Abstract
The study presents proof-of-concept for development of an impedimetric biosensor for ultrasensitive glycoprofiling of PSA. The biosensor exhibits three unique characteristics: 1) analysis of PSA with LOD down to 4 aM; 2) analysis of a glycan part of PSA with LOD down to 4 aM level and 3) both assays (i.e. PSA quantification and PSA glycoprofiling) can be performed on the same interface due to label-free format of analysis.
Introduction
There is a growing incidence and mortality of prostate cancer (PCa), which become the third leading cause of all cancer-related deaths amongst men in EU and estimated number of deaths caused by PCa was 92 000 in a single year 2012.1 Male androgenic hormones, particularly dihydrotestosterone, play a key role in the origin and progression of PCa. Other factors include race, lifestyle and genetic predisposition.2 As symptoms in early stages of PCa can be mild or even absent, patients become aware of PCa when it develops into more advanced or aggressive form, when the curability is inefficient. Golden standard for PCa diagnosis for more than twenty years was analysis of a prostate specific antigen (PSA) in serum. However, the serological level of PSA varies to high extent with age and ethnicity and test itself can provide variable results leading into false positive/negative results. Due to low sensitivity, specificity and prognostic value of PSA biomarker, in 2012 the US Preventive Services Task Force advised against analysis of PSA for routine screening of PCa.3 Thus, new and more reliable methods for PCa diagnosis are needed. Biomarker discovery in the field of PCa diagnosis and prognosis is therefore focused on novel targets such as circulating microRNAs4, gene fusions5, exosomes6 or changes in the glycan structure7 of PSA.
Monitoring of aberrant protein glycosylation for identification of various types of cancers8 and as targets for personalised medicine9 is gaining a momentum in recent years with at least 13 glycoprotein-based cancer biomarkers approved by the US Food and Drug Administration (FDA)8b. An increased attention to study correlation between cancer disease progression and changes in the glycan composition is due to involvement of glycan biorecognition in a wide range of cellular processes10 with the potential to use such knowledge for development of advanced therapeutic/diagnostic tools to treat numerous diseases in the future.11 The most frequently applied instrumental tool for structural analysis of glycans is mass spectrometry, which can be further combined/integrated with a diverse range of chromatographic and electrophoretic techniques.12 Even though instrumental machinery applied for glycan analysis can provide details about glycan microheterogenity, such approach is not suitable for routine and high-throughput glycan analyses. An alternative way of glycan analysis is to use naturally occurring proteins designed by nature to recognise glycans – lectins. Lectins are proteins recognising free or bound mono- and oligosaccharides with an affinity site being a shallow groove or a pocket present on the lectin surface. The main interaction between glycan and lectin involving four main amino acids (asparagine, glycine/arginine and aromatic amino acids) is triggered via hydrogen and hydrophobic bond, while electrostatic interactions are responsible for binding of negatively charged glycans containing sialic acids.13 Lectins, can in some cases recognise different linkages two carbohydrates are linked together within a glycan moiety (i.e. α2-3 linked vs. α2-6 linked sialic acid to galactose), what is not a trivial task for instrumental-based approach.11a, 12b Moreover, lectins can detect glycans, which are still attached to the protein backbone or present on the surface of the cells, thus, a simplified analysis protocol can be used.13 Additional advantage of using lectins is their ability to “sense” subtle changes in the glycan structure, as proved in the study, where 26 different lectins recognising N-acetylgalactosamine moiety were applied in glycoprofiling of samples from patients suffering from breast cancer.14 Lectins applied in a microarray format despite providing high throughput of analysis cannot detect low level of glycans (LOD in sub nM level) and there is a requirement to use a label.15 Thus, other detection platforms, which can offer high sensitivity of detection working in a label-free mode of operation are intensively sought.7b, 13, 16
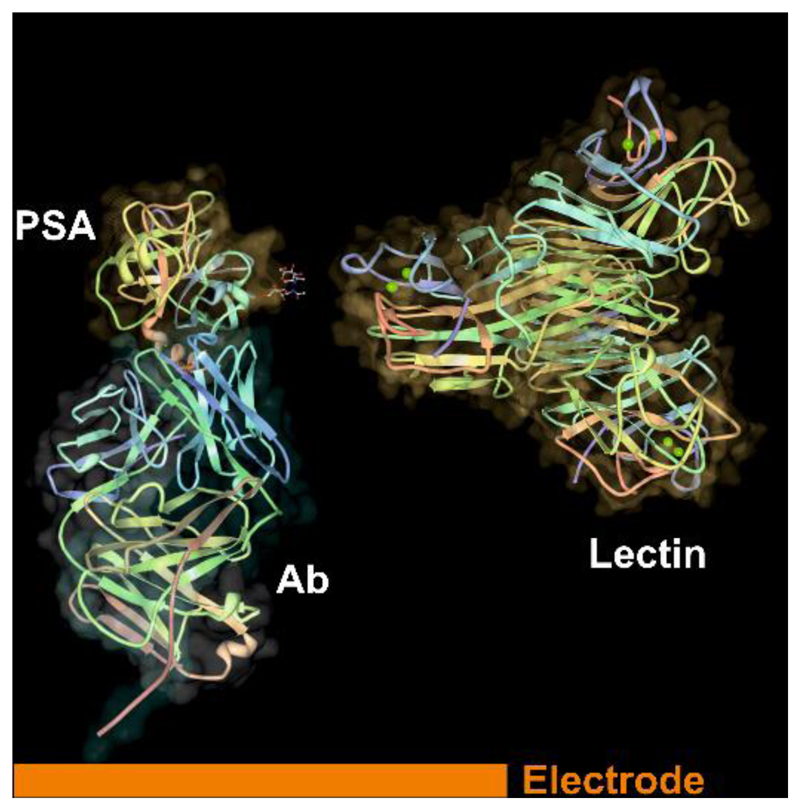
Electrochemical impedance spectroscopy (EIS) is one of label-free methods, which can allow detection of analytes down to a single molecule level.13, 17 The method is based on application of a small sinusoidal perturbation to an electrode and the output from the measurement is charge transfer resistance (Rct) of the electrode. After exposure of the biosensor to the analyte, Rct changes, what can be used for quantification of analyte concentration. The first EIS-based biosensor to evaluate glycan-lectin interactions was prepared by Joshi´s group18 and since then an increased interest to apply such approach especially in combination with immobilised lectins for analysis of intact glycoproteins and even various types of cancerous cells can be seen.13, 19 From our recent comprehensive review it is clear that glycoprofiling of PSA by electrochemical-based biosensor has not been done.13 Thus, in this work, the first lectin-based impedimetric biosensor for PSA glycoprofiling was developed. Moreover, label-free mode of operation not only allows to glycoprofile PSA, but also to detect its concentration in an ultrasensitive fashion (i.e. down to aM concentration level). This is possible by immobilisation of an antibody against PSA on the electrode surface, then incubation of the biosensor with PSA takes place and the final step is interaction of lectin with the biosensor via a glycan moiety present on PSA (Fig. 1).
Figure 1.
Scheme of a biosensor with immobilised antibody (Ab, only binding Fab fragment shown) on the modified gold electrode with formation of a complex with prostate specific antigen (PSA, pdb file 2ZCH) and a final incubation of the biosensor with lectin (pdb file 1DQ1) recognising glycan part of PSA. The structure of proteins was visualised using a Deep View/Swiss-Pdb Viewer.
Results and discussion
Concentration of an antibody
The first parameter being optimised was concentration of an antibody applied for its covalent immobilisation on a mixed SAM. When 6.7 nM stock solution of an antibody was incubated with activated SAM layer the density of antibody on the surface was so high, the biosensor could not detect any PSA upon incubation with the immunosensor (data not shown). This is why much lower concentration of an antibody was used in the subsequent experiments during Ab immobilisation (13 pM and 130 pM) to optimise this parameter. The results showed that 13 pM antibody solution applied for Ab immobilisation was not sufficient to immobilise Ab at density required for sensitive analysis of PSA since the slope of the calibration curve was 9.6-fold lower compared to the biosensor prepared by immobilisation of Ab from 130 pM stock solution (Fig. S1). Moreover, the biosensor constructed by immobilisation of Ab from 130 pM stock solution provided much wider linear response compared to the one constructed by immobilisation of Ab from 13 pM stock solution. This is why in the subsequent experiments the biosensor was prepared by immobilisation of Ab from its 130 pM stock solution.
Composition of SAM
In preliminary experiments various types of thiols with a concentration of 1 mM (being a typical concentration for SAMs formation20) for deposition of a mixed SAM were tested including besides aliphatic thiols such as MUA and MCH also thiols containing oligoethylene functional groups (i.e. –O-CH2-CH2-), but such mixed SAMs exhibited high initial Rct values, not favourable for construction of an impedimetric biosensor working in a sandwich configuration.
Quality of various SAMs could be examined using CV in the presence of a redox probe potassium hexacyanoferrate(III) (ferricyanide) and using EIS20, and such experiments are shown in Fig. S2 and Fig. S3. A redox process is observable on an Au electrode modified by pure MCH SAM or on the surface modified by a mixed SAM composed of MUA to MCH ratio of 1:33, but at lower dilution of MUA within MUA and MCH mixture capacitive behaviour start to be dominant indicating absence of pinholes (Fig. S2).21 Results obtained by CV are consistent with data obtained by EIS showing that deposition of SAM increases peak separation (or diminishes any redox process) and Rct compared to a bare Au electrode and that an increased proportion of MUA within a mixed SAM also resulted in high peak separation and value of Rct. Moreover a reductive desorption to strip SAM from the gold electrode by application of a negative voltage, run according to our previous study22 indicated that density of thiols within a mixed SAM were 4.4 molecules nm-2 for a mixed SAM composed of MUA:MCH=1:33 or 4.5 molecules nm-2 for a mixed SAM composed of MUA:MCH=1:3, suggesting presence of highly dense SAM layers with packing density comparable to a full monolayer based on (√3 × √3)R30° thiolate-gold structure (i.e. 4.5 molecules nm-2 23 or 4.7 molecules nm-2 24).
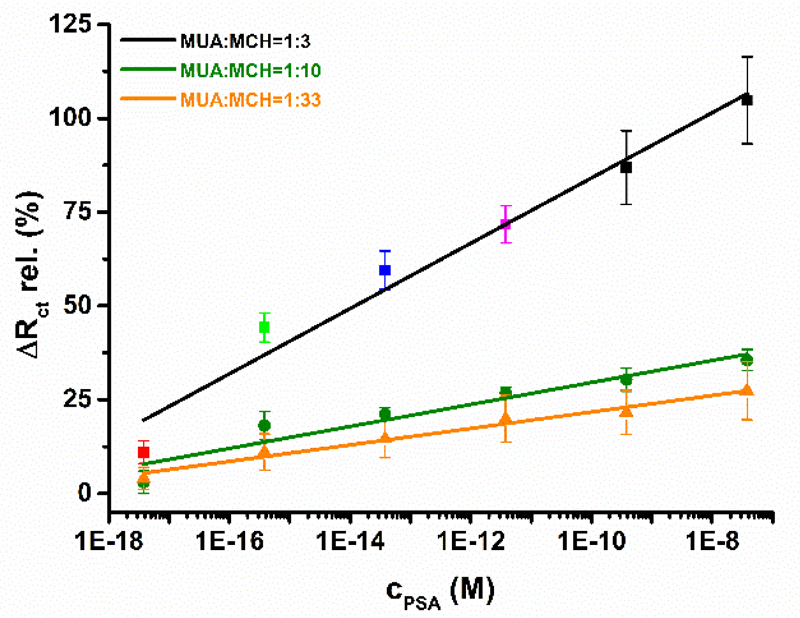
Since density of immobilised ligands on the biosensor surface influence biorecognition properties to high extent, the second optimisation parameter was composition of a mixed SAM, since this influences density of immobilised antibodies on the surface. Sensitivity of PSA detection was the highest with the immunosensor prepared on a mixed SAM with MUA:MCH ratio of 1:3 with a value of (8.7 ± 0.8) % decade-1, followed by the biosensor developed on a mixed SAM with MUA:MCH ratio of 1:10 with s=(2.9 ± 0.4) % decade-1 and the least sensitive PSA biosensor was the one based on a mixed SAM with MUA:MCH ratio of 1:33 with s=(2.2 ± 0.2) % decade-1 (Fig. 2). In all cases linear range for PSA detection spun 10 concentration orders of magnitude. For the following studies biosensor constructed on gold electrode modified by a mixed SAM with MUA:MCH ratio of 1:3 was selected (see Nyquist plots applied for construction of a calibration curve in Fig. S4).
Figure 2.
Calibration curves for analysis of PSA by the immunosensors differing in the composition of mixed SAM (i.e. with MUA:MCH ratio of 1:3; 1:10 and 1:33).
Fitting data into an equivalent circuit
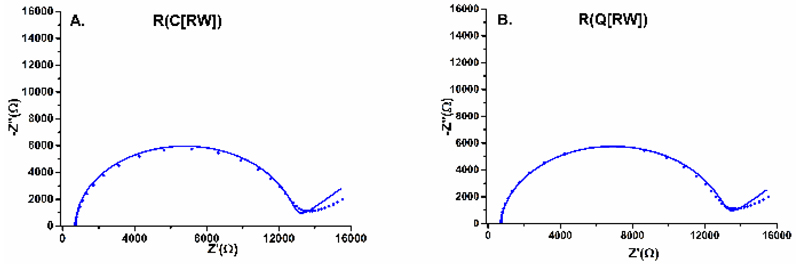
The next optimisation step was to find proper equivalent circuit to fit data obtained from EIS measurements represented in a Nyquist diagram. Randles circuit i.e. R(C[RW]) is the most frequently applied to describe interfacial properties on the electrode surface, but some phenomena (i.e. dipole/ion migration through the SAM), which can occur on the interface are not described by such circuit.25 This is why two different equivalent circuits i.e. R(C[RW]) or R(Q[RW]) were applied for fitting of a distinct set of data obtained in the preliminary phase of the study. Solid electrodes (which are not ideally polarizable, compared to liquid ones like mercury) don´t usually retain purely capacitive electrochemical double-layer. The lack of homogeneity during impedance studies is modelled with so-called constant-phase element (CPE), represented by the Q symbol. The impedance of CPE is described as
where n is an empirical constant. It is interesting to point out that when n approaches 1, the CPE behaves as an ideal capacitor, and on the contrary, when n = 0, it becomes a pure resistor. Thus for not ideally spherical semicircles, using the R(Q[RW]) model yields lesser fitting errors and more accurate Rct values. The results really showed that equivalent circuit R(Q[RW]) was better to fit EIS data with more reliable identification of Rct values, as can be seen in Fig. 3, Table S1 and Table S2 and this is why in the subsequent experiments this equivalent circuit was applied for data fitting.
Figure 3.
EIS data fitting for Nyquist plots with two different equivalent circuits applied i.e. R(C[RW]) (A) or R(Q[RW]) (B) as indicated in the figures. Experimental points obtained are shown as symbols and line represent fit of data by a particular equivalent circuit.
Choice of proper lectin for glycoprofiling
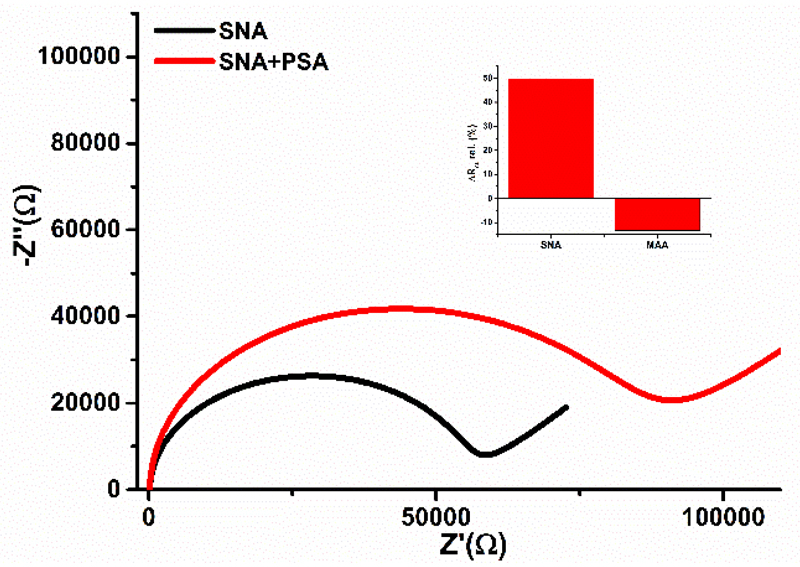
Two different lectins SNA and MAA able to detect terminal sialic acid were chosen to investigate binding of lectins to glycan part present on the surface of PSA. From Fig. 4 it can be seen that SNA lectin can specifically recognise glycan part of PSA with significant increase of Rct, while instead of increase of Rct after interaction of PSA with immobilised MAA lectin, a small decrease of Rct is observed (Fig. 4). Thus for further studies, SNA lectin was applied to prove if in-situ glycoprofiling of PSA in a sandwich configuration is possible. Further, concentration of SNA needed to complete a sandwich configuration was optimised, as well. Preliminary results indicate that concentration of SNA of 0.05 or 0.1 mg mL-1 resulted only in a moderate change of relative ∆Rct ~13-14%, but incubation of the biosensor with 0.5 mg mL-1 SNA concentration resulted in change of relative ∆Rct of (20 ± 1)%. This is why in the subsequent experiments this concentration of SNA was applied. PSA applied in this study was from a healthy donor, since it contains α-2,6 linked sialic acid.26
Figure 4.
Incubation of PSA on the biosensor surface modified by Sambucus nigra agglutinin (SNA, recognising α-2,6 linked sialic acid - SA or by Maackia amurensis agglutinin (MAA, recognising α-2,3 linked SA).
AFM visualisation of the surfaces
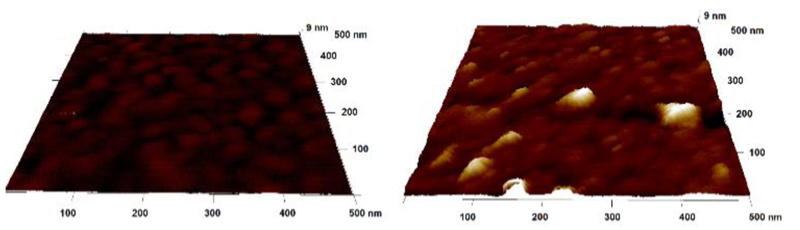
AFM was applied to see differences in the surface morphology after each surface patterning step. The results indicate surface roughness Rq=(0.91 ± 0.02) nm for a mixed SAM layer and such surface roughness increased to a value of (1.45 ± 0.28) after immobilisation of Ab and then decreased to a value of (0.71 ± 0.13) after incubation of the biosensor surface with PSA. Typical AFM images of the gold surface patterned by a mixed SAM and after incubation with anti-PSA antibody are shown in Fig. 5.
Figure 5.
AFM image of a mixed SAM layer (on left) and after covalent antibody immobilisation (on right). Note: it is possible to see Y-shaped antibody feature with two Fab fragments close to the figure edge on right.
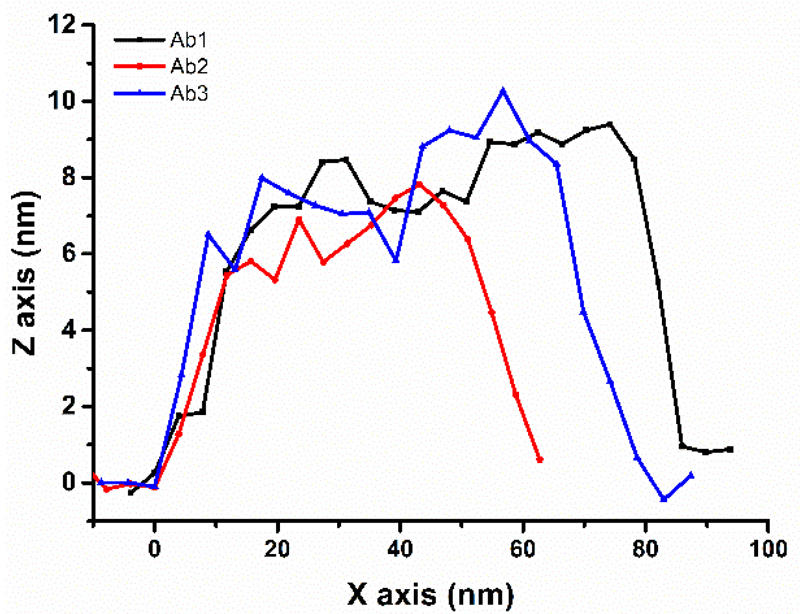
Analysis of individual spots revealed that average height of immobilised antibody is (8.8 ± 0.9) nm with a possibility to see extra space between Fab fragments (i.e. indicating that Y shape of Ab can be seen in Fig. 5 on right). Such height of antibody is in a good agreement with a previous study showing a value of (8.7 ± 0.1) nm27 and indicates attachment of the antibody on the surface preferentially via side on orientation of the antibody on the surface (Fig. 6)28, also in agreement with the size of an antibody of 14.3 × 7.7 × 4.0 nm.29 The size of PSA-Ab complex only slightly increased to a value of (10 ± 1) nm after PSA modified surface was exposed to Ab, while the size of individual PSA molecules was (4.1 ± 0.5) nm (Fig. S5).
Figure 6.
Height profile of individual Abs from AFM images.
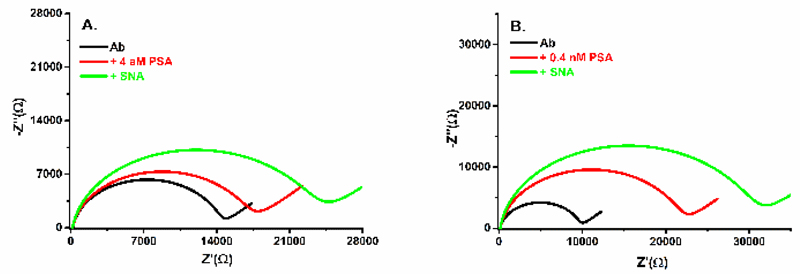
Finally it was demonstrated that the biosensor is able to glycoprofile PSA even though it was incubated with high concentration of PSA (i.e. 0.4 nM, clinically relevant concentration), when density of protein within two proteins layers (Ab and PSA) is quite high. This is possible only due to low initial density of Ab immobilised on the biosensor from 130 pM stock solution surface allowing to sense the 3rd protein layer. The final glycoprofiling of PSA is shown in Fig. 7B. From data presented in Fig. 2, limit of detection (LOD) was calculated taking into account 3×average SD as noise of assays.30 LOD for PSA analysis was 4 aM or 5 aM for the biosensor constructed on SAM layer with ratio MUA/MCH of 1:3 or 1:10, respectively. RSD for PSA analysis by the immunosensor based on SAM with MUA:MCH ratio of 1:3 was from 6.9% to 11.3% with an average RSD of (9.1 ± 1.7)%. An average RSD for analysis of PSA by the biosensors based on SAM composed of MUA:MCH ratio of 1:10 and 1:33 was much higher than this value due to lower sensitivity of detection of PSA. It is worth noting that such RSDs are not relative standard errors of PSA analysis, but rather represent reproducibility of the biosensor construction, since each calibration curve was constructed by an independent biosensor device. When, the biosensor was incubated with the plain buffer with 5 consecutive incubations for 20 min, RSD of the assay itself was only 1.0%. Glycoprofiling of PSA in a sandwich configuration revealed that SNA lectin is able to detect glycans of PSA, when PSA was incubated with the biosensor device from a stock PSA solution of 4 aM (see Fig. 7A). From Fig. 7 it can be concluded that the immunosensor is able to detect PSA with concentration of 4 aM (Fig. 7A) and much larger response is obtained after PSA addition with concentration of 0.4 nM (Fig. 7B) and a final glycoprofiling of PSA by SNA was possible for both cases with further increased response after addition of SNA lectin. Finally, the biosensor was incubated with human serum of a healthy man, which was measured before and after spiking with PSA added with a final concentration of 10 ng mL-1 and such result indicate a recovery index of 95.6%.
Figure 7.
Nyquist plots for the biosensor with immobilised anti-PSA antibody (Ab), after addition of PSA (+PSA) and after addition of SNA lectin (+SNA). Two different concentrations of PSA of 4 aM (A) or 0.4 nM (B) were incubated with the biosensor device and final in-situ glycoprofiling of PSA´s glycan was done by incubation with SNA lectin.
Immunosensing of PSA
PSA is one of the most popular analyte to be detected by newly developed biosensor devices. Not only affinity based interaction between PSA and its antibody were applied for PSA quantification, but also the function of PSA as a protease to cleave peptides.
Label-based methods with fluorescent principle of analysis can detect PSA down to pM level31, but 1 fM32 or 3 aM33 LOD were reported, as well; FRET allowed detection of PSA down to 50 pM34; electroluminescent detection platform can detect PSA down to 25 fM35 or 130 aM36. Other label-based methods using various (redox, nanoparticles or enzymes) labels can offer LOD for PSA due to amplification strategies down to 32 aM37 or even 3 aM.38
Label-free methods relying on localised surface plasmon resonance offer LOD down to 3 fM39 or 130 aM40; surface plasmon resonance imaging down to 3 pM41; surface-enhanced Raman scattering down to 313 fM42 and with other optical method down to 300 aM43. Microcantilever based biosensors provide LOD for PSA in low pM concentration level44.
Label-free electrochemical methods are extremely sensitive methods for analysis of a wide range of analytes relying on diverse range of detection principles.13, 45 EIS for example was applied for PSA analysis with LOD down to pM46 or fM47 level. Thus, to our knowledge, our approach using controlled interfacial SAM layer for immobilisation of Ab against PSA is the most sensitive EIS-based device for analysis of PSA described so far. Field-effect transistor (FET) sensing allow to detect PSA with LOD of 10 nM48, but more frequently LOD down to aM were achieved.49 Imprinting sensors could detect PSA with LOD of 8 aM.50
From all affinity based biosensor devices the most sensitive are the ones based on plasmonic ELISA with LOD down to zM level for PSA.40, 51 Analysis of enzymatic activity of PSA was applied for determination of PSA employing various techniques with LOD down to 24 fM.35, 52.
Thus, it can be concluded that our immunosensor for detection of PSA based on EIS is the most sensitive impedimetric biosensor described so far with LOD comparable to the most sensitive detection schemes for analysis of PSA.
Antibody-assisted lectin glycoprofiling of PSA
Application of a sandwich configuration is not a new approach, since it was introduced in 2007 by Haab´s group for analysis of two cancer biomarkers other than PSA.53 The main principle behind is to use lectin- or antibody-based fluorescent microarrays with subsequent incubation of the microarray with PSA and a final incubation with lectins (i.e. Ab-PSA-lectin)53 or with antibodies (i.e. lectin-PSA-Ab).54 Surface plasmon field-enhanced fluorescence spectroscopy is more sensitive with LOD of 630 fM32b compared to traditional fluorescent microarray with LOD in the range 1-44 pM (Ab-PSA-lectin)54c, 55 or 470 pM-6.3 nM (lectin-PSA-Ab)54. Physiological PSA level in human blood is below 130 pM.
From these studies it can be concluded that other assay formats than fluorescent microarray-based ones need to be applied to reliably glycoprofile PSA in the physiological concentration window and that for this purpose mainly Ab-PSA-lectin sandwich configuration is more suitable compared to lectin-PSA-Ab format of analysis. Moreover, Ab-PSA-lectin sandwich configuration has additional advantage since PSA could be fished out from complex sample in a direct way by incubation with a sample, while in case of lectin-PSA-Ab format of analysis, PSA has to be separated from sample prior to glycoprofiling. Finally it has to be pointed out to the fact that since all these methods for glycoprofiling of PSA are label-based methods, for quantification of PSA level in sample additional experiments needed to be performed by PSA quantification done using secondary antibodies in a microarray format53, by immunoblotting54a or by standard PSA assay kit54b, 54c.
EIS-based glycoprofiling in a sandwich configuration Ab-PSA-lectin developed in this study could detect glycans from PSA stock solution of 4 aM, indicating that such biosensor is much more sensitive compared to previously described approaches. In order to compete with high-throughput sandwich-based glycoprofiling in a fluorescent microarray format, some degree of multiplexing of impedimetric analysis has to be performed, what is a feasible task.7b, 13, 17 Since this contribution is proof of the concept study, the current format of SNA glycoprofiling has to be extended by application of other relevant lectins besides SNA26 in order to test diagnostic potential of this approach.
Experimental
Chemicals
Mouse monoclonal IgG antibody against PSA (Ab10187) was purchased from Abcam (UK), free-PSA (PSA) purified from human seminal fluid was obtained from Fitzgerald Industries International (USA). 11-mercaptoundecanoic acid (MUA), 6-mercapto-1-hexanol (MCH), ethanolamine, hydrogen peroxide (30% w/w), phosphate buffered saline (PBS) tablets, potassium chloride, potassium hexacyanoferrate(III), potassium hexacyanoferrate(II) trihydrate, sodium hydroxide, sulphuric acid (95.0 – 98.0%), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) were purchased from Sigma Aldrich (USA). Sambucus nigra agglutinin type I (SNA-I recognising α-2,6 linked sialic acid) lectin from elderberry was purchased from EY Laboratories (USA) and Maackia amurensis agglutinin (MAA recognizing α-2,3 linked sialic acid) lectin was obtained from Vector Laboratories (USA). Ethanol for UV/VIS spectroscopy (ultrapure) was purchased from Slavus (Slovakia). PBS solution (10 mM, pH 7.4) was prepared by dissolving 1 tablet in 200 mL of ultra-pure deionized water (DW). All solutions were filtered prior to use (0.2 μm sterile filters) and prepared in ultra-pure DW. Working solution of PSA and anti-PSA were prepared by dilution in PBS (10 mM, pH 7.4).
Pre-treatment of gold electrodes and surface functionalization
Briefly, polycrystalline gold disk electrodes (d=1.6 mm, BASi, USA) were extensively cleaned according to a previously described protocol22, namely by electrochemical reductive desorption under anaerobic conditions, mechanical polishing, chemical treatment with hot piranha solution, electrochemical polishing and gold oxide stripping procedure. Firstly, previously bound thiol molecules were desorbed from the surface using cyclic voltammetry (CV) measurements in 0.1 M NaOH (100 scans from -500 mV to -1500 mV at a scan rate of 100 mV s-1) under N2 atmosphere. Next, the electrodes were mechanically polished on a polishing pad using both 1 μm and 0.3 μm micropolish alumina (Buehler, USA) for 5 min each, followed by ultrasonic cleaning in ultra-pure DW to remove residual alumina particles. In the further step gold electrodes were dipped into a fresh piranha solution (mixture of H2O2 and H2SO4 in the ratio 1:3, caution: handle with special care) for approx. 15 min and sonicated in ultra-pure DW for 5 min. Afterwards, electrochemical polishing was carried out by performing 50 scans of the potential between -200 mV to +1500 mV in a fresh 0.1 M H2SO4 solution with final gold oxide stripping procedure (20 scans run from +750 mV to +200 mV at a scan rate of 100 mV s-1). Immediately after treatment procedure, the electrodes were rinsed thoroughly with DW and ultra-pure ethanol and then dried in a flow of nitrogen gas with subsequent surface functionalization using self-assembled monolayer (SAM) formation. The SAM was fabricated by immersion in a fresh mixed solution of 1 mM MUA and 1 mM MCH in ethanol with various ratio MUA to MCH (1:3; 1:10; 1:33.3) overnight at room temperature in the dark.
Preparation of lectin biosensor assay
After SAM surface functionalisation, terminal carboxyl groups of MUA were sequentially modified with an aqueous solution containing 1:1 mixture of 0.2 M EDC and 0.05 M NHS for 15 min. Thereafter, lectins (SNA-I, MAA) were covalently immobilised on the activated surface from a 40 μL stock solution (0.5 mg mL-1 in PBS) by 30 min immersion at room temperature with further gentle PBS rinsing. In the final step, an incubation of PSA analyte (40 μL) was performed for 30 min.
Fabrication of a sandwich immunosensor
Besides lectin biosensor fabrication, we prepared a sandwich immunosensor with final glyco-profiling. After SAM formation, monoclonal antibody (anti-PSA) was covalently attached on NHS-activated carboxyl groups by incubation with 40 μL of 2 or 20 ng mL-1 for 30 min. The remaining NHS-active esters sites were passivated by blocking with 1 M ethanolamine for 30 min. The following step was capturing of PSA glycoprotein for 30 min with a final incubation of electrodes with SNA lectin (40 μL droplet of 0.5 mg mL-1) for 30 min to glycoprofile a disease specific glycoprotein. After each immobilisation step, electrodes were gently washed with a PBS solution.
Electrochemical measurements and apparatus
A laboratory potentiostat/galvanostat PGSTAT 128N (Metrohm Autolab, the Netherlands) controlled by NOVA software 1.10 was used to record the changes in the impedance (the charge-transfer resistance, Rct) as a result of binding event taking place at the electrode surface. A conventional three-electrode cell system comprising of a gold working electrode, auxiliary Pt electrode and Ag/AgCl reference electrode was utilised in all experiments. All electrochemical measurements were performed at laboratory ambient room temperature (RT, ~ 25 °C).
All EIS characterisations were carried out in a fresh and filtered electrolyte containing 5 mM ferri/ferrocyanide ([Fe(CN)6]3–/4–) and 0.1 M KCl. Impedance measurements were recorded across a 0.1 Hz - 100 kHz frequency range scanning 50 different frequencies applying a 200 mV a.c. voltage. The acquired data were plotted in the form of complex plane diagrams (Nyquist plots) with ideal Randles-Erschler equivalent circuits (R(C[RW]), R(Q[RW])) employed for data fitting. Each experiment was conducted at least in triplicate (±SD) with an independent biosensor device. Cyclic voltammetry experiments with ferricyanide as a redox probe were carried out with a three electrode system in 0.1 M KCl with 5 mM ferricyanide in a potential window from -400 mV to +400 mV at a scan rate of 100 mV-1. Reductive desorption of thiols for calculation of their coverage was run in 100 mM NaOH deoxygenated by purging of N2 through the electrolyte for 15 min with N2 stream over the electrolyte during measurements carried in the potential window from -500 mV to -1,500 mV at a scan rate of 100 mV-1 (the 1st scan was used for calculations). Electrochemical surface area was calculated from electrochemical polishing procedure by integration of the charge passed through the electrode upon reduction of gold oxide.22
Atomic force microscopy (AFM) analysis
Monitoring of various biosensor interfaces was executed using continuous high-speed peak force tapping mode atomic force microscopy (ScanAsyst, Bruker, USA). All AFM experiments were performed with a Bioscope Catalyst instrument and Olympus IX71 microscope controlled by NanoScope 8.15 software in air mode. Square-shape gold chips (10x10 mm) were scanned with silicon nitride ScanAsyst-air probe (Bruker, USA) directly controlled using a continuous feedback loop. The AFM images were finally processed in NanoScope Analysis 1.40 software.
Conclusions
The most sensitive impedimetric biosensor for PSA analysis with extremely low LOD was described here. Furthermore, LOD of 4 aM for impedimetric PSA analysis is comparable or better than for other detection platforms published so far. The biosensor was successfully applied in glycoprofiling of PSA by application of SNA lectin. Moreover, the current label-free impedimetric detection scheme allows to perform PSA quantification and PSA glycoprofiling on the same interface, what has not been described so far. Label-free mode of detection has additional advantage that Ab-PSA and PSA-lectin biorecognition is not compromised by presence of a label. Further benefit of the current biosensing protocol is a minute consumption (130 pM with an incubation volume of 40 μL i.e. 5.2 fmol or 0.78ng) of precious and expensive anti-PSA antibody
Supplementary Material
Acknowledgements
The financial support received from the Slovak Scientific Grant Agency VEGA 2/0162/14 and 1/0229/12; and from the Slovak Research and Development Agency APVV 0282-11 is acknowledged. This report was made possible by a NPRP award [NPRP grant no. 6-381-1-078] from the Qatar National Research Fund (a member of The Qatar Foundation). The statements made herein are solely the responsibility of the authors. The research leading to these results received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement no. 311532 and this work has received funding from the European Union’s Seventh Framework Program for research, technological development and demonstration under grant agreement no. 317420.
References
- 1.(a) Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JWW, Comber H, Forman D, Bray F. Eur J Cancer. 2013;49:1374. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]; (b) Ferlay J, Soerjomataram I. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. [Accessed. 28/4/2014]; http://globocan.iarc.fr.
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S. CA: Cancer J Clin. 2012;62:220. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Moyer VA. Ann Intern Med. 2012;157:120. doi: 10.7326/0003-4819-157-2-201207170-00459. [DOI] [PubMed] [Google Scholar]
- 4.Giglio S, Nunzio CD, Cirombella R, Volinia S, Luciani E, Tubaro A, Vecchione A. Cancer Res. 2014;74:1475. [Google Scholar]
- 5.Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, de Reijke TM, Vergunst H, Kil P, Knipscheer BC. Eur Urol. 2014;65:534. doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Pasa-Tolic L, Jenster G. PLoS One. 2013;8:e82589. doi: 10.1371/journal.pone.0082589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Gilgunn S, Conroy PJ, Saldova R, Rudd PM, O’Kennedy RJ. Nat Rev Urol. 2013;10:99. doi: 10.1038/nrurol.2012.258. [DOI] [PubMed] [Google Scholar]; (b) Klukova L, Bertok T, Kasak P, Tkac J. Anal Methods. 2014;6:4922. doi: 10.1039/C4AY00495G. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pihíková D, Kasák P, Tkac J. Open Chem. 2015;13:636. doi: 10.1515/chem-2015-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Belicky S, Tkac J. Chem Pap. 2015;69:90. doi: 10.1515/chempap-2015-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Badr HA, Alsadek DM, Darwish AA, Elsayed AI, Bekmanov BO, Khussainova EM, Zhang X, Cho WC, Djansugurova LB, Li CZ. Expert Rev Proteom. 2014;11:227. doi: 10.1586/14789450.2014.897611. [DOI] [PubMed] [Google Scholar]; (b) Kim EH, Misek DE. Int J Proteom. 2011;2011:601937. doi: 10.1155/2011/601937. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mechref Y, Hu Y, Garcia A, Hussein A. Electrophoresis. 2012;33:1755. doi: 10.1002/elps.201100715. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Svarovsky SA, Joshi L. Anal Methods. 2014;6:3918. [Google Scholar]; (e) Wu L, Qu X. Chem Soc Rev. 2015;44:2963. doi: 10.1039/c4cs00370e. [DOI] [PubMed] [Google Scholar]; (f) Kim Y, Varki A. Glycoconjugate J. 1997;14:569. doi: 10.1023/a:1018580324971. [DOI] [PubMed] [Google Scholar]; (g) Welinder C, Baldetorp B, Borrebaeck C, Fredlund BM, Jansson B. Glycobiology. 2011;21:1097. doi: 10.1093/glycob/cwr048. [DOI] [PubMed] [Google Scholar]
- 9.Padler-Karavani V. Cancer Lett. 2014;352:102. doi: 10.1016/j.canlet.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 10.(a) Reichardt NC, Martín-Lomas M, Penadés S. Chem Soc Rev. 2013;42:4358. doi: 10.1039/c2cs35427f. [DOI] [PubMed] [Google Scholar]; (b) Park S, Gildersleeve JC, Blixt O, Shin I. Chem Soc Rev. 2013;42:4310. doi: 10.1039/c2cs35401b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cecioni S, Imberty A, Vidal S. Chem Rev. 2015;115:525. doi: 10.1021/cr500303t. [DOI] [PubMed] [Google Scholar]; (d) Varki A, Chrispeels MJ. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; 1999. [PubMed] [Google Scholar]
- 11.(a) Alley WR, Mann BF, Novotny MV. Chem Rev. 2013;113:2668. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Burton DR, Poignard P, Stanfield RL, Wilson IA. Science. 2012;337:183. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Science. 2014;343:37. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]; (d) Macauley MS, Arlian BM, Rillahan CD, Pang P-C, Bortell N, Marcondes MCG, Haslam SM, Dell A, Paulson JC. J Biol Chem. 2014;289:35149. doi: 10.1074/jbc.M114.606517. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney MC, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, et al. Nature. 2015;520:109. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]; (f) Bournazos S, Klein F, Pietzsch J, Seaman Michael S, Nussenzweig Michel C, Ravetch Jeffrey V. Cell. 2014;158:1243. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Medina RA, García-Sastre A. Nat Rev Microbiol. 2011;9:590. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.(a) Mariño K, Bones J, Kattla JJ, Rudd PM. Nat Chem Biol. 2010;6:713. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]; (b) Mechref Y, Novotny MV. Chem Rev. 2002;102:321. doi: 10.1021/cr0103017. [DOI] [PubMed] [Google Scholar]; (c) Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, Aebersold R, Watts JD. Nat Biotechnol. 2009;27:378. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Zhang H, Li XJ, Martin DB, Aebersold R. Nat Biotechnol. 2003;21:660. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 13.Paleček E, Tkáč J, Bartošík M, Bertók T, Ostatná V, Paleček J. Chem Rev. 2015;115:2045. doi: 10.1021/cr500279h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks S. Mol Biotechnol. 2009;43:76. doi: 10.1007/s12033-009-9184-6. [DOI] [PubMed] [Google Scholar]
- 15.(a) Gemeiner P, Mislovičová D, Tkáč J, Švitel J, Pätoprstý V, Hrabárová E, Kogan G, Kožár T. Biotechnol Adv. 2009;27:1. doi: 10.1016/j.biotechadv.2008.07.003. [DOI] [PubMed] [Google Scholar]; (b) Katrlik J, Svitel J, Gemeiner P, Kozar T, Tkac J. Med Res Rev. 2010;30:394. doi: 10.1002/med.20195. [DOI] [PubMed] [Google Scholar]
- 16.(a) Mu B, Zhang J, McNicholas TP, Reuel NF, Kruss S, Strano MS. Acc Chem Res. 2014;47:979. doi: 10.1021/ar400162w. [DOI] [PubMed] [Google Scholar]; (b) Reuel NF, Mu B, Zhang J, Hinckley A, Strano MS. Chem Soc Rev. 2012;41:5744. doi: 10.1039/c2cs35142k. [DOI] [PubMed] [Google Scholar]; (c) Reuel NF, Mu B, Zhang J, Hinckley A, Strano MS. Chem Soc Rev. 2012;41:5744. doi: 10.1039/c2cs35142k. [DOI] [PubMed] [Google Scholar]; (d) Hushegyi A, Tkac J. Anal Methods. 2014;6:6610. doi: 10.1039/C4AY00692E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hushegyi A, Bertok T, Damborsky P, Katrlik J, Tkac J. Chem Commun. 2015;51:7474. doi: 10.1039/c5cc00922g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Belle JT, Gerlach JQ, Svarovsky S, Joshi L. Anal Chem. 2007;79:6959. doi: 10.1021/ac070651e. [DOI] [PubMed] [Google Scholar]
- 19.(a) Santos A, Carvalho FC, Roque-Barreira M-C, Bueno PR. Biosens Bioelectron. 2014;62:102. doi: 10.1016/j.bios.2014.06.034. [DOI] [PubMed] [Google Scholar]; (b) Carvalho FC, Martins DC, Santos A, Roque-Barreira M-C, Bueno PR. Biosensors. 2014;4:358. doi: 10.3390/bios4040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaki NK, Vijayamohanan K. Biosens Bioelectron. 2002;17:1. doi: 10.1016/s0956-5663(01)00277-9. [DOI] [PubMed] [Google Scholar]
- 21.Wink T, van Zuilen SJ, Bult A, van Bennekom WP. Analyst. 1997;122:43R. doi: 10.1039/a606964i. [DOI] [PubMed] [Google Scholar]
- 22.Tkac J, Davis JJ. J Electroanal Chem. 2008;621:117. [Google Scholar]
- 23.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Chem Rev. 2005;105:1103. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 24.Herrwerth S, Eck W, Reinhardt S, Grunze M. J Am Chem Soc. 2003;125:9359. doi: 10.1021/ja034820y. [DOI] [PubMed] [Google Scholar]
- 25.Goes MS, Rahman H, Ryall J, Davis JJ, Bueno PR. Langmuir. 2012;28:9689. doi: 10.1021/la301281y. [DOI] [PubMed] [Google Scholar]
- 26.Vermassen T, Speeckaert MM, Lumen N, Rottey S, Delanghe JR. Clin Chim Acta. 2012;413:1500. doi: 10.1016/j.cca.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Dutta P, Sawoo S, Ray N, Bouloussa O, Sarkar A. Bioconjugate Chem. 2011;22:1202. doi: 10.1021/bc200073r. [DOI] [PubMed] [Google Scholar]
- 28.(a) Wadu-Mesthrige K, Amro NA, Garno JC, Xu S, Liu G-y. Biophys J. 2001;80:1891. doi: 10.1016/s0006-3495(01)76158-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Farris L, McDonald M. Anal Bioanal Chem. 2011;401:2821. doi: 10.1007/s00216-011-5365-9. [DOI] [PubMed] [Google Scholar]
- 29.Trilling AK, Beekwilder J, Zuilhof H. Analyst. 2013;138:1619. doi: 10.1039/c2an36787d. [DOI] [PubMed] [Google Scholar]
- 30.Bertok T, Sediva A, Filip J, Ilcikova M, Kasak P, Velic D, Jane E, Mravcova M, Rovensky J, Kunzo P, Lobotka P, et al. Langmuir. 2015;31:7148. doi: 10.1021/acs.langmuir.5b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song HY, Wong TI, Sadovoy A, Wu L, Bai P, Deng J, Guo S, Wang Y, Knoll W, Zhou X. Lab Chip. 2015;15:253. doi: 10.1039/c4lc00978a. [DOI] [PubMed] [Google Scholar]
- 32.(a) Liu D, Huang X, Wang Z, Jin A, Sun X, Zhu L, Wang F, Ma Y, Niu G, Hight Walker AR. ACS Nano. 2013;7:5568. doi: 10.1021/nn401837q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kaya T, Kaneko T, Kojima S, Nakamura Y, Ide Y, Ishida K, Suda Y, Yamashita K. Anal Chem. 2015;87:1797. doi: 10.1021/ac503735e. [DOI] [PubMed] [Google Scholar]
- 33.Ou L-J, Liu S-J, Chu X, Shen G-L, Yu R-Q. Anal Chem. 2009;81:9664. doi: 10.1021/ac901786m. [DOI] [PubMed] [Google Scholar]
- 34.Wegner KD, Jin Z, Linden S, Jennings TL, Hildebrandt N. ACS Nano. 2013;7:7411. doi: 10.1021/nn403253y. [DOI] [PubMed] [Google Scholar]
- 35.Qi H, Li M, Dong M, Ruan S, Gao Q, Zhang C. Anal Chem. 2014;86:1372. doi: 10.1021/ac402991r. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Chai Y, Yuan R, Wang H, Bai L, Liao N. Analyst. 2014;139:5209. doi: 10.1039/c4an01002g. [DOI] [PubMed] [Google Scholar]
- 37.(a) Zhang J, Ting BP, Khan M, Pearce MC, Yang Y, Gao Z, Ying JY. Biosens Bioelectron. 2010;26:418. doi: 10.1016/j.bios.2010.07.112. [DOI] [PubMed] [Google Scholar]; (b) Ting BP, Zhang J, Khan M, Yang YY, Ying JY. Chem Commun. 2009:6231. doi: 10.1039/b910730d. [DOI] [PubMed] [Google Scholar]
- 38.(a) Qin X, Xu A, Liu L, Deng W, Chen C, Tan Y, Fu Y, Xie Q, Yao S. Chem Commun. 2015;51:8540. doi: 10.1039/c5cc01439e. [DOI] [PubMed] [Google Scholar]; (b) Kosaka PM, Pini V, Ruz JJ, da Silva RA, Gonzalez MU, Ramos D, Calleja M, Tamayo J. Nat Nanotechnol. 2014;9:1047. doi: 10.1038/nnano.2014.250. [DOI] [PubMed] [Google Scholar]
- 39.Sanders M, Lin Y, Wei J, Bono T, Lindquist RG. Biosens Bioelectron. 2014;61:95. doi: 10.1016/j.bios.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 40.Liang J, Yao C, Li X, Wu Z, Huang C, Fu Q, Lan C, Cao D, Tang Y. Biosens Bioelectron. 2015;69:128. doi: 10.1016/j.bios.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 41.Malic L, Sandros MG, Tabrizian M. Anal Chem. 2011;83:5222. doi: 10.1021/ac200465m. [DOI] [PubMed] [Google Scholar]
- 42.Yang A-q, Wang D, Wang X, Han Y, Ke X-b, Wang H-j, Zhou X, Ren L. RSC Adv. 2015;5:38354. [Google Scholar]
- 43.Su L-C, Chen R-C, Li Y-C, Chang Y-F, Lee Y-J, Lee C-C, Chou C. Anal Chem. 2010;82:3714. doi: 10.1021/ac100071h. [DOI] [PubMed] [Google Scholar]
- 44.Wu GH, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Nat Biotechnol. 2001;19:856. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- 45.Luo X, Davis JJ. Chem Soc Rev. 2013;42:5944. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- 46.(a) Jolly P, Formisano N, Tkáč J, Kasák P, Frost CG, Estrela P. Sens Actuat B: Chem. 2015;209:306. [Google Scholar]; (b) Chiriaco MS, Primiceri E, Montanaro A, de Feo F, Leone L, Rinaldi R, Maruccio G. Analyst. 2013;138:5404. doi: 10.1039/c3an00911d. [DOI] [PubMed] [Google Scholar]
- 47.(a) Chornokur G, Arya SK, Phelan C, Tanner R, Bhansali S. J Sensor. 2011 [Google Scholar]; (b) Barton AC, Davis F, Higson SaP. Anal Chem. 2008;80:6198. doi: 10.1021/ac800491m. [DOI] [PubMed] [Google Scholar]
- 48.Gao N, Zhou W, Jiang X, Hong G, Fu T-M, Lieber CM. Nano Lett. 2015;15:2143. doi: 10.1021/acs.nanolett.5b00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.(a) Gao A, Lu N, Dai P, Fan C, Wang Y, Li T. Nanoscale. 2014;6:13036. doi: 10.1039/c4nr03210a. [DOI] [PubMed] [Google Scholar]; (b) Huang Y-W, Wu C-S, Chuang C-K, Pang S-T, Pan T-M, Yang Y-S, Ko F-H. Anal Chem. 2013;85:7912. doi: 10.1021/ac401610s. [DOI] [PubMed] [Google Scholar]; (c) Gong J-R. Small. 2010;6:967. doi: 10.1002/smll.200902132. [DOI] [PubMed] [Google Scholar]
- 50.Patra S, Roy E, Madhuri R, Sharma PK. Biosens Bioelectron. 2015;66:1. doi: 10.1016/j.bios.2014.10.076. [DOI] [PubMed] [Google Scholar]
- 51.(a) Liu D, Yang J, Wang H-F, Wang Z, Huang X, Wang Z, Niu G, Hight Walker A, Chen X. Anal Chem. 2014;86:5800. doi: 10.1021/ac500478g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) de la Rica R, Stevens MM. Nat Nanotechnol. 2012;7:821. doi: 10.1038/nnano.2012.186. [DOI] [PubMed] [Google Scholar]; (c) Rodríguez-Lorenzo L, de la Rica R, Álvarez-Puebla RA, Liz-Marzán LM, Stevens MM. Nat Mater. 2012;11:604. doi: 10.1038/nmat3337. [DOI] [PubMed] [Google Scholar]; (d) Howes PD, Rana S, Stevens MM. Chem Soc Rev. 2014;43:3835. doi: 10.1039/c3cs60346f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.(a) Suaifan GARY, Esseghaier C, Ng A, Zourob M. Analyst. 2012;137:5614. doi: 10.1039/c2an36243k. [DOI] [PubMed] [Google Scholar]; (b) Xie S, Zhang J, Yuan Y, Chai Y, Yuan R. Chem Commun. 2015;51:3387. doi: 10.1039/c4cc10363g. [DOI] [PubMed] [Google Scholar]
- 53.Chen SM, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Nat Methods. 2007;4:437. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- 54.(a) Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, Chiba Y, Narimatsu H, Hirabayashi J. Mol Cel Proteom. 2009;8:99. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]; (b) Meany DL, Hackler L, Jr, Zhang H, Chan DW. J Proteome Res. 2011;10:1425. doi: 10.1021/pr1010873. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li Y, Tao S-C, Bova GS, Liu AY, Chan DW, Zhu H, Zhang H. Anal Chem. 2011;83:8509. doi: 10.1021/ac201452f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meany DL, Zhang Z, Sokoll LJ, Zhang H, Chan DW. J Proteome Res. 2009;8:613. doi: 10.1021/pr8007539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.