Abstract
The monofunctional platinum anticancer agent phenanthriplatin generates covalent adducts with the purine bases guanine and adenine. Preferential nucleotide binding was investigated using a polymerase stop assay and linear DNA amplification with a 163-base pair DNA double helix. Like cisplatin, phenanthriplatin forms the majority of adducts at guanosine residues, but significant differences in both the number and position of platination sites emerge when comparing results for the two complexes. Notably, the monofunctional complex generates a greater number of polymerasehalting lesions at adenosine residues than does cisplatin. Studies with 9-methyladenine reveal that, under abiological conditions, phenanthriplatin binds to the N1 or N7 position of 9-methyladenine in approximately equimolar amounts. By contrast, comparable reactions with 9-methylguanine afforded only the N7-bound species. Both of the 9-methyladenine linkage isomers (N1 and N7) exist as two diastereomeric species arising from hindered rotation of the aromatic ligands about their respective platinum–nitrogen bonds. Eyring analysis of rate constants extracted from variable temperature NMR spectroscopic data revealed that the activation energies for ligand rotation in the N1 bound platinum complex and the N7 linkage isomers are comparable. Finally, a kinetic analysis indicated that phenanthriplatin reacts more rapidly by a factor of eight with 9-methylguanine than with 9-methyladenine, suggesting that the distribution of lesions formed on double-stranded DNA is kinetically controlled. In addition, implications for the potent anticancer activity of phenanthriplatin are discussed herein.
Keywords: platinum, anticancer, monofunctional, rotamer, nucleotide binding
TOC image
Purine Preference: Phenanthriplatin generates biologically relevant adducts at both adenosine and guanosine nucleotides. In contrast to reactions with 9-alkylguanine, phenanthriplatin generates a mixture of isomeric and diastereomeric species upon reaction with 9-methyladenine. Characterisation of the different adducts facilitates kinetic analysis into the relative rates of adduct formation.
Introduction
Following the great clinical success of the platinum anticancer drug cisplatin and its analogs, the field of metal-based anticancer drug discovery has turned toward non-classical drug candidates.[1–3] A wide variety of metal complexes differing from cisplatin both in metal identity[4–10] and metal center coordination environment[11–13] have shown promising anticancer activity and, in some cases, different mechanisms of action. Cisplatin and its classical derivatives kill cancer cells by binding to DNA, predominantly at the N7 position of guanine residues.[14–15] A number of different Pt–DNA adducts can form, but the most prevalent is the cis-1,2-[Pt(NH3)2d(GpG)] intrastrand cross-link.[16] The distortion of the DNA double helix induced by this cross-link[15] produces a number of downstream biological effects, the most notable of which is transcription inhibition.[17–18] Transcription inhibition can trigger the repair machinery of the cell and/or signal apoptotic cell death.
Monofunctional platinum complexes have emerged as a promising class of non-classical platinum drug candidates.[19–23] Unlike traditional bifunctional platinum agents, monofunctional complexes generate only a single covalent bond with nuclear DNA, and therefore do not typically bend or unwind the structure of the Watson-Crick duplex.[19,24] It is well established that the DNA distortion induced by bifunctional adducts plays an important role in their cellular processing. The failure of the monofunctional adducts to generate such distortions[19] may indicate an alternative mode of action for these complexes.
Detailed crystallographic and biochemical studies of monofunctional complexes of the form cis-[Pt(NH3)2(Am)Cl]+,[20] where Am is an N-heterocyclic ligand, indicate that, like cisplatin (Chart 1), these complexes initiate apoptosis in cultured mammalian cells as a result of transcription inhibition. The ability of monofunctional platinum adducts to inhibit the procession of RNA polymerase II (Pol II)[25] has been linked to both the stereochemistry[26] of the platinum complex and the steric bulk of the N-heterocyclic amine ligand.[20] These fundamental studies guided the design of phenanthriplatin (Chart 1), which displays significantly improved anticancer activity over its parent compound, pyriplatin (Chart 1).[19] Phenanthriplatin was previously shown to be a highly efficacious anti-cancer agent with a spectrum of activity complementary to that of cisplatin.[20]
Chart 1.
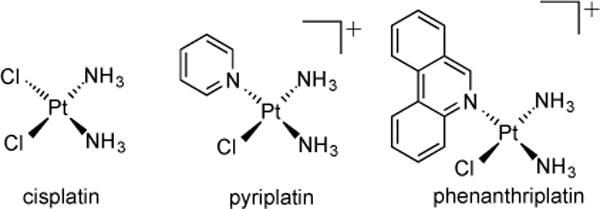
Structures of the classical platinum anticancer drug, cisplatin, and the non-classical platinum anticancer agents, pyriplatin and phenanthriplatin.
One common assumption of many mechanistic studies with monofunctional platinum complexes is that they, like bifunctional agents, predominantly form adducts by binding to the N7 position of guanosine, the most nucleophilic site on the DNA double helix. It is possible, however, that monofunctional agents might display a different nucleotide binding preference as compared to bifunctional agents.[27] In particular, binding to adenine, which bears the most potently nucleophilic sites after guanine, could afford unique biological consequences.[28] Such Pt-DNA adducts may avoid recognition by DNA repair enzymes or alter downstream biological pathways.[29–30] Furthermore, platinum adducts with guanine (Pt-NGua) might have different mutagenicity as compared with adenine (Pt-NAde) adducts.[31]
Several different strategies have been employed to rationally design platinum anticancer complexes that promote binding at adenosine over guanosine. Collins, Wheate, and coworkers have reported a series of multinuclear platinum complexes that pre-associate with the minor groove of DNA.[32–33] Binding to the N7-position of adenine is sterically favoured in the minor groove, whereas binding at the N7-position of guanine is preferred in the major groove of DNA. An alternative approach pioneered by Bierbach and coworkers employs a platinum complex with an appended 9-aminoacridine unit (Pt-ACRAMTU) that intercalates preferentially at GT/TC, TA/TA, and CG/CG base pair steps.[27] The unusually high number of adenine residues (~20%) platinated with this complex has been attributed to pre-association at the minor groove by intercalation.
We were interested to find that phenanthriplatin, which was not designed to have any preferential binding to adenine, forms a proportionally larger number of adducts with adenine than does cisplatin. Here we identify the nucleotide-binding specificity of phenanthriplatin in oligomeric double-stranded DNA, and compare the binding pattern to that of cisplatin. We show that, in addition to formation of monofunctional adducts at guanine, a significant proportion of adenine adducts are generated. The nature of these adenine adducts was elucidated by preparing small molecule models. Detailed characterization of these model compounds allowed an analysis of the kinetics of nucleotide binding under physiological conditions. The rates of purine platination mirror the distribution of adducts formed on double stranded DNA, suggesting that the platination pattern is kinetically controlled.
Results and Discussion
Gel Electrophoresis
Initial studies designed to probe the base binding preference of phenanthriplatin employed a 163 base pair fragment of DNA platinated at different platinum-to-nucleotide ratios (rf = 0.025, 0.05, 0.10, 0.15). Analysis of the platinated duplex using the polymerase stop assay[34–35] and slab gel electrophoresis (Figure 1A) revealed significant differences in both the number of adducts formed by, and the preferred binding sites of, cisplatin and phenanthriplatin.
Figure 1.
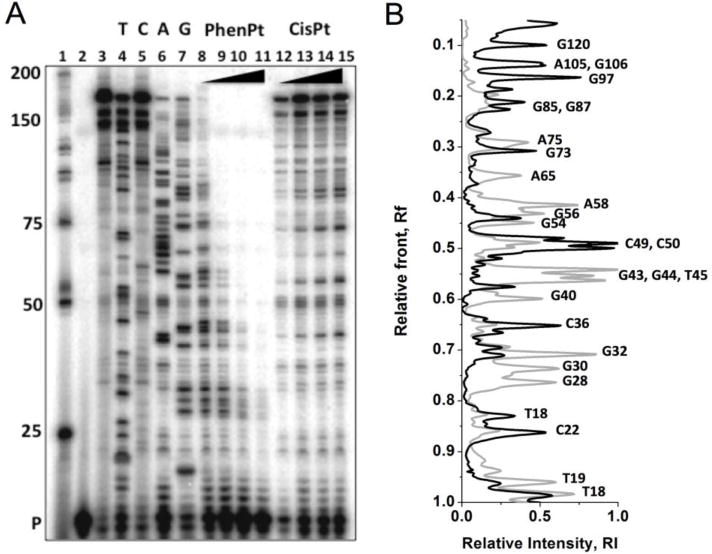
Autoradiograph of linear amplification experiment performed with a 163 bp sequence of DNA and 5′-GTAAG CGGCA GGGTC primer. Lane 1) Low molecular weight ladder; 2) unextended primer; 3) replication control; 4–7) dideoxy sequencing reactions; lanes labeled to correspond to platinated bases, T, C, A, G; 8–11) phenanthriplatin rf = 0.025, 0.05, 0.1, 0.15; 12–15) cisplatin rf = 0.025, 0.05, 0.1, 0.15. B) Overlaid densitometry traces for phenanthriplatin (grey) and cisplatin (black) highlighting differences in the position of the platination sites of the two platinum agents.
Increasing the platinum-to-nucleotide ratio from 0.025 to 0.15 decreased the efficiency of primer extension for both cisplatin and phenanthriplatin, with phenanthriplatin inhibiting replication to a greater extent. Analysis of the platination sites on the complimentary strand was limited because of the extended tract of consecutive guanines within the first 40 bases. This sequence strongly inhibited replication with both phenanthriplatin and cisplatin (Figure S1, Supporting Information). Comparison of the phenanthriplatin and cisplatin densitometry traces at rf values of 0.025 (Figure 1B), where platination sites could be determined along the full length of the DNA, indicates that both the number and positions of platination sites for the two complexes differ significantly.
The nucleotide binding preference for cisplatin has been studied extensively, with the 1,2-GG intrastrand cross-link having been identified as the most common lesion (47–65%) followed by the 1,2-AG cross-link (21–28%).[36] Theoretical studies have demonstrated that the preference for cisplatin to bind at guanine is both thermodynamically and kinetically controlled,[37] with regions containing multiple guanine residues being particularly favoured. In contrast, the nucleotide base preference of monofunctional platinum agents has yet to receive much attention. Conceptually, the reduced binding restrictions of monofunctional agents should give rise to more adducts than are present for classical bifunctional agents requiring two adjacent nucleotides for cross-linking. Furthermore, it has been proposed that the ability to platinate single base sites not favoured by bifunctional agents should give rise to unique biological consequences.[29]
Analysis of the phenanthriplatin lanes on the autoradiograph revealed 30 distinct binding sites, of which 19 are attributed to guanine binding and 8 to adenine. By contrast, 16 binding sites are attributed to cisplatin binding, 14 of which were determined to be guanine binding sites and only one an adenine site. The higher incidence of adenine binding when phenanthriplatin is employed is noteworthy, especially when considering the intensity of the stop sites at the 58A and 65A, bases that are directly adjacent to more nucleophilic guanine positions.
Structural characterisation of 9-methyladenine adducts of phenanthriplatin
Having observed that a higher percentage of adenine lesions are generated with phenanthriplatin than cisplatin, we were motivated to explore the formation of phenanthriplatin-adenine adducts in greater detail. Preliminary experiments employing 9-methyladenine as an analogue of adenosine revealed a mixture of species to form following incubation with phenanthriplatin.
Analysis of the products isolated from the reaction of 9-methyladenine and the triflate salt of phenanthriplatin by 1H NMR spectroscopy revealed the presence of four distinct species in solution at a ratio of 1.0:0.8:0.8:0.6 (Figure S2, Supporting Information). In order to identify these species, we employed preparative HPLC and were able to separate the two unique products observed by analytical HPLC in a preliminary experiment (Figure S3, Supporting Information). 1H NMR spectroscopic analysis of the isolated fractions confirmed that each of the two species identified in the HPLC generated two spectroscopic signatures on the NMR timescale (Figures S4–S5, Supporting Information). Furthermore, both fractions eluted by HPLC gave rise to the same fragmentation pattern and m/z values when analysed by ESI-MS (Figure S6, Supporting Information). The peak observed at 556.4+ m/z corresponds to the desired 9-methyladenine adduct of phenanthriplatin and is present in both fractions isolated by HPLC, which we assign as isomers.
Unlike 9-methylguanine, for which only the N7 platinum adduct is commonly reported,[38–43] both the N7 and N1 positions of adenine[44–48] frequently form adducts with platinum. We therefore inferred that the two species observed by HPLC correspond to N1 and N7 linkage isomers of the platinated nucleobase. Moreover, both of these linkage isomers can potentially exist as a mixture of rotational diastereoisomers, or rotamers, which arise from hindered rotation of the phenanthridine and adenine ligands about their Pt–N bonds in the square-planar platinum coordination sphere.[49] Neither coordinated phenanthridine nor 9-methyladenine ligands are symmetric about the platinum coordination plane, and both can be described as either M or P using the conventions of axial chirality.[50] In Chart 2 we depict both the linkage isomers and diastereomers. The adenine ring system is drawn as its M conformer, and the phenanthridine ring system as either M or P, where interconversion of M and P is achieved through rotation of the phenanthridine ring by 180°. Each of the structures enumerated in Chart 2 consequently has an enantiomeric pair (not shown) in which the 9-methyladenine ring is in the P conformation. Enantiomeric pairs are indistinguishable by NMR spectroscopy and are therefore not addressed explicitly in the following discussion.
Chart 2.
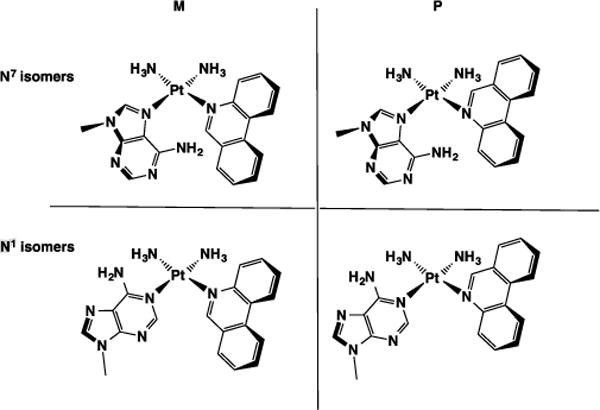
Linkage (N1 and N7) and diastereomeric isomers (M and P) of cis-[(Pt(NH3)2(9-methyladenine)(phenanthridine)]2+. Corresponding enantiomeric pairs are omitted for clarity
Previous small molecule studies indicated that cis-[Pt(NH3)2(phenanthridine)(9-alkylguanine)]2+ exists as a single diastereoisomer, with the selectivity being ascribed to a stabilizing hydrogen bond interaction between the guanine carbonyl oxygen atom and the N-H of the cis-coordinated ammine.[50–51] By contrast, variable temperature 1H NMR spectroscopic analysis of both HPLC fractions isolated following the reaction of phenanthriplatin with 9-methyladenine confirmed facile interconversion between the proposed diastereoisomers.
Further support for formation of a mixture of linkage isomers and diastereoisomers was obtained through a single-crystal X-ray diffraction study. X-ray quality crystals of the N1-Pt adduct of cis-[Pt(NH3)2(phenanthridine)(9-methyladenine)](OTf)2 were obtained by slow diffusion of diethyl ether into a DMF solution of the reaction mixture generated from the reaction of phenanthriplatin with 9-methyladenine (Figure 2).
Figure 2.
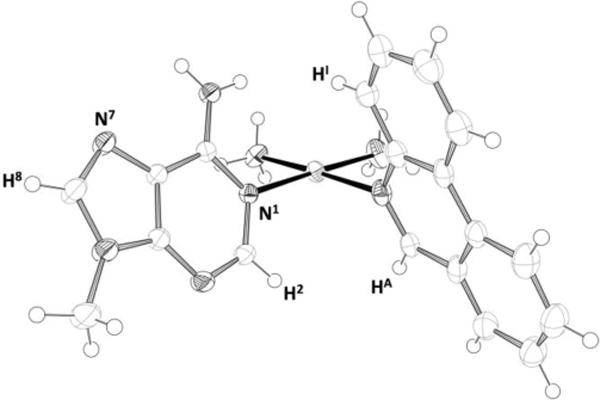
Molecular diagram of the N1, M complex based on the single-crystal X-ray structure, thermal ellipsoids drawn at the 50% probability level. Anions and solvent omitted for clarity. Colour code: N black, C light grey, H open circles, Pt mid grey. Key atom designations are given to aid with the following NMR discussion.
The N1, M rotameric isomer, (Chart 2), was the sole species that crystallized from the crude reaction mixture. In common with the previously reported cis-[Pt(NH3)2(phenanthridine)(9-ethylguanine)]2+ structure, the N-heterocyclic ligands are not strictly perpendicular to the platinum coordination plane. Instead, the phenanthridine ring is canted toward the adenine ring forming dihedral angles of 75.2(8)° and 65.4(9)°, respectively, with respect to the platinum coordination plane. The distance between the ammine and exocyclic amino group is 3.296(5) Å, a value comparable to the 3.19(1) Å distance previously measured for the carbonyl oxygen atom and the ammine ligand in the N7 bound 9-ethylguanine-phenanthriplatin crystal structure.[51]
Crystallisation of the N7 adduct was achieved by diffusion of diethyl ether into a DMF solution of the HPLC-purified N7 isomeric product. The structure confirmed that the platinum centre is coordinated at the N7 position of 9-methyladenine and that exchange of triflate for trifluoroacetate counterions had occurred during chromatographic purification. The structure is disordered with the phenanthridine ring assuming two different orientations with respect to the 9-methyladenine ligand. The latter is also disordered but maintains the same planar chirality about the Pt–NAde bond in the two portions of the disorder (Figure 3). The extensive use of restraints and the low quality of the diffraction data prohibit extensive discussion of the precise bond lengths and angles. It is possible, nonetheless, to make certain generalizations about the complex.
Figure 3.
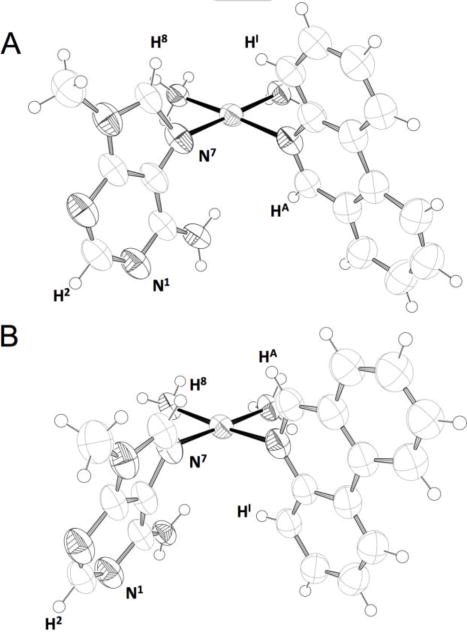
Molecular diagrams of the of the N7, M (A) and N7, P (B) complexes based on the single-crystal X-ray structure. Structural interconversion between the two diastereoisomers occurs via a 180° rotation of the phenanthridine ring, while maintaining the orientation of the 9-methyladenine ring. Thermal ellipsoids are drawn at the 50% probability level, anions and solvent is omitted for clarity. Colour code: N black, C light grey, H open circles, Pt mid grey. Key atom designations are given to aid the accompanying NMR spectroscopic discussion.
The disorder of the phenanthridine ring involves approximately 180° rotation about the Pt-Nphen bond. The planar chirality[50] that exists about this bond arise from asymmetry of the phenanthridine ligand with respect to the platinum coordination plane and is inverted by the rotation. Disorder of the adenine ligand is not a consequence of rotation; both diastereomeric forms of this adduct are present within the crystal lattice. The phenanthridine disorder is analogous to that previously reported for the phenanthriplatin-N7Gua adduct.[51] The adenine and phenanthridine ligands are both approximately perpendicular to the platinum coordination plane, forming dihedral angels of 89.6(7)° and 88.4(9)°, respectively. These values differ significantly from those previously reported for the 9-ethylguanine adduct of phenanthriplatin bonded through the N7 position and might reflect the decreased hydrogen bond affinity of the exocyclic amino group of the adenine ring for the neighbouring ammine ligand, as compared with the carbonyl oxygen atom of the guanine ring system. A dihedral angle of 67(1)° for the 9-ethylguanine ring in [Pt(NH3)2(phenanthridine)(9-ethylguanine)]2+ was previously reported, and the phenanthridine ring is perpendicular to the platinum coordination plane in this structure.
Complete assignment of the 1H NMR spectra of both the linkage isomers and their corresponding diastereoisomers was possible (Figures S7–S13, Supporting Information) through careful consideration of 2D NMR data of the isolated HPLC fractions. Through-space NOE correlations between protons on the cis coordinated phenanthridine and 9-methyladenine residues proved especially insightful in distinguishing diastereoisomers.
Complete assignment of fraction 1, the first to elute by HPLC (retention time, tret = 29.6 min), by 1H NMR spectroscopy was made possible through the observation of a single through-space interaction between the H8 proton (see Figure S4, Supporting Information for atom labels) of 9-methyladenine and proton HI of the phenanthridine ring. The observation that the H8 proton interacts with the adjacent phenanthridine proton confirms the Pt-N7 connectivity of this isomer. For an N1-Pt adduct the H8 proton would be directed outwards from the platinum center, as shown in Figure 2, prohibiting a through-space interaction with the phenanthridine ring. The distance measured between H8 and HI in the crystal structure of the N1-Pt adduct is 7.05 Å, which is greater than the 5 Å limit typically observed for NOE interactions. Although an H8⋯HI correlation was observed for the major diastereoisomer (N7,M), no comparable cross-peaks were observed for the minor diastereoisomer (N1,P) (Figure S10, Supporting Information).[52] The inability to observe this interaction, or any other structurally informative cross-peaks, for the minor diastereoisomer most likely arises from the reduced concentration of this species in solution relative to the major diastereoisomer, coupled with the dynamic interconversion of the N7, M and P diastereoisomers.
Analysis of the second fraction collected by HPLC (tret = 30.9 min) supported assignment of this species as a mixture of N1-Pt diastereoisomers (Figures S14–S19, Supporting Information). Characteristic NOESY cross-peaks were observed for both the M and P diastereoisomers of the N1-Pt mixture (Figure 4). The N1,M species was identified by the cross-peak between the H2-Ade resonance and the HA resonance of the corresponding phenanthridine, while the N1,P species was characterized by the corresponding H2⋯HI cross-peak.
Figure 4.
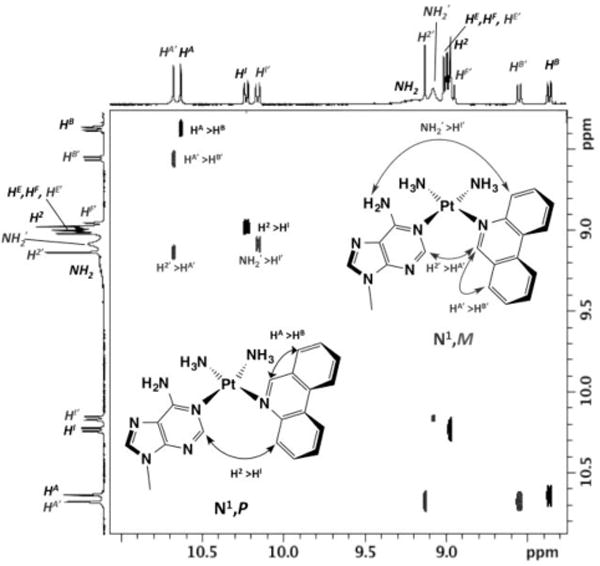
Partial 1H NOESY spectrum (400 MHz, 298 K, [D7] DMF) showing informative NOE cross-peaks allowing assignment of the 1H NMR resonances corresponding to the N1 diastereomeric complexes. Resonances in the 1H projections and NOE cross-peaks are designated H and H′ according to the two complexes N1,M (H′) and N1,P (H).
A detailed investigation of the rate and energetics of diastereomer interconversion was also undertaken. 1H NMR spectral lineshapes across a range of temperatures were simulated, giving access to temperature-dependent rate constants (Figures S20–S24, Supporting Information), which were subjected to Eyring analysis. Linear plots of ln(k/T) versus 1/T (Figures S22 and S25, Supporting Information) facilitated calculation of the enthalpy (ΔH‡) and entropy (ΔS‡) of activation, as well as subsequent calculation of the free energy of activation (ΔG‡) for rotation about the platinum-Nphen bond. The results are summarized in Table 1.
Table 1.
Summary of enthalpy, entropy and free energy of activation values obtained from Eyring analysis of VT-NMR data.
| ΔH‡ (kJ mol−1) | ΔS‡ (J mol−1 K−1) | ΔG‡ 298(kJ mol−1) | |
|---|---|---|---|
| N7 isomer | 57.5 ± 2.4 | −32.8 ± 7.5 | 67.3 ± 2.4 |
| N1 isomer | 44.8 ± 1.8 | −82.4 ± 5.4 | 69.4 ± 1.8 |
The activation energies determined for interconversion of one rotamer to the other are very similar for the N7 and N1 isomers. Furthermore, both values, 67.3 ± 2.4 kJ mol−1 and 69.4 ± 1.8 kJ mol−1, lie within the range that has been reported for platinum complexes containing two N-heterocyclic ligands.[53–57]
Kinetic Analysis of Phenanthriplatin Adduct Formation
Finally, the relative rates at which 9-methyladenine and 9-methylguanine react with phenanthriplatin were investigated. By analogy to the known substitution reaction mechanism of cisplatin,[58] the initial step for reaction of phenanthriplatin with purine bases was proposed to be aquation, as illustrated in equation 1.
Equation 1: Speciation during the reaction of phenanthriplatin with 9-alkylpurine bases. Phen = phenanthridine; N7Gua = N7 bonded 9-methylguanine adduct; N7Ade = N7 bonded 9-methyladenine adduct; N1Ade = N1 bonded 9-methyladenine adduct.
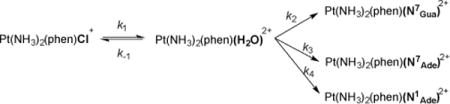 |
Reactions were performed in triplicate at 37 °C in pH 7.4 buffered solution containing 24 mM HEPES and 10 mM NaCl. Aliquots of the reaction mixture that contained 1 mM total platinum concentration were removed at various time points and analysed by HPLC (Figures S26–S29, Supporting Information).
In an initial control reaction containing no nucleophile, the rate and position of the equilibrium (Keq = k1/k−1) between phenanthriplatin and its aquated form were established. After 4 h at 37 °C the ratio of phenanthriplatin to aquated phenanthriplatin stabilized and Keq was determined to be 0.43.
For the reaction containing excess 9-methylguanine and 9-methyladenine, the rate of the forward and reverse reactions in the aquation step were almost twice as large, promoted by consumption of aquated phenanthriplatin. The equilibrium position, Keq = 0.41, however, was comparable to that in the control reaction (Figure S28, Supporting Information).
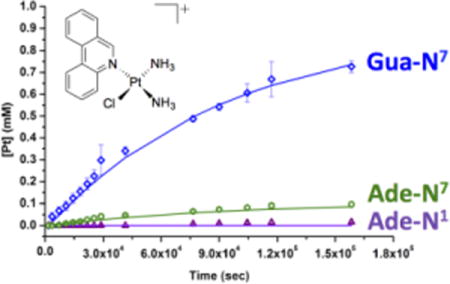
The relative rates of formation of the 9-methylguanine and -adenine adducts were next determined. In the case of 9-methyladenine, formation of both the N7 and N1 isomers was observed by HPLC. Formation of the N7 bound 9-methylguanine adduct proceeded fastest, at a rate of 0.36 ± 0.02 × 10−4 s−1 (Figure 5). By contrast, the 9-methyladenine adducts formed significantly more slowly, with the N7 bound adduct having a formation rate constant of 0.042 ± 0.005 × 10−4 s−1, an 8.6 fold decrease compared to that of 9-methylguanine. Under the experimental conditions, the N1 adduct was not generated to any considerable extent. There was no measurable quantity detected up to 8 h, and less than 2% of this product was generated over the course of the reaction. The composition of the reaction mixture after 44 h was determined to be 13% phenanthriplatin, 5% aquated phenanthriplatin, 73% phenanthriplatin-N7Gua, 8% phenanthriplatin-N7Ade, and <2% phenanthriplatin-N1Ade.
Figure 5.
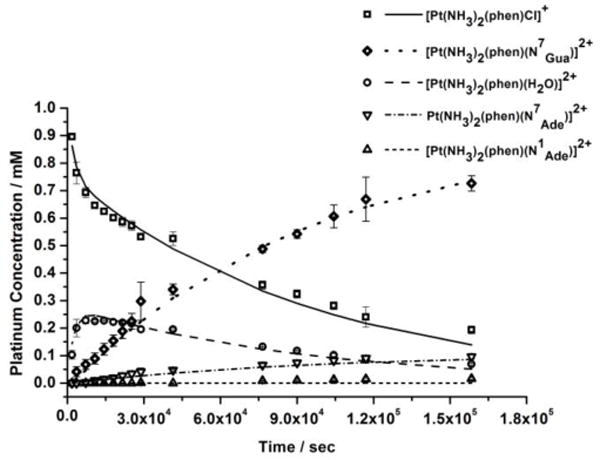
Kinetic data for the reaction of phenanthriplatin with nucleobases 9-methyladenine and 9-methylguanine. Consumption of [Pt(NH3)2(phen)Cl]+ is shown with squares and the aquated intermediate [Pt(NH3)2(phen)(H2O)]2+ with circles. Adduct formation with 9-methylguanine (N7), 9-methyladenine (N7) and 9-methyladenine is depicted with diamonds and triangles.
The systematic decrease in experimentally determined reaction rates for phenanthriplatin with N7-9-methylguanine > N7-9-methyladenine > N1-9-methyladenine nicely reflects the relative nucleophilicity of these species. Furthermore, the final population of the reaction mixture correlates well with the proportion of adenine and guanine adducts observed in the gel electrophoresis experiments.
Conclusions
The number and position of phenanthriplatin binding sites differ notably from those of cisplatin when visualised using linear amplification and gel electrophoresis methodologies. The importance of phenanthriplatin adducts formed with adenine is noteworthy. Complexes able to generate platinum adducts that differ from the major 1,2-d(GpG) intrastrand cross-link formed by cisplatin have the potential to elude DNA repair through failure of repair enzymes to recognize the lesion,[29–30] and they may display altered mutagenicity.[31] Experiments exploring the mixture of species generated upon reaction of 9-methyladenine and phenanthridine confirm the identity of isomeric and diastereomeric products. Four discrete 9-methyladenine-phenanthriplatin adducts were characterized by solution NMR spectroscopy. Two of the adducts can be generated from the other two by rotation of the phenanthridine ring about the Pt–N bond. The free energy of activation for this rotation was calculated by using VT NMR data. Finally the rate of formation of 9-methyadenine (N1 and N7 bonded) and 9-methylguanine adducts was determined under biologically relevant conditions. The rate of adduct formation for 9-methylguanine is more than 8-fold greater than adduct formation with 9-methyladenine, and no significant proportion of 9-methyl adenine bound in the N1 position formed over the course of the reaction. Together these results indicate that, whereas formation of guanine adducts is kinetically favoured over adenine, a significant proportion of phenanthriplatin is expected to react at the N7 position of adenine on DNA in cells.
Experimental Section
General
Oligonucleotides were obtained from Sigma Aldrich and Integrated DNA Technologies (IDT). T4 PNK and Taq DNA polymerase were supplied from New England Biolabs. Individual dideoxy nucleotides and deoxynucleotide mix were supplied by Affymatix. [γ32P]-ATP was supplied by Perkin Elmer. 9-Methyladenine was supplied by Santa Cruz Biotech and silver triflate by Strem Chemicals, Inc. Triflate and nitrate salts of [Pt(NH3)2(phenanthridine)Cl)]+ were synthesized as described previously.[20,51] DNA sequencing gels were dried using a Labconco gel drier and imaged with a Storm 840 Phophorimager system from Amersham Biosciences. Data analysis was performed using the Quantity One Software (Bio-Rad). NMR spectra were acquired on a Bruker AVANCE-400 NMR spectrometer with a Spectro Spin superconducting magnet in the Massachusetts Institute of Technology, Department of Chemistry Instrument Facility (MIT DCIF). Chemical shifts are referenced to solvent signals (1H and 13C) or an external solution of K2PtCl4 in D2O (195Pt δ = −1628 ppm). Deuterated solvents were purchased from Cambridge Isotope Laboratory (Andover, MA). Electrospray ionization mass spectrometry (ESI-MS) spectra were obtained on an Agilent Technologies 1100 Series liquid chromatography-mass spectrometer, high resolution spectra (HRMS) were obtained on a Bruker Daltonics APEXIV 4.7 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer (MIT DCIF). HPLC purifications were carried out on an Agilent Technologies 1200 Series HPLC system fitted with a Zorbax-C18 column (21.2 × 250 mm).
Platination of 163-bp Oligonucleotide
Amplification of pBR322 in DH5α cells followed by sequential digestion with Hpa II then Hae III yielded a 163 bp oligonucleotide as described previously.[59] Purified 163-bp DNA (2 pmol, 40 nM) was incubated with either phenanthriplatin or cisplatin (rf (platinum-to-nucleotide) = 0.025, 0.05, 0.1, 0.15) in 1 mM sodium phosphate, pH 7.4, 3 mM NaCl at 37 °C for 20 h. Following reaction, ethanol precipitation of the sample was performed by addition of NaOAc (22 μL, 3 M) and 170 μL ice cold ethanol, and incubation at −80 °C for 2 h. Following precipitation samples were centrifuged at 4 °C, 13000 rpm, for 30 min, and then the supernatant was decanted and washed twice with ice-cold ethanol. After isolation, samples were stored at −20 °C.
Linear Amplification
One pmol of primer was 5´-labeled in a final volume of 20 μL with 70 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 5 mM dithiothreitol, 10 units of polynucleotide kinase, and 10 μCi of [γ-32P]ATP at 37 °C for 30 min. A 1 μL aliquot of this reaction mixture (0.05 pmol primer) was added to 0.2 pmol of platinated ds-DNA and 1 unit of Taq DNA Polymerase in 400 μM dNTP mix, 10 mM Tris-HCl, pH 8.3, 1.5 mM MgCl2, 50 mM KCl, and 5 mM dithiothreitol, in a final volume of 10 μL. Dideoxy sequencing reactions were performed in parallel using the same primers and DNA strand in the absence of platinum. The dNTP mix concentration was reduced to 2.5 μM with one of ddT (1 mM), ddA (500 μM), ddC (500 μM), or ddG (25 μM) added as the chain terminator. Linear PCR was performed following the cycle 95 °C (2 min), [95 °C (1 min), 50 °C(Primer 1)/57°C Primer 2 (2 min), and 72 °C (2 min) × 10] then 95 °C for a further 10 min before samples were stored on ice. After amplification, samples and dideoxy sequencing reactions (2 μL) were mixed with 2 μL loading dye (95% formamide, 0.5% bromophenol blue, 0.5% xylene cyanol, 10 mM EDTA) and 2 μL diH2O. Samples were then heated at 95 °C for 2 min and cooled on ice before being loaded on a 0.4 mm 8% polyacrylamide/7M urea sequencing gel, which had been pre-electrophoresed (constant 60 W) for 30 min. Samples were then run for 1 h and 5 min (constant 60 W) before being dried on 3 MM paper with a vacuum gel drier (2 h at 70 °C). The dried gel was exposed to a phosphor storage screen overnight before being imaged with a phosphorimager. Densitometry was performed using the quantification tools supplied with the Quantity One software (Bio-Rad). Relative intensities were calculated by integration of the peak area between 10 and 120 bps. The background was determined at the lowest point and subtracted across the entire lane. Relative intensities are reported as a fraction of the total lane intensity.
Synthesis of cis-[Pt(NH3)2(9-methyladenine)(phenanthriidine)]·2OTf/TFA (N1 and N7 isomers)
AgOTf (43.8 mg, 0.17 mmol) in 1 mL DMF was added dropwise to a stirring solution of [Pt(NH3)2(phenanthridine)Cl]OTf (100 mg, 0.17 mmol) dissolved in 6 mL of DMF. The reaction was sealed, protected from light, and stirred at room temperature overnight. After filtration through Celite, 6-amino-9-methylpurine (25.4 mg, 0.17 mmol) was added to the reaction mixture and the solution was stirred at room temperature under protection from light for an additional 7 h. DMF was removed from the sample under vacuum yielding an orange oil, which was taken up in acetone (2 mL) and filtered through Celite yielding a pale yellow solution. Purification of the title compound was obtained through dropwise addition of the acetone solution to a rapidly stirring solution of diethyl ether (50 mL). The off-white solid that precipitated was isolated by filtration, dissolved in acetone (2 mL), and precipitated from ether a second time. The isolated off-white solid, consisting of a mixture of linkage and diastereomeric isomers, was collected by filtration and dried under vacuum (70.1 mg, 47%). 19F NMR (376 MHz, 298 K, [D7]DMF): δ = −78.5 (s, OTf−). The N1 and N7 isomeric species were separated by preparative reverse phase HPLC (details in Supporting Information), facilitating assignment of two distinct species by 1H NMR spectroscopy, defined as fractions 1 and 2 based on the order of elution.
Fraction 1 (29.6 min): 1H NMR (400 MHz, 298 K, [D7]DMF): N7,M: δ = 10.39 (s, 1H, HA), 10.17 (d, 1H J = 8.3 Hz, HI), 9.25 (s, 1H, H8), 8.97 (m, 2H, HE and HF overlap with N7, P), 8.55 (s, 2H, NH2), 8.36 (d, 1H, J = 8.3 Hz, HB overlap with N7, P), 8.29 (s, 1H, H2), 8.20 (m, 1H, HC overlap with N7,P), 8.13 (t, 1H, J = 7.4 Hz, HH), 8.00 (m, 2H, HD and HG overlap with N7,P), 5.50 (br. s, 6H, 2 × NH3 overlap with N7,P), 3.80 (s, 3H, CH3); N7,P : 10.57 (s, 1H, HA), 9.97 (d, 1H, J = 8.2 Hz, HI), 9.51 (s, 1H, H8), 8.97 (m, 2H, HE and HF overlap with N7,M), 8.51 (d, 1H, J = 8.7 Hz, HB), 8.38 (s, H2, NH2 overlap with N7,M), 8.28 (s, 1H, H2), 8.20 (m, 1H, HD overlap with N7,M), 8.00 (m, 3H, HC, HG and HH overlap with N7,M), 5.50 (br. s, 6H, 2 × NH3 overlap with N7,M), 3.82 (s, 3H, CH3); 13C NMR (126 MHz, 298 K, [D7]DMF): δ = 161.3, 160.9, 159.9, 159.6, 154.6, 154.3, 154.2, 149.8, 145.8, 144.8, 142.8, 142.4, 141.5, 135.0 × 2, 132.8, 131.1, 130.8, 130.7, 130.2, 129.6, 129.5, 129.4, 128.7, 128.5, 127.2, 126.8, 126.2, 124.0, 123.0 × 2, 122.9, 119.0, 116.2, 116.1, 116.0, 29.8 × 2; 19F NMR (376 MHz, 298 K, [D7]DMF): δ = −75.0 (s, TFA−); 195Pt (85.8 MHz, D2O, 298 K): δ = −2443.2 (br. m.); HRMS (ESI+) m/z calc (C19H21N8Pt)+ 556.1488, found 556.1649; (C19H18N7Pt)+ 539.1223, found 539.1449; (C19H15N6Pt)+ 522.0957, found 522.1084. Fraction 2 (30.9 min): 1H NMR (400 MHz, 298 K, [D7]DMF): N1, P: δ = 10.63 (s, 1H, HA), 10.22 (d, 1H, J = 8.2 Hz, HI), 9.20 (br. S, 2H, NH2), 9.00 (d, 1H, J = 8.3 Hz, HE overlap with N1,M), 8.99 (d, 1H, J = 8.4 Hz, HF), 8.97 (s, 1H, H2), 8.36 (d, 1H, J = 8.1 Hz, HB), 8.21 (s, 1H, H8), 8.19 (m, 2H, HH and HD overlap with N1,M), 7.99 (m, 2H, HC and HG overlap with N1,M), 3.72 (s, 3H, CH3); N1,M: 10.67 (s, 1H, HA), 10.16 (d, 1H, J = 8.3 Hz, HI), 9.13 (s, 1H, H2), 9.08 (br. S, 2H, NH2), 9.00 (d, 1H, J = 8.3 Hz, HE overlap with N1,P), 8.96 (d, 1H, J = 7.6 Hz, HF), 8.55 (d, 1H, J = 7.7 Hz, HB), 8.20 (s, 1H, H8), 8.19 (m, 1H, HD overlap with N1,P), 7.99 (m, 3H, HC, HH and HG overlap with N1,P), 3.74 (s, 3H, CH3); 13C NMR (126 MHz, 298K, [D7]DMF) δ = 161.9, 160.9, 156.3, 156.1, 153.8, 152.9, 150.7, 148.2, 148.0, 143.7, 142.5, 134.9 × 2, 132.7, 132.6, 130.7, 130.6, 130.5, 130.2, 129.6, 129.5 × 2, 129.3, 128.6, 127.0, 126.6, 126.5, 126.3, 126.0, 125.9, 125.6, 124.0, 123.7, 122.9, 122.8, 119.7, 30.0 × 2; 19F NMR (376 MHz, 298 K, [D7]DMF): δ = −74.7 (s, TFA−); 195Pt (85.8 MHz, D2O, 298 K): δ = −2598.5 (s), −2516.6 (s); HRMS (ESI+) m/z calc (C19H21N8Pt)+ 556.1488, found 556.1554; (C19H18N7Pt)+ 539.1223, found 539.1316; (C19H15N6Pt)+ 522.0957, found 522.1003.
Line Shape analysis of Variable Temperature 1H NMR data
1H NMR spectra were collected at 5 °C increments for fractions 1 and 2 across the range 25–70 °C. Decomposition of fraction 1 was observed above 65 °C preventing observation of complete peak coalescence. The line shapes of proton HA, which was well resolved and separated from other resonances at room temperature, were simulated and fit to the experimental data by altering the rate constant for ring rotation. These rate constants were then used to construct Eyring plots giving the enthalpy (ΔH‡) and entropy (ΔS‡) of activation for rotation of the phenanthridine ring. These values allowed subsequent calculation of the free energy of activation for this process using the Gibbs free energy equation. Errors were obtained by standard error propagation.
Kinetic Analysis
Reactions containing 1 mM platinum, were incubated in buffered solution (24 mM HEPES buffer, 10 MM NaCl, pH 7.4) at 37 °C. Aliquots were removed periodically across the course of 44 h (1.6 × 105 sec) and analysed by HPLC. Peak areas monitored at 260 nm were calculated by integration and normalized relative to standard peak areas for 1 mM solutions. Competition reactions performed in the presence of 9-alkylpurine bases contained 3 mM 9-methyladenine (3 eq.) and 3 mM 9-methylguanine (3 eq.). Reactions were performed in triplicate. Data was fit to the model shown in equation 1 (Scheme S1, Supporting Information) using Dynafit, and was found to be internally consistent. Holding the platinum concentration at a constant value and fitting only rates gave an equivalent result to that obtained when both the platinum concentration and rates were allowed to vary.
Supplementary Material
Acknowledgments
This work is supported by the NCI under grant CA034992. G.Y.P. received support from a Misrock Fellowship. We thank Dr. M. L. Zastrow and Dr. S. Becker for helpful discussions.
Footnotes
Supporting Information for this article is given via a link at the end of the document. CCDC 1431871 and 1431872 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data%5Frequest/cif Included in the Supporting Information are coloured versions of Figures 2, 3, and 4 depicted here.
References
- 1.Quiroga AG. Curr Top Med Chem. 2011;11:2613–2622. doi: 10.2174/156802611798040723. [DOI] [PubMed] [Google Scholar]
- 2.Johnstone TC, Wilson JJ, Lippard SJ. Inorg Chem. 2013;52:12234–12249. doi: 10.1021/ic400538c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero-Canelon I, Sadler PJ. Inorg Chem. 2013;52:12276–12291. doi: 10.1021/ic400835n. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Sadler PJ. Acc Chem Res. 2014;47:1174–1185. doi: 10.1021/ar400266c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suntharalingam K, Awuah SG, Bruno PM, Johnstone TC, Wang F, Lin W, Zheng YR, Page JE, Hemann MT, Lippard SJ. J Am Chem Soc. 2015;137:2967–2974. doi: 10.1021/ja511978y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suntharalingam K, Johnstone TC, Bruno PM, Lin W, Hemann MT, Lippard SJ. J Am Chem Soc. 2013;135:14060–14063. doi: 10.1021/ja4075375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnstone TC, Suntharalingam K, Lippard SJ. Philos Trans R Soc, A. 2015;373:1–12. doi: 10.1098/rsta.2014.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hearn JM, Romero-Canelon I, Munro AF, Fu Y, Pizarro AM, Garnett MJ, McDermott U, Carragher NO, Sadler PJ. Proc Natl Acad Sci U S A. 2015;112:E3800–E3805. doi: 10.1073/pnas.1500925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubey A, Min JW, Koo HJ, Kim H, Cook TR, Kang SC, Stang PJ, Chi KW. Chem - Eur J. 2013;19:11622–11628. doi: 10.1002/chem.201300870. [DOI] [PubMed] [Google Scholar]
- 10.Munteanu CR, Suntharalingam K. Dalton Trans. 2015;44:13796–13808. doi: 10.1039/c5dt02101d. [DOI] [PubMed] [Google Scholar]
- 11.Cook TR, Vajpayee V, Lee MH, Stang PJ, Chi KW. Acc Chem Res. 2013;46:2464–2474. doi: 10.1021/ar400010v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollis LS, Amundsen AR, Stern EW. J Med Chem. 1989;32:128–136. doi: 10.1021/jm00121a024. [DOI] [PubMed] [Google Scholar]
- 13.Bierbach U, Hambley TW, Farrell N. Inorg Chem. 1998;37:708–716. [Google Scholar]
- 14.Jamieson ER, Lippard SJ. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 15.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 16.Fichtinger-Schepman AMJ, Van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J. Biochemistry. 1985;24:707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 17.Todd RC, Lippard SJ. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorenson CM, Eastman A. Cancer Res. 1988;48:4484–4488. [PubMed] [Google Scholar]
- 19.Lovejoy KS, Todd RC, Zhang S, McCormick MS, D’Aquino JA, Reardon JT, Sancar A, Giacomini KM, Lippard SJ. Proc Natl Acad Sci U S A. 2008;105:8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park GY, Wilson JJ, Song Y, Lippard SJ. Proc Natl Acad Sci U S A. 2012;109:11987–11992. doi: 10.1073/pnas.1207670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaucheron F, Malinge JM, Blacker AJ, Lehn JM, Leng M. Proc Natl Acad Sci U S A. 1991;88:3516–3519. doi: 10.1073/pnas.88.9.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, Wang X, Zhu C, Song Y, Wang J, Li Y, Guo Z. Dalton Trans. 2011;40:10376–10382. doi: 10.1039/c1dt10555h. [DOI] [PubMed] [Google Scholar]
- 23.Cerasino L, Intini FP, Kobe J, de Clercq E, Natile G. Inorg Chim Acta. 2003;344:174–182. [Google Scholar]
- 24.Bauer C, Peleg-Shulman T, Gibson D, Wang AHJ. Eur J Biochem. 1998;256:253–260. doi: 10.1046/j.1432-1327.1998.2560253.x. [DOI] [PubMed] [Google Scholar]
- 25.Kellinger MW, Park GY, Chong J, Lippard SJ, Wang D. J Am Chem Soc. 2013;135:13054–13061. doi: 10.1021/ja405475y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D, Zhu G, Huang X, Lippard SJ. Proc Natl Acad Sci U S A. 2010;107:9584–9589. doi: 10.1073/pnas.1002565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budiman ME, Alexander RW, Bierbach U. Biochemistry. 2004;43:8560–8567. doi: 10.1021/bi049415d. [DOI] [PubMed] [Google Scholar]
- 28.Guddneppanavar R, Saluta G, Kucera GL, Bierbach U. J Med Chem. 2006;49:3204–3214. doi: 10.1021/jm060035v. [DOI] [PubMed] [Google Scholar]
- 29.Budiman ME, Bierbach U, Alexander RW. Biochemistry. 2005;44:11262–11268. doi: 10.1021/bi050745n. [DOI] [PubMed] [Google Scholar]
- 30.Brabec V. Progress in Nucleic Acid Research and Molecular Biology. Vol. 71. Academic Press; 2002. pp. 1–68. [DOI] [PubMed] [Google Scholar]
- 31.Comess KM, Burstyn JN, Essigmann JM, Lippard SJ. Biochemistry. 1992;31:3975–3990. doi: 10.1021/bi00131a013. [DOI] [PubMed] [Google Scholar]
- 32.Wheate NJ, Evison BJ, Herlt AJ, Phillips DR, Collins JG. Dalton Trans. 2003:3486–3492. [Google Scholar]
- 33.Grant Collins J, Wheate NJ. J Inorg Biochem. 2004;98:1578–1584. doi: 10.1016/j.jinorgbio.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 34.Murray V. Nucleic Acids Res. 1989;17:8889. doi: 10.1093/nar/17.21.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto AL, Lippard SJ. Proceedings of the National Academy of Sciences. 1985;82:4616–4619. doi: 10.1073/pnas.82.14.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman SE, Lippard SJ. Chemical Reviews. 1987;87:1153–1181. [Google Scholar]
- 37.Baik MH, Friesner RA, Lippard SJ. J Am Chem Soc. 2003;125:14082–14092. doi: 10.1021/ja036960d. [DOI] [PubMed] [Google Scholar]
- 38.Müller B, Shen W-Z, Sanz Miguel PJ, Albertí FM, van der Wijst T, Noguera M, Rodríguez-Santiago L, Sodupe M, Lippert B. Chem - Eur J. 2011;17:9970–9983. doi: 10.1002/chem.201101148. [DOI] [PubMed] [Google Scholar]
- 39.Frommer G, Mutikainen I, Pesch FJ, Hillgeris EC, Preut H, Lippert B. Inorg Chem. 1992;31:2429–2434. [Google Scholar]
- 40.Longato B, Bandoli G, Trovo G, Marasciulo E, Valle G. Inorg Chem. 1995;34:1745–1750. [Google Scholar]
- 41.Brandi-Blanco P, Sanz Miguel PJ, Müller B, Gil Bardají E, Willermann M, Lippert B. Inorg Chem. 2009;48:5208–5215. doi: 10.1021/ic900176c. [DOI] [PubMed] [Google Scholar]
- 42.Frommer G, Schoellhorn H, Thewalt U, Lippert B. Inorg Chem. 1990;29:1417–1422. [Google Scholar]
- 43.Raudaschl-Sieber G, Schoellhorn H, Thewalt U, Lippert B. J Am Chem Soc. 1985;107:3591–3595. [Google Scholar]
- 44.Ibáñez S, Mihály B, Sanz Miguel PJ, Steinborn D, Pretzer I, Hiller W, Lippert B. Chem - Eur J. 2015;21:5794–5806. doi: 10.1002/chem.201406378. [DOI] [PubMed] [Google Scholar]
- 45.Martin RB. Acc Chem Res. 1985;18:32–38. [Google Scholar]
- 46.Arpalahti J, Lehikoinen P. Inorg Chim Acta. 1989;159:115–120. [Google Scholar]
- 47.Arpalahti J, Klika KD, Sillanpaa R, Kivekas R. Dalton Trans. 1998:1397–1402. [Google Scholar]
- 48.Lüth MS, Freisinger E, Kampf G, Garijo Anorbe M, Griesser R, Operschall BP, Sigel H, Lippert B. J Inorg Biochem. 2015;148:93–104. doi: 10.1016/j.jinorgbio.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Carlone M, Marzilli LG, Natile G. Eur J Inorg Chem. 2005;2005:1264–1273. [Google Scholar]
- 50.Johnstone TC, Lippard SJ. J Am Chem Soc. 2014;136:2126–2134. doi: 10.1021/ja4125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregory MT, Park GY, Johnstone TC, Lee YS, Yang W, Lippard SJ. Proc Natl Acad Sci U S A. 2014;111:9133–9138. doi: 10.1073/pnas.1405739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.The terms major and minor diastereomer refer to the relative proportion of one of the rotameric species of a given linkage isomer, as assessed by 1H NMR spectral integration.
- 53.Davies MS, Diakos CI, Messerle BA, Hambley TW. Inorg Chem. 2001;40:3048–3054. doi: 10.1021/ic001278v. [DOI] [PubMed] [Google Scholar]
- 54.Cramer RE, Dahlstrom PL. J Am Chem Soc. 1979;101:3679–3681. [Google Scholar]
- 55.Li D, Bose RN. J Chem Soc, Dalton Trans. 1994:3717–3721. [Google Scholar]
- 56.Stang PJ, Olenyuk B, Arif AM. Organometallics. 1995;14:5281–5289. [Google Scholar]
- 57.Rochon FD, Beauchamp AL, Bensimon C. Can J Chem. 1996;74:2121–2130. [Google Scholar]
- 58.Davies MS, Berners-Price SJ, Hambley TW. Inorg Chem. 2000;39:5603–5613. doi: 10.1021/ic000847w. [DOI] [PubMed] [Google Scholar]
- 59.Tullius TD, Lippard SJ. Proc Natl Acad Sci U S A. 1982;79:3489–3492. doi: 10.1073/pnas.79.11.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


