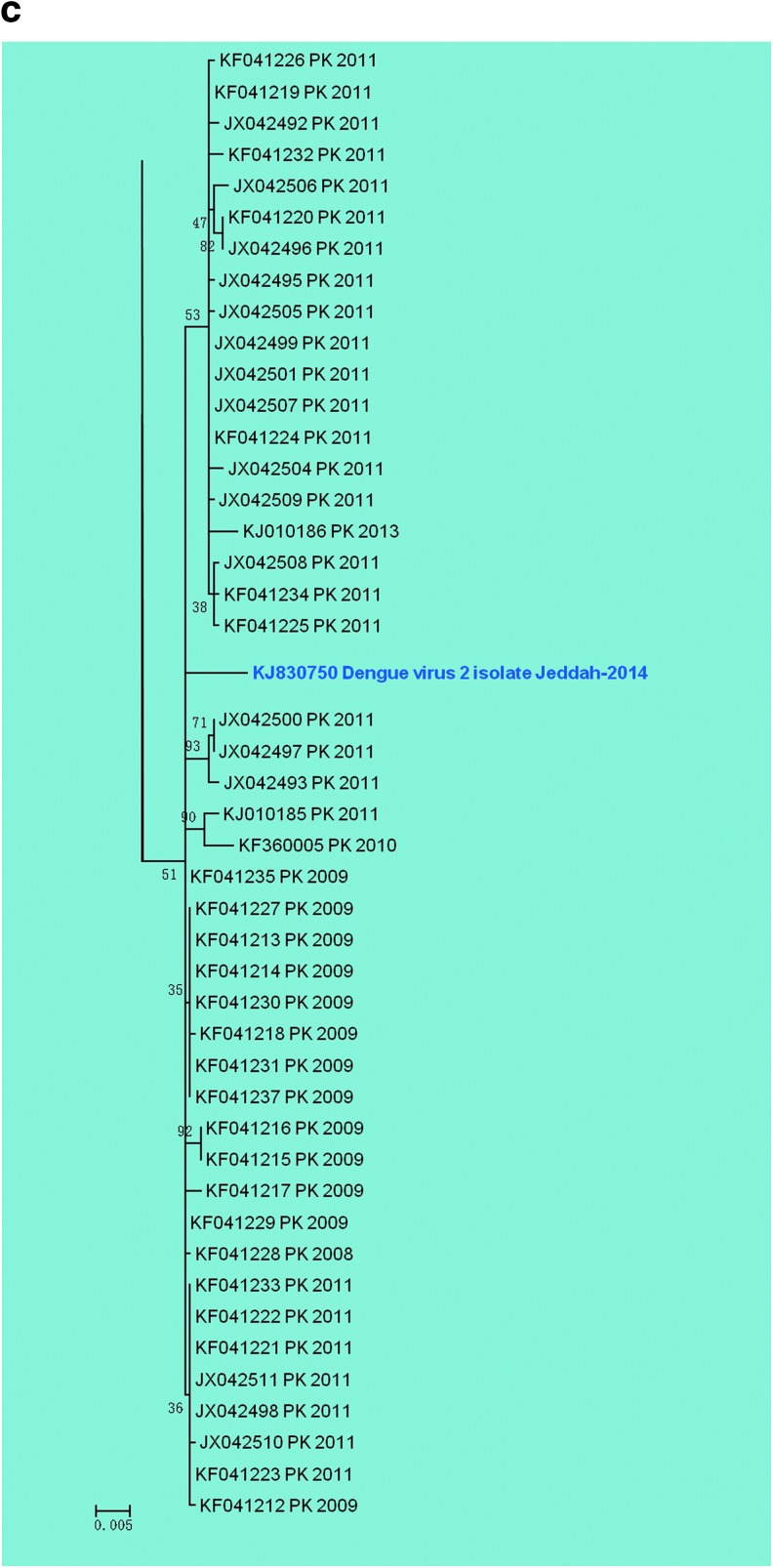
Further phylogenetic analysis of the E gene indicated that there were at least three previous distinct introductions of DENV-2 strains in Saudi Arabia (Fig. 3). A strain isolated in 1994 (Fig. 3a) was clearly distinct since it grouped with strains from Africa (Somalia, Burkina Faso, and Ghana). This strain in particular showed the highest divergence from the DENV-2-Jeddah-2014 isolate with nine aa changes in the E protein (Table 2). Strains isolated in 1994 and in 2004 (Fig. 3b, gray box) were closely related to strains present in China, Australia, India, and Pakistan. A second cluster of strains isolated in 1992 and 1994 (Fig. 3b, green box) was most closely related to strains from India, Singapore, and Sri Lanka although there was also one strain from Uganda in this group.
FIG. 3.
Phylogenetic clustering of DENV-2 isolates from Jeddah in the period between 1992 and 2014 based on E gene sequences. (a) Clustering of one isolate from 1994 highlighted in the pink box. (b) Clustering of isolates from 1994 to 2004 in the gray box and isolates from 1994 in the green box. (c) Clustering of the isolate from 2014 in the blue box
Discussion
DENVs are highly diverse viruses and this is mainly because of the high replication rate, nonproofreading nature of the viral RNA polymerase, and the immune pressure imposed by hosts (Twiddy et al. 2003). This high diversity has been suggested to affect disease severity. Therefore, it is critical to study viral evolution and diversity to better understand their association with disease severity in different geographical areas. In Saudi Arabia, DENVs seem to be limited to the southwestern part due to its proximity to Yemen, a country that is endemic for dengue infection, as well as the western region particularly the two holy cities of Makkah and Al-Madinah together with Jeddah (Fakeeh and Zaki 2001, 2003, Ayyub et al. 2006, Khan et al. 2008, Zaki et al. 2008, Ahmed 2010, Madani et al. 2013, El-Badry et al. 2014, Azhar et al. 2015). While this distribution clearly suggests a role of pilgrims in the spread of the disease at least in the western part of the country, such conclusion requires more molecular epidemiological studies to provide convincing evidence.
Previous reports have suggested that DENV-2 virus is the predominant serotype in Saudi Arabia particularly in western Saudi Arabia since 1992 (Fakeeh and Zaki 2001, 2003, Zaki et al. 2008). In addition, they showed that all isolated DENV-2 strains in Saudi Arabia are from the Cosmopolitan genotype (Zaki et al. 2008). In the current study, we also show that the DENV-2-Jeddah-2014 isolate belongs to the Cosmopolitan genotype where it is most genetically related to isolates from Pakistan circulating from 2008 to 2013 indicating that the DENV-2 Cosmopolitan genotype is still the predominant and the only reported DENV-2 genotype in Saudi Arabia.
Based on sequence similarities, phylogenetic analysis, and the efficiency of Cosmopolitan genotype to circulate throughout the world, our results also provide an evidence for the role of pilgrims in the importation of DENVs into Saudi Arabia from endemic areas. All reported DENV-2 isolates, including the one in this study, seem to be introduced into Saudi Arabia with pilgrims or expatriates coming from countries providing high numbers of pilgrims attending either Hajj or Umrah pilgrimages. Interestingly, Indonesia, Pakistan, and India ranked as the three countries providing the highest number of pilgrims for Hajj, with respective numbers at 293,000, 189,500, and 151,000 pilgrims, that is, a total of 634,000 (45%) over a total of 1,400,000 foreign pilgrims (www.english.alarabiya.net/en/perspective/features/2014/10/19/Saudi-Arabia-hosted-25m-hajj-pilgrims-in-past-10-years.html). Furthermore, recombination events have high probability in regions where multiple strains are introduced, as in the situation at Jeddah where millions of Muslims come to the city from DENV endemic areas on their way to the holy places in Makkah and Al-Madinah. While our recombination analysis of the DENV-2-Jeddah-2014 isolate showed no recombination event with closely related isolates, the availability of such full genome sequences allows testing these probable recombination events in future studies.
Although the topology of the E gene tree demonstrates that all strains from Saudi Arabia belong to the Cosmopolitan genotype, it is possible to identify at least four different introductions especially all strains that belong to four distinct sublineages (Fig. 2). One introduction occurred in 1994 from Africa (Fig. 3a). The same year strains originating from the Indian subcontinent and from Asia were established in Saudi Arabia representing at least two independent introductions (Fig. 3b). Similarly, the DENV-2-Jeddah-2014 isolate represents a more recent introduction (Fig. 3c) as it clusters with recent isolates from Pakistan.
Detailed analysis of the E protein showed three unique aa changes T226I, I322V, and E360G in the DENV-2-Jeddah-2014 isolate E protein compared to previous ones from Saudi Arabia (Table 2). Of particular interest is the E360G mutation that is also observed in comparison to the closest match to DENV-2-Jeddah-2014 isolate (i.e., KF041237 from Pakistan). While it is not clear if these mutations have any implication on virulence or viral fitness, they clearly require further analysis.
DENV-2 genotypes differ in virulence and it is not known whether currently circulating DENV-2 strains in Saudi Arabia are of low or high virulence. Based on the analysis of E gene sequences, Twiddy et al. (2002) reported that the Cosmopolitan genotype of DENV-2 is under positive selection at least in three aa positions (52, 129, and 390) within the E protein. Amino acid at position 129 has been suggested to be involved in the acidic pH conformational change responsible for fusion peptide exposure on the viral surface (Roehrig et al. 1994). Similarly, aa at position 390 is part of domain III in the E protein, which is believed to be involved in viral binding to the cell receptor (Sánchez and Ruiz 1996). Thus, it was suggested that substitutions at these two positions (I129V and N390S) may affect viral tropism and virulence (Twiddy et al. 2002). Indeed, it has been shown that N390S mutation affects virulence in mice (Sánchez and Ruiz 1996, Leitmeyer et al. 1999). Interestingly, most DENV-2 strains from Saudi Arabia, including DENV-2-Jeddah-2014 isolate, possessed both substitutions (I129V and N390S), as shown in Table 2. The combined outcome of these two aa substitutions on viral fitness and virulence is not clear and certainly requires further investigation.
Conclusions
This study demonstrates that DENV-2-Jeddah-2014 isolate belongs to the Indian subcontinent lineage of the Cosmopolitan DENV-2 genotype, which predominantly circulates causing disease in the Kingdom. Phylogenetic analysis of the E gene for DENV-2-Jeddah-2014 isolate indicates that it is slightly divergent from the previously reported E sequences from Jeddah (1994–2004). Our analysis also provides molecular evidence of multiple introductions of DENV-2 in Saudi Arabia in the periods 1992–1994, 1994–2004, and 2014. Although analysis of the few reported sequences from Saudi Arabia represents a limitation in this study, our analysis clearly highlights the importance of the continuous monitoring through larger molecular epidemiology surveillance studies to help undertake effective control and management strategies in the Kingdom. Screening of the pilgrims coming to perform Hajj and Umrah might help preventing the introduction of new DENV strains, which is expected to increase the burden of the disease.
Acknowledgment
This work was supported by grants funded from the Deanship of Scientific Research (DSR), King Abdulaziz University (Jeddah, Saudi Arabia) under grant nos. RG/34/2 and 543/141/1432. The authors therefore acknowledge with thanks DSR technical and financial support.
Author Disclosure Statement
No competing financial interests exist.
References
- Ahmed MM. Clinical profile of dengue fever infection in King Abdul Aziz University Hospital Saudi Arabia. J Infect Dev Ctries 2010; 4:503–510 [DOI] [PubMed] [Google Scholar]
- Alikhan M, Al Ghamdi K, Mahyoub JA. Aedes mosquito species in western Saudi Arabia. J Insect Sci 2014; 14:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, et al. . Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anoop M, Issac A, Mathew T, Philip S, et al. . Genetic characterization of dengue virus serotypes causing concurrent infection in an outbreak in Ernakulam, Kerala, South India. Indian J Exp Biol 2010; 48:849–857 [PubMed] [Google Scholar]
- Ayyub M, Khazindar AM, Lubbad EH, Barlas S, et al. . Characteristics of dengue fever in a large public hospital, Jeddah, Saudi Arabia. J Ayub Med Coll Abbottabad 2006; 18:9–13 [PubMed] [Google Scholar]
- Azhar EI, Hashem AM, El-Kafrawy SA, Abol-Ela S, et al. . Complete genome sequencing and phylogenetic analysis of dengue type 1 virus isolated from Jeddah, Saudi Arabia. Virol J 2015; 12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christenbury JG, Aw PP, Ong SH, Schreiber MJ, et al. . A method for full genome sequencing of all four serotypes of the dengue virus. J Virol Methods 2010; 169:202–206 [DOI] [PubMed] [Google Scholar]
- Drosten C, Göttig S, Schilling S, Asper M, et al. . Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Micobiol 2002; 40:2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Badry AA, El-Beshbishy HA, Al-Ali KH, Al-Hejin AM, et al. . Molecular and seroprevalence of imported dengue virus infection in Al-Madinah, Saudi Arabia. Comp Clin Pathol 2014; 23:861–868 [Google Scholar]
- Fakeeh M, Zaki AM. Virologic and serologic surveillance for dengue fever in Jeddah, Saudi Arabia, 1994–1999. Am J Trop Med Hyg 2001; 65:764–767 [DOI] [PubMed] [Google Scholar]
- Fakeeh M, Zaki AM. Dengue in Jeddah, Saudi Arabia, 1994–2002. Dengue Bull 2003; 27:13–18 [Google Scholar]
- Figueiredo RM, Naveca FG, Oliveira CM, BastosMde S, et al. . Co-infection of dengue virus by serotypes 3 and 4 in patients from Amazonas, Brazil. Rev Inst Med Trop Sao Paulo 2011; 53:321–323 [DOI] [PubMed] [Google Scholar]
- Green S, Rothman A. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr Opin Infect Dis 2006; 19:429–436 [DOI] [PubMed] [Google Scholar]
- Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev 1998; 11:480–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal EA, Putnak JR. The dengue viruses. Clin Microbiol Rev 1990; 3:376–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NA, Azhar EI, El-Fiky S, Madani HH, et al. . Clinical profile and outcome of hospitalized patients during first outbreak of dengue in Makkah, Saudi Arabia. Acta Trop 2008; 105:39–44 [DOI] [PubMed] [Google Scholar]
- Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol 2008; 62:71–92 [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, et al. . Dengue virus structural differences that correlate with pathogenesis. J Virol 1999; 73:4738–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, et al. . Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 1999; 73:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani TA, Abuelzein ETME, Al-Bar HMS, Azhar EI, et al. . Outbreak of viral hemorrhagic fever caused by dengue virus type 3 in Al-Mukalla, Yemen. BMC Infect Dis 2013; 13:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medlock JM, Hansford KM, Versteirt V, Cull B, et al. . An entomological review of invasive mosquitoes in Europe. Bull Entomol Res 2015; 25:1–27 [DOI] [PubMed] [Google Scholar]
- Nunes MRT, Palacios G, Faria NR, Sousa EC Jr., et al. . Air travel is associated with intracontinental spread of dengue virus serotypes 1–3 in Brazil. PLoS Negl Trop Dis 2014; 8:e2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pem-Novosel I, Vilibic-Cavlek T, Gjenero-Margan I, Kaic B, et al. . Dengue virus infection in Croatia: Seroprevalence and entomological study. New Microbiol 2015; 38:97–100 [PubMed] [Google Scholar]
- Rico-Hesse R. Dengue virus virulence and transmission determinants. Curr Top Microbiol Immunol 2010; 338:45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehrig JT, Risi PA, Brubaker JR, Hunt AR, et al. . T-helper cell epitopes on the E-glycoprotein of dengue 2 Jamaica virus. Virology 1994; 198:31–38 [DOI] [PubMed] [Google Scholar]
- Sánchez IJ, Ruiz BH. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J Gen Virol 1996; 77:2541–2545 [DOI] [PubMed] [Google Scholar]
- San Martín JL, Brathwaite O, Zambrano B, Solórzano JO, et al. . The epidemiology of dengue in the Americas over the last three decades: A worrisome reality. Am J Trop Med Hyg 2010; 82:128–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, et al. . MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, et al. . Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 2002; 298:63–72 [DOI] [PubMed] [Google Scholar]
- Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol 2003; 20:122–129 [DOI] [PubMed] [Google Scholar]
- Wang E, Ni H, Xu R, Barrett AD, et al. . Evolutionary relationships of endemic/epidemic and sylvatic dengue viruses. J Virol 2000; 74:3227–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Vasilakis N. Molecular evolution of dengue viruses: Contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol 2009; 9:523–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization.Dengue and severe dengue. Fact sheet no. 117, 2012. Available at www.who.int/mediacentre/factsheets/fs117/en/
- Zaki A, Perera D, Jahan SS, Cardosa MJ. Phylogeny of dengue viruses circulating in Jeddah, Saudi Arabia: 1994 to 2006. Trop Med Int Health 2008; 13:584–592 [DOI] [PubMed] [Google Scholar]