Abstract
The applicability of next generation DNA sequencing (NGS) methods for water quality assessment has so far not been broadly investigated. This study set out to evaluate the potential of an NGS-based approach in a complex catchment with importance for drinking water abstraction. In this multicompartment investigation, total bacterial communities in water, faeces, soil, and sediment samples were investigated by 454 pyrosequencing of bacterial 16S rRNA gene amplicons to assess the capabilities of this NGS method for (i) the development and evaluation of environmental molecular diagnostics, (ii) direct screening of the bulk bacterial communities, and (iii) the detection of faecal pollution in water. Results indicate that NGS methods can highlight potential target populations for diagnostics and will prove useful for the evaluation of existing and the development of novel DNA-based detection methods in the field of water microbiology. The used approach allowed unveiling of dominant bacterial populations but failed to detect populations with low abundances such as faecal indicators in surface waters. In combination with metadata, NGS data will also allow the identification of drivers of bacterial community composition during water treatment and distribution, highlighting the power of this approach for monitoring of bacterial regrowth and contamination in technical systems.
Keywords: DNA extraction, faecal pollution, microbial source tracking, molecular diagnostics, next generation sequencing, water quality assessment
Introduction
During the last 30 years, molecular biological methods have contributed vastly to our understanding of ecosystems and their functions (Zinger et al. 2012). In the field of microbial water quality assessment, detection methods targeting nucleic acids have expanded our view on the microbial world beyond the minority of bacterial taxa cultivable by classical microbiological methods. Microscopy-based fluorescence in situ hybridisation (FISH) (DeLong et al. 1989) and related techniques allow the detection of single cells in their native habitat (Amann et al. 1995) and the investigation of the distribution and dynamics of specific bacterial populations in natural and engineered systems with relevance for water quality (Farnleitner et al. 2005; Wilhartitz et al. 2007). Polymerase chain reaction (PCR)-based methods allow for the highly specific and sensitive detection and amplification of genes in environmental samples (Bej et al. 1990) and the investigation of complete microbiomes including viruses and phages (Schwab et al. 1993). Based on PCR amplification, various typing methods such as denaturing gradient gel electrophoresis (DGGE) (Muyzer et al. 1993) or terminal restriction fragment length polymorphism (T-RFLP) (Cancilla et al. 1992) have been developed to characterise marker gene communities and thus the corresponding bacterial, archaeal, protozoan, or viral populations.
All these molecular biological methods are based on the utilisation of DNA sequence information to target the respective desired nucleic acid (DNA or RNA). During the last 30 years, Sanger DNA sequencing was the method of choice for obtaining sequence information from target cells. Despite a high degree of automatisation, especially during the sequencing of the human genome, this approach is time consuming and laborious, especially in terms of sample preparation (gene cloning) (Venter et al. 2001). These limitations became particularly evident in metagenomic studies conducted with Sanger approaches (Venter et al. 2004). The advent of next generation sequencing (NGS) platforms, which started in 2005 (Margulies et al. 2005) with the introduction of the 454 GS 20 sequencer, revolutionised DNA sequencing by allowing massively parallel sequencing with millions of reactions running in the same experiment. In the course of the last decade, numerous other platforms have been introduced (e.g. Illumina, Pacific Biosciences, Ion Torrent, SOLiD), yielding up to 600 gigabases of sequence information and up to 4 billion sequence reads per instrument run, usually with relatively short read length of about 150 bases (Quail et al. 2012). The use of multiplex identifiers for sample-specific labelling of nucleic acids allows the analysis of hundreds of samples in parallel in a single instrument run (Hamady et al. 2008).
Since their inception, NGS approaches have been used extensively for the detailed investigation of the microbial consortia of the human microbiome (Hamady & Knight 2009), marine ecosystems (Sogin et al. 2006), or environmental microbiomes in general (Shokralla et al. 2012). Two main approaches are used in these studies: (1) the elucidation of microbial community structure in an environmental sample by deep amplicon sequencing, i.e. the in-depth sequencing of PCR amplicons of a marker gene (most often the 16S rRNA gene); and (2) the metagenomic analysis of the complete DNA or RNA content of an environmental sample, referred to as metagenomics or metatranscriptomics, respectively. The second approach allows surveying for the presence of gene families with distinct metabolic potential in a community (‘What can the community do?’), while amplicon sequencing permits making of a detailed census of the microbial communities with unprecedented resolution (‘Who is there?’). In the water sector, applications of NGS are currently rather limited and there are no studies assessing the applicability of these methods in water quality investigations in general. This lack of investigations is most probably due to the novelty and technical challenges associated with NGS methods (molecular biological and bioinformatic know-how). Also, molecular methods are just at the beginning of being broadly applied in the field of water quality. To remedy this lack of information, this study was initiated to assess the potential applicability and the limitations of 16S rRNA gene amplicon sequencing for water quality assessment in general and the specific detection of faecal pollution in particular. A complex river backwater catchment, which serves multiple purposes (drinking water source, recreation, national park), was selected as the model catchment to sample the compartments of surface water, sediment, and soil, and supplemented by faecal sampling. The research questions of this study were: (i) Is the used NGS approach useful in the development and evaluation of molecular tools for the detection of microbial pollution (e.g. faecal pollution, source tracking)? (ii) Can NGS tools serve as affordable, direct molecular monitoring tools for bulk bacterial communities and changes in their composition from source to tap? (iii) What is the potential of NGS methods for the detection of faecal pollution in environmental waters?
Methods
Sampling, sample processing, and DNA extraction
Samples (n = 29) of different types were collected between June 2010 and May 2011 (Table 1). The main sampling area was the backwater catchment area of the porous groundwater well aquifer (PGWA), where surface water (n = 11), soil (n = 2), sediment (n = 2), and animal faeces (n = 5) were sampled. This riverine wetland is a national park and an important water resource located to the north of the Danube River at the south-eastern border of the city of Vienna, Austria. In addition, faecal samples were collected from a broad range of vertebrate animals at the Vienna Zoo, Austria (n = 9). For soil and sediment samples, material from three cores, taken within an area of 1 m2, was pooled. All samples were aseptically collected in sterile 1,000 ml glass bottles (surface water) or 50 ml plastic vials (soil, sediment, faeces) and kept cool and dark during transport to the laboratory. Samples were stored at ‒20 °C until DNA from soil, sediment, and faecal samples (each approximately 250 mg) was extracted using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, USA) in combination with bead-beating. In order to test the effect of modifications in the DNA extraction procedure, an additional experiment was performed, in which the method was modified by adding glass beads to the kit’s extraction tubes before bead-beating. The DNA of the nine faecal samples from the zoo was extracted both with and without these additional glass beads, totalling 18 DNA extracts. The water samples (250 ml) were filtered immediately after arrival in the laboratory on 0.2 μm polycarbonate membrane filters (Millipore, Bedford, MA). These were stored at ‒20 °C for 6 days until DNA extraction using an adapted CTAB (cetyltrimethyl ammonium bromide) protocol including bead-beating and phenol/chloroform according to Griffiths et al. (2000). The recovered DNA (n = 38) was redissolved in 50 μl of sterile TRIS buffer (10 mM, pH 8). An overview of the samples is given in Table 1. DNA filtration and extraction blanks were included as controls. All DNA extracts were stored at ‒80 °C until analysis within less than 4 months.
Table 1.
Overview of samples (n = 29)
| Sample | Sample type | Source |
|---|---|---|
| W.2016 | Surface water | PGWAa |
| W.1B | Surface water | PGWAa |
| W.87 | Surface water | PGWAa |
| W.2011 | Surface water | PGWAa |
| W.51 | Surface water | PGWAa |
| W.2007 | Surface water | PGWAa |
| W.2003 | Surface water | PGWAa |
| W.42 | Surface water | PGWAa |
| W.60 | Surface water | PGWAa |
| W.48 | Surface water | PGWAa |
| W.2017 | Surface water | PGWAa; Danube River |
| S.87 | Sediment | PGWAa |
| SI.2016 | Sediment | PGWAa |
| B.87 | Soil | PGWAa |
| B.2011 | Soil | PGWAa |
| F.wildboar | Faeces | PGWAa; Sus scrofa (Wild boar) |
| F.reddeer | Faeces | PGWAa; Cervus elaphus (Red deer) |
| F.roedeer | Faeces | PGWAa; Capreolus capreolus (Roe deer) |
| F.fallowdeer | Faeces | PGWAa; Dama dama (Fallow deer) |
| F.mouflon | Faeces | PGWAa; Ovis orientalis musimon (European mouflon) |
| F.iguanab | Faeces | Zoo; Cyclura cornuta (Rhinoceros iguana) |
| F.skinkb | Faeces | Zoo; Corucia zebrata (Solomon Islands skink) |
| F.agamidb | Faeces | Zoo; Pogona barbata (Eastern bearded dragon) |
| F.tortoise1b | Faeces | Zoo; Geochelone elegans (Indian star tortoise) |
| F.tortoise2b | Faeces | Zoo; Malacochersus tornieri (Pancake tortoise) |
| F.tortoise3b | Faeces | Zoo; Testudo sp. (Tortoise) |
| F.meerkatb | Faeces | Zoo; Suricata suricatta (Meerkat) |
| F.orangutanb | Faeces | Zoo; Pongo pygmaeus/abelii (Bornean/Sumatran orangutan) |
| F.lemurb | Faeces | Zoo; Lemur catta (Ring-tailed lemur) |
Porous groundwater well aquifer (PGWA) area.
DNA extraction of these nine samples both with and without additional glass beads to test the effect of modifications in the DNA extraction procedure (resulting in 18 DNA extracts).
PCR amplification, amplicon processing, and pyrosequencing
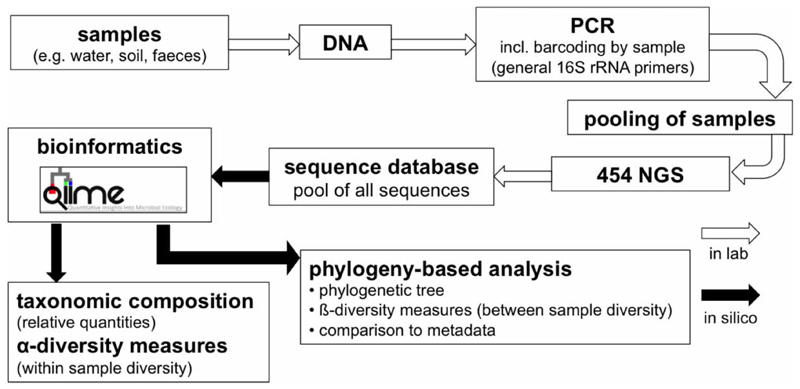
The DNA extracts were used as templates in PCR to amplify the variable regions V1–V2 of the 16S rRNA gene for 25 cycles. All reactions were run in triplicate with the bacterial-specific primers S-D-Bact-0008-a-S-20 (5′-AGAGTTTGATCCTGGCTCAG-3′, as described by Edwards et al. (1989) and S-D-Bact-0338-a-A-19 (5′-TGCTGCCTCCCGTAGGAGT-3′, as described by Etchebehere & (Tiedje 2005)), the latter equipped with a distinct 12-nucleotide error-correcting Golay barcode for each extract as a multiplex tag (Hamady et al. 2008). The nomenclature for the PCR primers was standardised according to Alm et al. (1996). Amplicons were visualised on a 0.8% agarose gel. All samples gave positive results; all controls (filtration, extraction, PCR) were negative and thus not analysed further. Subsequently, the sample amplicons (n = 38) were purified, pooled in equimolar amounts and sent to Selah Clinical Genomic Center, formerly EnGenCore (Columbia, SC, USA) for 454 pyrosequencing (titanium chemistry) (Figure 1).
Figure 1.
NGS pipeline followed in this study in the laboratory and in silico.
Sequence analysis
Sequence analysis was performed using the software package Quantitative Insights Into Microbial Ecology, QIIME (Caporaso et al. 2010). Raw sequences were quality filtered and assigned to the samples according to their barcodes. The remaining flowgrams were denoised to reduce sequencing noise. After removing the primer sequences, chimeric sequences identified by de novo (abundance-based) and reference-based chimera detection with UCHIME were filtered out (Edgar et al. 2011).
The remaining sequences were binned into operational taxonomic units (OTUs) using USEARCH, with a minimum pairwise identity of 97%. Greengenes OTUs (97%; version August 2013) were specified as a reference database at the previous two steps. Rare OTUs represented by less than four sequences were filtered out. Samples yielding less than 1,994 sequences (i.e. fourth smallest number of sequences per sample) were not subjected to further analyses (n = 3). The most abundant sequence in each OTU was chosen as a representative and aligned using PyNAST and the Green-genes reference alignment (DeSantis et al. 2006) trimmed to the V1–V2 region of the 16S rRNA gene with a minimum percent identity of 75%. The hypervariable regions were filtered out with the V1–V2 trimmed version of the lanemask, and a phylogenetic tree was constructed using FastTree (Price et al. 2009). Taxonomy was assigned with the Ribosomal Database Project classifier with a minimum confidence of 80% and the Greengenes taxonomy (August 2013). A total of 1,994 sequences were randomly selected from each sample for further analyses (rarefaction). In order to compare the bacterial communities between the samples, we calculated the pairwise unweighted UniFrac distance metric (Lozupone & Knight 2005) and clustered the resulting matrix using principal coordinate analysis to visualise the phylogenetic relatedness of the bacterial communities (Figure 1). Sequence data from this project is available in the Sequence Read Archive of the National Center for Biotechnology Information under the study accession number SRP055404.
Results and Discussion
This study set out to assess the suitability of an amplicon sequencing approach targeting bacterial 16S rRNA genes using 454 pyrosequencing for the evaluation and development of molecular biological methods in water quality testing as well as a direct tool for monitoring water quality. The test sample set comprised water, sediment, soil, and faecal samples from a backwater study area influenced by the river Danube, as well as faecal samples from various zoo animals. Sequencing yielded 240,944 raw sequence reads assigned to the 38 DNA samples, which were reduced to 136,821 high quality sequences by quality filtering. Subsequent identification and removal of chimeric sequences and rare OTUs further decreased the number to 126,720 reads. The samples W.42, W.1B, and F.orangutan yielded less than 1,994 filtered sequences and were excluded from further analysis.
NGS as a tool for the development and evaluation of molecular detection methods?
Using NGS to evaluate DNA extraction bias
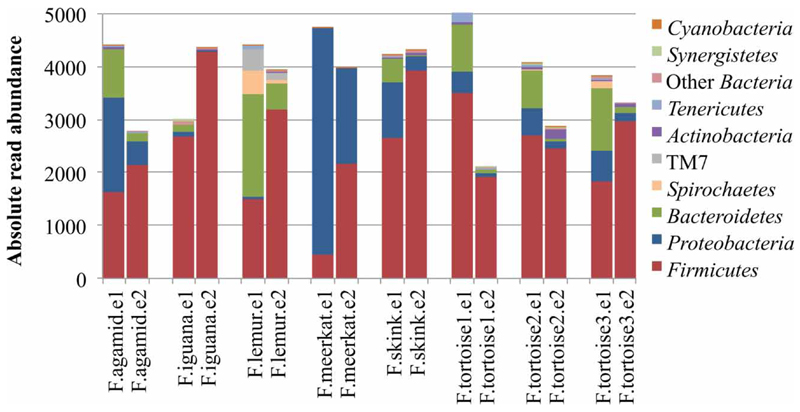
DNA isolation is a (highly) critical step, especially in the application of (semi-)quantitative molecular biological methods on environmental samples. Inappropriate DNA extraction efficiency will bias all subsequent analysis, preventing meaningful biological insights (Feinstein et al. 2009). In this study a commercial kit (MoBio PowerSoil DNA Isolation Kit) was used to extract DNA from faecal samples. To test the effect of different DNA extraction procedures on the community composition detected in the DNA extract, the extraction protocol was modified by adding glass beads to the extraction vials before the initial bead-beating step. This leads to higher mechanical stress on particles, cells, and molecules. Figure 2 shows that the change in procedure led to a distinct shift in the detected community composition on the level of bacterial phyla. The abundance in terms of read number of the dominant phyla Bacteroidetes and Proteobacteria decreased significantly across all samples. Conversely, members of the phylum Firmicutes became much more dominant in the extracts obtained with the harsher extraction, often reaching proportions of greater than 90% of the total community. These results might indicate that the Gram-positive and often spore-forming Firmicutes are more efficiently lysed with the modified procedure, leading to an elevated representation in the results. However, the shift could also be explained by increased shearing of DNA from less resilient bacterial clades, destroying their DNA and making it unavailable for downstream analysis. This experiment demonstrates that the chosen NGS approach was sensitive enough to detect dramatic changes in the composition of DNA extracts caused by relatively minor changes in extraction procedures. The observed effects are most likely to cause biases in all kinds of downstream analysis such as PCR-based methods, and are particularly critical for quantitative approaches (Feinstein et al. 2009).
Figure 2.
Phylum-level bacterial community composition of faecal samples extracted in parallel with the original DNA extraction procedure (e1) and applying the modified, harsher extraction procedure (e2). Results are given in absolute read numbers per sample. Phyla represented by ≤3 sequences are not shown.
NGS as a tool for evaluation of existing molecular methods
The high-resolution, sequence-based picture of community composition in the samples is also useful for the evaluation of existing PCR-based methods targeting dominant populations of the investigated marker gene. The used NGS approach provides a sample-specific ‘sequence database’ that might be searched for binding sites of PCR primers and probes or FISH probes, giving an indication whether the targets of the assays are present and abundant in a sample or a group of samples. Among other applications, this allows in silico evaluation of the source-sensitivity of microbial source tracking assays (Newton et al. 2011) or assays for faecal indication (Vierheilig et al. 2012).
NGS as a tool for the development of novel molecular methods
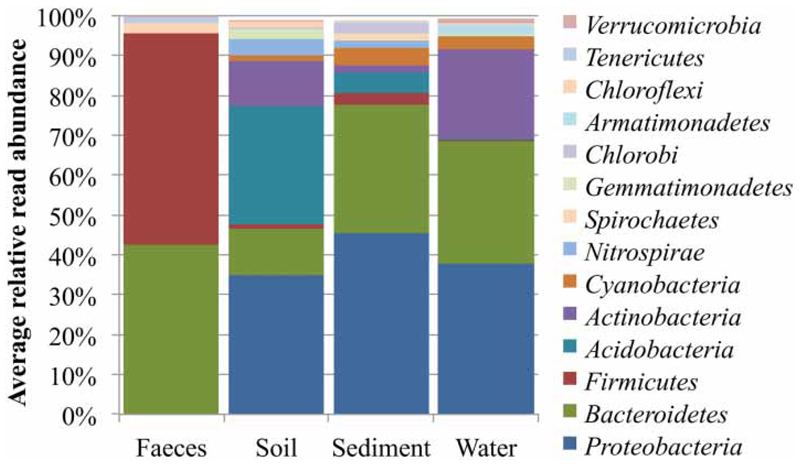
An extensive sample-derived sequence database is ideally suited for the development of novel methods targeting bacterial populations that are represented in the respective sample-derived NGS database. NGS results reveal the relative abundances of the dominant bacterial populations in each sample and thereby give a semi-quantitative indication of potential target populations to inform assay design. Figure 3 shows the bacterial phyla abundances found in the different sample types investigated in this study. It becomes evident that different habitats were dominated by distinct bacterial populations. While faecal communities were dominated by Firmicutes and (to a lesser degree) Bacteroidetes and Proteobacteria, soil samples were dominated by Proteobacteria and Acidobacteria (Figures 2 and 3). Sediment and water communities were more similar to each other and mainly contained members of the phyla Proteobacteria, Bacteroidetes, and significant populations of Cyanobacteria. It should be mentioned that the taxonomic composition of a sample can often be resolved down to the level of bacterial genera. Depending on the read length and quality (Kuczynski et al. 2011). Bacterial populations that are characteristic for a group of samples and highly abundant in that group are thereby considered as ideal targets for molecular diagnostics (Eren et al. 2014). Assay design (primers, probes) can be directly based on the sequence information retrieved by the NGS approach, and a preliminary testing of assay specificity against non-target samples can be performed in silico, as mentioned above (Newton et al. 2011). Alternative methods for the characterisation of bacterial community structures such as DGGE and T-RFLP in fact also allow the highlighting of target populations, but have much lower resolution and do not directly provide sequence information. In contrast, the unprecedented depth and information density provided by NGS approaches form a much more stable basis for state-of-the-art assay development.
Figure 3.
Bacterial phyla found in the faeces (n = 5), soil (n = 2), sediment (n = 2) and water (n = 9) samples from the PGWA study area. Results are average abundances as a percentage of the complete community. Phyla that could be detected with an average abundance of <1% are not shown.
NGS as a tool for microbial water quality monitoring?
NGS for the characterisation of microbial diversity
The high-resolution insight into community composition provided by deep amplicon sequencing makes it a valuable tool for monitoring of microbial communities in natural as well as technical aquatic systems (Lin et al. 2012). It is particularly suited for the investigation of temporal or spatial changes in the community composition as well as for the identification of drivers triggering changes along environmental gradients (Fierer et al. 2012).
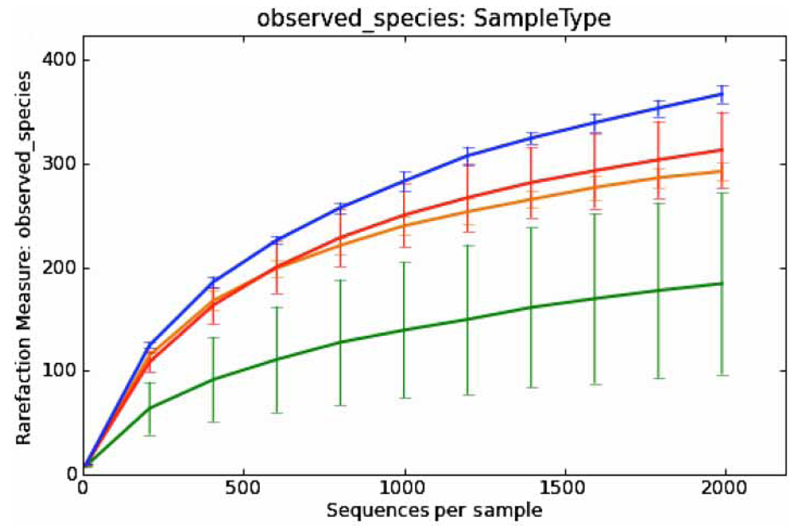
To assess whether the applied sequencing depth in this study (minimum of 1,994 sequence reads per sample) was sufficient to give a representative impression of the community in the samples, α-diversity measures were estimated by rarefaction analysis (Figure 4). The analysis made evident that the complete diversity was unveiled in none of the four sample types: faeces, soil, sediment, and water. Diversity was much lower in water samples than in faeces, soil, and sediment, indicating that for this habitat lower sequencing depth might be sufficient. This finding is in accordance with literature data showing that soil habitats have very high bacterial diversity when compared to water or intestinal systems (Ley et al. 2008). One of the strengths of NGS approaches is that sequencing depth and effort can and indeed have to be adapted to the complexity of the investigated environment in order to ensure efficient use of resources and provide meaningful results.
Figure 4.
Rarefaction analysis estimating average α-diversity by counting the observed species in the samples of sediment (n = 2), soil (n = 2), faeces (n = 5) and water (n = 9). Error bars denote the standard deviation.
NGS revealing bulk microbial community structure and dynamics
As shown in Figure 3 the taxonomic composition of the microbial community can be derived directly from the NGS results. That in itself can give crucial insights into the constitution and status of the investigated sample by identifying signature taxa or monitoring quantitative shifts between dominant taxa with known traits. Beyond and independent of taxonomic identification, a deep amplicon sequencing database allows the investigation of the relatedness of communities in different samples based on the phylogenetic history of their members.
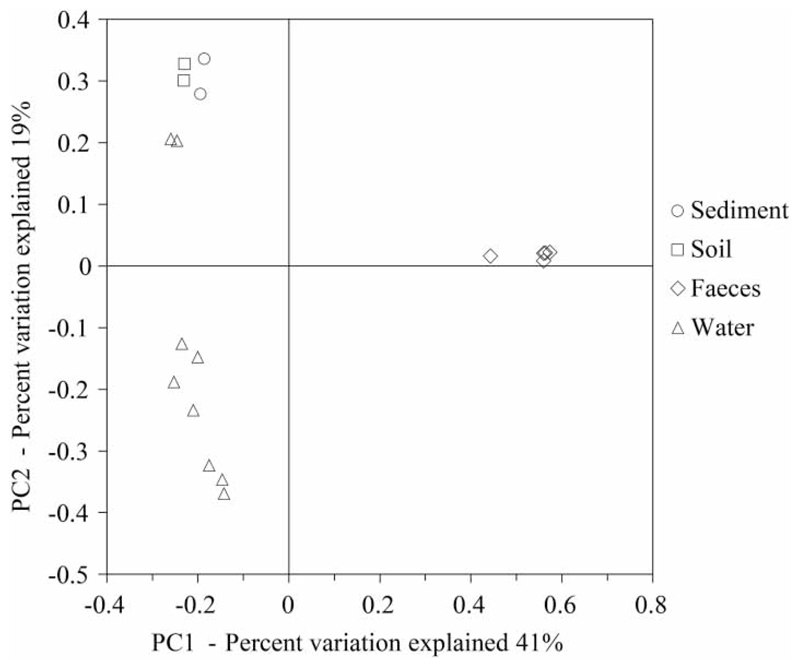
In order to investigate the diversity between samples (β-diversity) in this study, the sequence reads of each sample were aligned to a 16S rRNA gene reference alignment, from which a phylogenetic tree was constructed. This tree, representing the phylogenetic composition of the samples, was used to calculate the so-called UniFrac metric, which serves as a distance measure for β-diversity, i.e. a measure for assessing how closely related two communities are in terms of shared evolutionary ancestry of their constituents. The resulting UniFrac distance matrix was subjected to cluster analysis to visualise which communities are more closely related and which are more distinct (Figure 5). Faecal communities were clearly set apart from other sample types while soil and sediment communities were closely related. This is not surprising because the study area is a backwater area regularly inundated during flooding. Interestingly, two of the water samples exhibited communities more closely related to soil and sediment samples, namely the Danube River water sample W.2017 and the PGWA sample W.2016, which was taken directly adjacent to the Danube in a branch of the river. All other water-sampling sites are only connected to the river during flood events. Taken together, these results demonstrate that the chosen NGS approach is indeed able to resolve spatial heterogeneities in community composition of the dominant bacterial populations in a sample and will therefore be a useful tool for the monitoring of both spatial and temporal changes in community composition in the environment. This has been demonstrated previously in several studies which successfully monitored microbial community changes in drinking water treatment and distribution systems (Hong et al. 2010; Pinto et al. 2014) and wastewater (Ye et al. 2011; Shanks et al. 2013).
Figure 5.
Visualisation of a principle coordinate analysis of the β-diversity (between sample diversity) as calculated by the phylogeny-based, unweighted UniFrac metric. In this analysis, samples with phylogenetically more similar communities cluster more closely together.
NGS for the detection of faecal pollution in water
In contrast to bulk bacterial community analysis, other investigators suggested the use of NGS-derived community signatures as a tool for identifying faecal pollution sources in water (Unno et al. 2010; Newton et al. 2013). Other scientists proposed the use of NGS approaches for the direct detection of pathogens in wastewater treatment plants (Ye & Zhang 2011; Cai & Zhang 2013). These applications highlight one of the basic restrictions of deep amplicon sequencing of total bacterial communities, which is the problem of relative abundances of target and background populations. Wastewater or faecal bacterial communities become rapidly diluted when entering environmental waters. In addition, pathogens constitute only a very minor portion of wastewater bacterial communities in the first place. To exemplify this, we searched for the commonly used faecal indicator Escherichia coli in the sequencing results of this study. Although E. coli can be detected by cultivation in high abundances in most faecal samples (Farnleitner et al. 2010) and was consistently cultivated in the water samples included in this study (concentrations ranging from 7 to >300 colony forming units per 100 ml), we were unable to find a single sequence read related to that species or even the genus Escherichia in the entire dataset. Faecal indicators and, to an even greater degree, pathogens are quantitatively very minor constituents even of faecal communities. These results highlight that NGS amplicon pyrosequencing using general bacterial primers is indeed able to detect abundant bulk populations in a community (e.g. Bacteroidetes or Firmicutes in faeces) but, at the applied sequencing depth, is not able to detect very low abundant populations that often are of relevance for the microbiological quality assessment of water (faecal indicators and pathogens) (Cai & Zhang 2013). Ipso facto, it is evident that the dilution in water resources limits the capability of any NGS method to find these target populations in a background of autochthonous, i.e. ‘native’ populations (Farnleitner et al. 2005; Ye & Zhang 2011). One way to circumvent this problem is to use group-specific primers instead of general primers targeting most bacteria (Unno et al. 2012), although this sacrifices the general overview and broad focus provided by total community analysis. Another possibility is the substantial increase of sequencing depth by at least two orders of magnitude. Novel, very recently emerging sequencing technologies and platforms (Liang & Zhang 2015) might offer sequencing depths that are also able to detect (very) low abundant populations of interest.
Other possible methodical restrictions that should be considered are the relatively short read length of current NGS methods and the sequencing error rate that might suggest a level of diversity that is actually not present in the sample (‘rare biosphere problem’)(Reeder & Knight 2009). These problems can be overcome by conservative and careful data analysis and interpretation. Furthermore, one also has to keep in mind that results of NGS amplicon sequencing do not provide quantitative concentrations but relative quantities as related to the total gene community. Additionally, all biases associated with the application of PCR methods also apply to deep amplicon sequencing (von Wintzingerode et al. 1997). In contrast, this is not the case when applying metagenomic sequencing approaches, which do not employ gene-specific primers for DNA amplification and therefore avoid the respective biases (Cai & Zhang 2013). However, these approaches require much higher sequencing effort and bioinformatic analysis resources. With NGS sequencing services getting cheaper by the month, concomitant with increasing sequencing yield and quality, the main technical bottleneck will be the handling of the enormous amounts of data provided by these methods. Today there are precious few ready-made tools for data analysis available, and bioinformatics expertise is in short supply. This topic highlights the necessity to formulate clear hypotheses and research questions before starting an NGS-based investigation.
The currently used applications of NGS in water quality monitoring are still rather demanding in terms of necessary expertise and equipment, i.e. they require the availability of a molecular biological laboratory for sample processing. Although sequencing facilities offer full service packages for amplicon sequencing, metagenomic sequencing, and even preliminary bioinformatic analysis of the results, the costs of NGS analysis remain rather high and the high-throughput NGS methods are not well suited to small-scale investigations. Despite current attempts to establish NGS-based analysis pipelines to the needs of the water industry (Unno et al. 2012), it remains one of the main challenges of the coming years to make these technologies accessible also to facilities for practical application in the water sector.
Conclusions
The results of this study demonstrate that deep amplicon sequencing of the 16S rRNA marker gene using NGS methods could be a valuable tool for many applications in water quality monitoring. It is useful for the development and evaluation of molecular diagnostic tools to detect abundant bacterial indicators in water resources (McLellan & Eren 2014). But for the detection of pathogens or faecal indicators with low abundances in the environment, however, amplicon sequencing of bulk bacterial communities is not sufficiently sensitive at the applied sequencing depth. Deeper sequencing as provided by new NGS approaches and technologies might prove up to this task. However, the results of the present study demonstrate that this method is indeed capable of unveiling the dominant or bulk bacterial communities in water samples. The approach, applied on environmental water samples in this study, can be directly translated to the monitoring of bacterial communities in water treatment plants and distribution systems, where it can provide unprecedented insights into efficiency of measures and biostability of water (Roeselers et al. 2015). At present, NGS approaches remain mainly a highly powerful research tool with the potential to fundamentally revolutionise our knowledge about microbial content and dynamics in water resources and treatment. In order to translate the novel methods and findings into useful and accessible solutions for the practitioner in the water field, research and future development will have to supply standardised laboratory procedures and, in particular, data analysis pipelines and software tools as well as specialised sequence databases tailored to the requirements of water quality assessment.
Acknowledgements
This study was financed by the Austrian Science Fund (FWF) project P22032 granted to Georg Reischer. Georg Reischer is a recipient of an APART Fellowship of the Austrian Academy of Sciences. Further support came from the FWF project P23900 granted to Andreas Farnleitner and the FWF DKplus ‘Vienna Doctoral Programme on Water Resource Systems’ (W1219-N22) granted to Günter Blöschl and Andreas Farnleitner. This study is a joint publication of the Interuniversity Cooperation Centre Water & Health (www.waterandhealth.at).
Contributor Information
J. Vierheilig, Research Group Environmental Microbiology and Molecular Ecology, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria; Centre for Water Resource Systems (CWRS), Vienna University of Technology, Karlsplatz 13/222, A-1040 Vienna, Austria
D. Savio, Research Group Environmental Microbiology and Molecular Ecology, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria; Centre for Water Resource Systems (CWRS), Vienna University of Technology, Karlsplatz 13/222, A-1040 Vienna, Austria
R. E. Ley, Department of Microbiology, Cornell University, Ithaca, NY 14853, USA
R. L. Mach, Gene Technology Group, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria
A. H. Farnleitner, Research Group Environmental Microbiology and Molecular Ecology, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria; Interuniversity Cooperation Centre Water & Health, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria
G. H. Reischer, Research Group Environmental Microbiology and Molecular Ecology, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria; Interuniversity Cooperation Centre Water & Health, Institute for Chemical Engineering, Vienna University of Technology, Gumpendorfer Straße 1a, A-1060 Vienna, Austria
References
- Alm EW, Oerther DB, Larsen N, Stahl DA, Raskin L. The oligonucleotide probe database. Applied and Environmental Microbiology. 1996;62(10):3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiology Reviews. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bej AK, Steffan RJ, Dicesare J, Haff L, Atlas RM. Detection of coliform bacteria in water by polymerase chain-reaction and gene probes. Applied and Environmental Microbiology. 1990;56(2):307–314. doi: 10.1128/aem.56.2.307-314.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Zhang T. Detecting human bacterial pathogens in wastewater treatment plants by a high-throughput shotgun sequencing technique. Environmental Science & Technology. 2013;47(10):5433–5441. doi: 10.1021/es400275r. [DOI] [PubMed] [Google Scholar]
- Cancilla MR, Powell IB, Hillier AJ, Davidson BE. Rapid genomic fingerprinting of Lactococcus lactis strains by arbitrarily primed polymerase chain-reaction with P-32 and fluorescent labels. Applied and Environmental Microbiology. 1992;58(5):1772–1775. doi: 10.1128/aem.58.5.1772-1775.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Wickham GS, Pace NR. Phylogenetic stains – ribosomal RNA-based probes for the identification of single cells. Science. 1989;243(4896):1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Haas B, Clemente J, Quince C, Knight R. UCHIME Improves sensitivity and speed of chimera detection. Bioinformatics. 2011;15(27):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards U, Rogall T, Blocker H, Emde M, Bottger EC. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Research. 1989;17(19):7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. A single genus in the gut microbiome reflects host preference and specificity. The ISME Journal. 2014;9(1):90–100. doi: 10.1038/ismej.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchebehere C, Tiedje J. Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor. Applied and Environmental Microbiology. 2005;71(9):5642–5645. doi: 10.1128/AEM.71.9.5642-5645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnleitner AH, Wilhartitz I, Ryzinska G, Kirschner AK, Stadler H, Burtscher MM, Hornek R, Szewzyk U, Herndl G, Mach RL. Bacterial dynamics in spring water of alpine karst aquifers indicates the presence of stable autochthonous microbial endokarst communities. Environmental Microbiology. 2005;7(8):1248–1259. doi: 10.1111/j.1462-2920.2005.00810.x. [DOI] [PubMed] [Google Scholar]
- Farnleitner AH, Ryzinska-Paier G, Reischer GH, Burtscher MM, Knetsch S, Kirschner AKT, Dirnböck T, Kuschnig G, Mach RL, Sommer R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. Journal of Applied Microbiology. 2010;109:1599–1608. doi: 10.1111/j.1365-2672.2010.04788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein LM, Sul WJ, Blackwood CB. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Applied and Environmental Microbiology. 2009;75(16):5428–5433. doi: 10.1128/AEM.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. The ISME Journal. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Applied and Environmental Microbiology. 2000;66(12):5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Research. 2009;19(7):1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nature Methods. 2008;5(3):235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong PY, Hwang CC, Ling FQ, Andersen GL, LeChevallier MW, Liu WT. Pyrosequencing analysis of bacterial biofilm communities in water meters of a drinking water distribution system. Applied and Environmental Microbiology. 2010;76(16):5631–5635. doi: 10.1128/AEM.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Current Protocols in Bioinformatics. 2011;36(10.7):10.7.1–10.7.20. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews Microbiology. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Zhang PM. Nanopore DNA sequencing: Are we there yet? Science Bulletin. 2015;60(3):296–303. [Google Scholar]
- Lin X, McKinley J, Resch C, Kaluzny R, Lauber C, Fredrickson J, Knight R, Konopka A. Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. The ISME Journal. 2012;6(9):1665–1676. doi: 10.1038/ismej.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. Unifrac: a new phylogenetic method for comparing microbial communities. Applied and Environmental Microbiology. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan SL, Eren AM. Discovering new indicators of fecal pollution. Trends in Microbiology. 2014;22(12):697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Applied and Environmental Microbiology. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, VandeWalle JL, Borchardt MA, Gorelick MH, McLellan SL. Lachnospiraceae and Bacteroidales alternative fecal indicators reveal chronic human sewage contamination in an urban harbor. Applied and Environmental Microbiology. 2011;77(19):6972–6981. doi: 10.1128/AEM.05480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, Bootsma MJ, Morrison HG, Sogin ML, McLellan SL. A microbial signature approach to identify fecal pollution in the waters off an urbanized coast of Lake Michigan. Microbial Ecology. 2013;65(4):1011–1023. doi: 10.1007/s00248-013-0200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto AJ, Schroeder J, Lunn M, Sloan W, Raskin L. Spatial-temporal survey and occupancy-abundance modeling to predict bacterial community dynamics in the drinking water microbiome. Mbio. 2014;5(3):e01135–14. doi: 10.1128/mBio.01135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Molecular Biology and Evolution. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M, Smith M, Coupland P, Otto T, Harris S, Connor T, Bertoni A, Swerdlow H, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13(1):341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Knight R. The ‘rare biosphere’: a reality check. Nature Methods. 2009;6(9):636–637. doi: 10.1038/nmeth0909-636. [DOI] [PubMed] [Google Scholar]
- Roeselers G, Coolen J, van der Wielen PW, Jaspers MC, Atsma A, de Graaf B, Schuren F. Microbial biogeography of drinking water: patterns in phylogenetic diversity across space and time. Environmental Microbiology. 2015;17(7):2505–2514. doi: 10.1111/1462-2920.12739. [DOI] [PubMed] [Google Scholar]
- Schwab KJ, Deleon R, Sobsey MD. Development of PCR methods for enteric virus detection in water. Water Science and Technology. 1993;27(3–4):211–218. [Google Scholar]
- Shanks OC, Newton RJ, Kelty CA, Huse SM, Sogin ML, McLellan SL. Comparison of the microbial community structures of untreated wastewaters from different geographic locales. Applied and Environmental Microbiology. 2013;79(9):2906–2913. doi: 10.1128/AEM.03448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Molecular Ecology. 2012;21(8):1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno T, Jang J, Han D, Kim JH, Sadowsky MJ, Kim OS, Chun J, Hur HG. Use of barcoded pyrosequencing and shared OTUs to determine sources of fecal bacteria in watersheds. Environmental Science & Technology. 2010;44(20):7777–7782. doi: 10.1021/es101500z. [DOI] [PubMed] [Google Scholar]
- Unno T, Di D, Jang J, Suh Y, Sadowsky M, Hur H. Integrated online system for a pyrosequencing-based microbial source tracking method that targets Bacteroidetes16S rDNA. Environmental Science & Technology. 2012;46(1):93–98. doi: 10.1021/es201380c. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, et al. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, Fouts DE, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304(5667):66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vierheilig J, Farnleitner AH, Kollanur D, Blöschl G, Reischer GH. High abundance of genetic Bacteroidetes markers for total fecal pollution in pristine alpine soils suggests lack in specificity for feces. Journal of Microbiological Methods. 2012;88(3):433–435. doi: 10.1016/j.mimet.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: Pitfalls of PCR-based rRNA analysis. FEMS Microbiology Reviews. 1997;21(3):213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- Wilhartitz I, Mach RL, Teira E, Reinthaler T, Herndl GJ, Farnleitner AH. Prokaryotic community analysis with CARD-FISH in comparison with FISH in ultra-oligotrophic ground- and drinking water. Journal of Applied Microbiology. 2007;103(4):871–881. doi: 10.1111/j.1365-2672.2007.03319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Zhang T. Pathogenic bacteria in sewage treatment plants as revealed by 454 pyrosequencing. Environmental Science & Technology. 2011;45(17):7173–7179. doi: 10.1021/es201045e. [DOI] [PubMed] [Google Scholar]
- Ye L, Shao MF, Zhang T, Tong AHY, Lok S. Analysis of the bacterial community in a laboratory-scale nitrification reactor and a wastewater treatment plant by 454-pyrosequencing. Water Research. 2011;45(15):4390–4398. doi: 10.1016/j.watres.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Zinger L, Gobet A, Pommier T. Two decades of describing the unseen majority of aquatic microbial diversity. Molecular Ecology. 2012;21(8):1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]