Abstract
This was a detailed investigation of the seasonal occurrence, dynamics, removal and resistance of human-associated genetic Bacteroidetes faecal markers (GeBaM) compared with ISO-based standard faecal indicator bacteria (SFIB), human-specific viral faecal markers and one human-associated Bacteroidetes phage in raw and treated wastewater of municipal and domestic origin. Characteristics of the selected activated sludge wastewater treatment plants (WWTPs) from Austria and Germany were studied in detail (WWTPs, n = 13, connected populations from 3 to 49000 individuals), supported by volume-proportional automated 24-h sampling and chemical water quality analysis. GeBaM were consistently detected in high concentrations in raw (median log10 8.6 marker equivalents (ME) 100 ml−1) and biologically treated wastewater samples (median log10 6.2–6.5 ME 100 ml−1), irrespective of plant size, type and time of the season (n = 53–65). GeBaM, Escherichia coli, and enterococci concentrations revealed the same range of statistical variability for raw (multiplicative standard deviations s* = 2.3–3.0) and treated wastewater (s* = 3.7–4.5), with increased variability after treatment. Clostridium perfringens spores revealed the lowest variability for raw wastewater (s* = 1.5). In raw wastewater correlations among microbiological parameters were only detectable between GeBaM, C. perfringens and JC polyomaviruses. Statistical associations amongst microbial parameters increased during wastewater treatment. Two plants with advanced treatment were also investigated, revealing a minimum log10 5.0 (10th percentile) reduction of GeBaM in the activated sludge membrane bioreactor, but no reduction of the genetic markers during UV irradiation (254 nm). This study highlights the potential of human-associated GeBaM to complement wastewater impact monitoring based on the determination of SFIB. In addition, human-specific JC polyomaviruses and adenoviruses seem to be a valuable support if highly specific markers are needed.
Keywords: HF183 TaqMan, BacHUM, AllBac, Escherichia coli, Clostridium perfringens, Enterococci, JC polyomavirus (JCPyV), Human adenovirus (HAdV), Bacteriophage thetaiotaomicron
1. Introduction
Contamination of aquatic systems by wastewater of human origin can pose a serious threat to public health because it frequently contains high numbers of intestinal pathogens (Stevens et al., 2009). Appropriate disposal systems combined with efficient faecal pollution monitoring techniques for municipal and domestic wastewater are thus essential for safeguarding our water resources. Wastewater treatment plants (WWTPs) based on primary (mechanical), secondary (biological), and tertiary (enhanced biological and chemical) treatment are designed to remove organic carbon (C), nitrogen (N) and phosphorus (P) out of wastewater to a great extent. Although providing a first essential barrier, conventional WWTPs are not built to sufficiently remove microbial faecal loads to support the safe use of effluent wastewater for human related activities, such as recreational purposes or irrigation. Disinfection of wastewater effluents has not yet become a common standard in most regions of the world, and such advanced treatment is often restricted to the discharge of wastewater into sensitive aquatic areas. Rainfall events may also lead to a bypass of WWTPs (i.e. combined sewer overflows) and the contamination of water resources with raw wastewater (Molina et al., 2014; Shibata et al., 2014; Tryland et al., 2014).
Routine monitoring of microbial faecal pollution in the aquatic environment is still based on the selective cultivation of standard faecal indicator bacteria (SFIB), including Escherichia coli and intestinal enterococci (ISO, 2005). Without doubt, water quality testing based on the application of SFIB has contributed to a fundamental improvement in water safety management since the end of the 19th century (Tallon et al., 2005). However, the application of SFIB has also recently been subjected to increasing criticism (Ishii and Sadowsky, 2008). Several studies suggested that SFIB in aquatic habitats also originate from non-enteric compartments, such as soil, sediment and algae (Byappanahalli et al., 2012; Desmarais et al., 2001; Whitman et al., 2003). In addition, typing of isolated SFIB strains, to recover information about their origin, requires the formation of unrealistically large catchment-specific strain libraries and does currently not provide a feasible option for monitoring applications (Domingo et al., 2007). These limitations obviously call for additional indicators and tools to complement the existing standard methods to obtain a more detailed and certain view on the existing faecal pollution patterns to support MST and risk assessment (Harwood et al., 2014).
Amongst the vast number of alternative parameters (Hagedorn et al., 2011; Wuertz et al., 2011), PCR-based assays for the analysis of human-associated genetic Bacteroidetes faecal markers (GeBaM) have gained increasing popularity in the field of faecal pollution analysis and microbial source tracking (MST) during recent years (Harwood et al., 2014). Quantitative PCR (qPCR)-based GeBaM assays for general-, human-, wastewater-, or animal-associated faecal sources have been developed (Kildare et al., 2007; Layton et al., 2006; Reischer et al., 2006; Shanks et al., 2009). Several evaluation studies including various aquatic environments successfully demonstrated the value of GeBaM diagnostics (Boehm et al., 2009; Reischer et al., 2011; Ridley et al., 2014; Riedel et al., 2014; Sauer et al., 2011; Tambalo et al., 2012). However, the application of qPCR-based GeBaM assays is not yet standardized. It requires careful study design and background information on the catchment to create unbiased results and to recognize methodical limits (Boehm et al., 2013; Reischer et al., 2011).
A useful parameter for the analysis of general- or host associated microbial faecal pollution in water has to fulfil several basic performance criteria, including source-sensitivity and source-specificity (Wuertz et al., 2011). Considerable effort has been dedicated to sensitivity and specificity testing of GeBaM qPCR assays during recent years, most frequently based on individual sampling strategies covering various sources of animal and human excreta or wastewater (Ahmed et al., 2013; Boehm et al., 2013; Keity et al., 2012; Reischer et al., 2013, 2011; Riedel et al., 2014; Shanks et al., 2009). Emphasis has also been put on sampling techniques, DNA extraction, and PCR quantification procedures (Cankar et al., 2006; Karlen et al., 2007; Shanks et al., 2012; Siefring et al., 2008; Stoeckel et al., 2009). However, information on the occurrence of GeBaM in wastewater regarding the characteristics of the disposal system (combined and separate sewer systems), its seasonal variability, and its relationship to standard and alternative faecal indicators is scarce (Srinivasan et al., 2011).
The aim of this study was to investigate the prevalence and abundance of human-associated GeBaM by qPCR determination in raw and treated wastewater of well-characterized municipal wastewater treatment plants over one year. Emphasis was put on the selection of municipal WWTPs with primary, secondary, and tertiary treatment, as such systems are representative for the situation in Austria and the Central European Region (CER). Small domestic WWTP (dWWTPs) were also included in our investigation, as they are frequently implemented in remote areas, where the connection to municipal sewer systems is not possible. Although advanced treatment was not the main focus of this study, the investigation of UV disinfection at one selected WWTP was included, as such treatment is becoming increasingly important. The Taqman HF183 qPCR assay (Haugland et al., 2010) and the BacHUM qPCR assay (Kildare et al., 2007) were selected for the determination of human-associated GeBaM concentrations, following recommendations of recent evaluation studies (Boehm et al., 2013; Layton et al., 2013; Reischer et al., 2013). To support methodical cross-comparisons, cultivation-based SFIB using ISO standard methods and viral faecal markers for human-specific faecal pollution were simultaneously determined. Among these, JC polyomavirus (JCPyV) as well as human adenoviruses (HAdV), which have been used as human faecal viral indicators and highly specific MST tools (Bofill-Mas et al., 2000; Pina et al., 1998), and bacteriophages infecting Bacteroides thetaiotaomicron, which have been proposed as a human-associated faecal indicator, were tested. Raw and treated wastewater was also analysed by chemical standard methods to support treatment plant characterisation and comparison of elimination characteristics between microbial and chemical parameters.
2. Materials and methods
2.1. Selection criteria and parameters to characterize the sewer disposal systems and WWTPs
The Danube Region and other parts of the CER are defined as sensitive areas with respect to water bodies. In terms of nutrients, strict discharge limits for WWTPs according to the EU urban wastewater directive have been established (EC, 1991). Wastewater disposal is caused by very small (i.e., a few inhabitants) up to large treatment systems (>100 000 persons connected), as a mix of rural areas and large cities characterize this region. Activated sludge treatment is the common process to treat the wastewater. In Austria, WWTP effluent concentrations are restricted by certain removal performance targets related to the influent load (%) and by a maximum effluent concentration (mg L−1). These limits depend on the plant size (AEV, 1996). Only municipal WWTPs providing data for a basic characterization over the investigation period were selected. Essential information on WWTP design, including design capacity (p.e., population equivalent), actual average loading inhabitants connected, type of treatment (mechanical (M), carbon removal (C), nitrification (N), denitrification (D), phosphorus removal (P)), advanced treatment available (UV irradiation, membrane filtration), removal efficiency of nutrients (C, N and P), and sludge age, were required. To characterize the raw wastewater quantity and quality over the investigation period, volumetric flow rate (Q), chemical oxygen demand (COD) or biological oxygen demand (BOD), total nitrogen (TN), total phosphorus (TP), temperature (°C), and pH were requested. In addition to these parameters, ammonium (NH4), nitrate (NO3), and suspended solids (SS) were included for the treated wastewater. All of these flow and chemical data were provided on a daily basis because they were available from representative sampling by automated and cooled sampling devices (24 h proportional-flow sampling). All chemical information provided was cross-checked by our own investigations of the investigated samples (see below).
In contrast to municipal WWTP, data availability for dWWTPs (<150 p.e.) was very low. The basic requirements were information on the type of treatment system, the total number of persons connected, and the effluent concentrations of COD, NH4 and pH. Only information based on grab sample analysis was available.
2.2. Sampling for chemical and microbiological analysis
60 influent and 60 effluent 24 h volume-proportional composite samples were recovered by fixed installed and cooled (4 °C) automatic sampling devices (cf. supplemental material) from raw and treated municipal wastewater. Further background information on automated sampling in WWTP can also be found in Mayer et al. (Mayer et al., 2015). 2 L of samples were collected during the automated sampling process in sterilized 2 L glass bottles and immediately transferred to the laboratory at 5 ± 3 °C for further analysis within 8 h. To cover seasonal variations, samples were taken in 4- to 6-week intervals over one annual cycle. Following a homogenization by manual shaking, 1 L of the sample volume of each sample was used for chemical and microbiological analysis.
In summer and fall (2013) 16 grab samples were taken at the effluent discharge unit of the selected dWWTPs. 2 L of samples were collected in sterilized 2 L glass bottles and immediately transferred to the laboratory at 5 ± 3 °C for further analysis within 8 h. Following a homogenization by manual shaking, a volume of 1 L was used for chemical and microbiological investigations each.
To evaluate the effect of UV irradiation on GeBaM and other microbiological indicators, 10 additional pairs of 1 L grab samples (before and after the UV system) were taken at the WWTP5 throughout the season (UV-2000, Trojan Technologies, Canada; 48 UV lamps).
2.3. Chemical analysis
For COD, BOD5, TP, TN, pH, SS, and conductivity analysis, the first preparation step included the homogenization of the sample by manual agitation. A pre-filtration step, applying a 0.45 µm membrane (sterilized cellulose-nitrate filter), was needed to analyse the dissolved parameters PO4–P, NH4–N, NO2–N, NOx-N. All selected parameters were performed according to standardized methods, as given in detail in the additional material section (Table S2 additional material).
2.4. Quantification of genetic Bacteroidetes markers (GeBaM) by qPCR
For DNA extraction, polycarbonate membrane filtration (0.2 µm Millipore, Isopore Membrane Filter - GTTP) followed by phenol/chloroform extraction of 20 ml influent, 50 ml effluent and 1500 ml membrane-filtrated effluent, was used as previously described (Griffiths et al., 2000; Reischer et al., 2006). Every filtration event was monitored by a minimum of two blank filter controls and every extraction event (equals 11 samples) was also followed by a blank extraction control. The controls were processed in exactly the same way as the samples. Cells were lysed with a FastPrepR-24 Instrument (MP Biomedicals Inc., Irvine, USA) with a speed setting of 6 for 30 s. The extracted DNA was dissolved in 10 mM TRIS HCl pH = 8 and stored at −80 °C not longer than 21 days before PCR testing. Respective 16S-rRNA-gene markers for AllBac (Layton et al., 2006), BacHUM (Kildare et al., 2007), and HF183 TaqMan (Haugland et al., 2010) were quantified by qPCR. The rotor-discs and 96-well plates were loaded with the mastermix and the sample DNA using a Qiagility Roboter (Qiagen, Hilden, Germany). The measurements were subsequently performed on a Rotorgene Q Cycler (Qiagen, HILDEN, Germany). For the AllBac qPCR assay we used 2.5 µl of the respective DNA sample dilution, 600 nmol L−1 primer AllBac296f, 600 nmol L−1 primer AllBac412r, 25 nmol L−1 TaqMan MGB probe AllBac375Bhqr (Layton et al., 2006), 0.4 g L−1 bovine serum albumin (Roche Diagnostics, Mannheim, Germany), 7.5 µl of iQ Supermix (Bio-Rad, Hercules, USA) in a total reaction volume of 15 µl; additionally, 5 mmol L−1 MgCl2 was added to obtain a total Mg2+ concentration of 8 mmol L−1 (Layton et al., 2006). For the BacHUM assay we used 2.5 µl of the respective DNA sample dilution, 400 nmol L−1 primer BacHUM-160f, 400 nmol L−1 primer BacHUM-241r, 80 nmol L−1 TaqMan MGB probeeBacHUM-193p (Kildare et al., 2007), 0.4 g L−1 bovine serum albumin, and 7.5 µl of iQ Supermix in a total reaction volume of 15 ml. For the HF183 TaqMan assay, we used 2.5 µl of the respective DNA sample-dilution, 100 nmol L−1 primer HF183, 100 nmol L−1 primer BFD REV, 80 nmol L−1 TaqMan MGB probe eBFD FAM (Haugland et al., 2010) 0.4 g l L−1 bovine serum albumin, and 7.5 ml of iQ Supermix in a total reaction volume of 15 ml. The PCR program for the AllBac assay was initial 95 °C for 3 min and for the cycles of 95 °C for 30 s and 60 °C for 45 s; for the BacHUM assay, initial 95 °C for 3 min and for the cycles of 95 °C for 15 s and 60 °C for 1 min; for the HF183 TaqMan assay, initial 95 °C for 3 min and for the cycles of 95 °C for 15 s and 60 °C for 30 s. The real-time data were collected during the primer annealing step at 60 °C. Quantification was based on plasmid standard dilutions and given as molecular equivalent targets per volume (ME vo−1) as previously described (Reischer et al., 2006). The respective plasmid stock (cf. supplemental material) for each assay was diluted in an unspecific 500 ng ml−1 poly(dI-dC) (Roche Diagnostics, Mannheim, Germany) background to avoid adsorption of plasmid DNA to reaction vials at low plasmid concentrations. A total of at least seven tenfold serial dilutions of plasmid standard (100–106 gene copies) were run in each qPCR experiment.
Each DNA sample was analysed in two dilution steps (10- and 100-fold dilution) with each dilution in duplicate to check for a possible PCR inhibition. In case the quotient of the concentration ratio (after correction with the dilution factor) was in between the range of 0.5–2, inhibition was assumed to be absent (i.e. correspondence of results from different dilutions). No signs of PCR inhibition could be detected for any of the applied qPCR assays. Inhibition was also analysed by dilution plot analysis (cf. supplemental material) in which the concentrations of the two dilutions are shown on the x vs. y axes. Dilution plot analysis resulted in a very good correlation (R2) and a 1:1 slope amongst the concentrations, as revealed for the different dilutions (cf. Fig. S1, supplementary material). Every qPCR run also included a no template control in duplicates (only mastermix, primers and probe) to monitor for possible contaminations. All qPCR runs in this study revealed a calculated PCR efficiency of between 90% and 105% and coefficient of determination (R2) higher than 0.99 for the calibration curve. The no-template controls were consistently negative. The range of quantification was linked to the calibration curve covering concentrations in between 102 - 107 targets per reaction. To introduce a process control for the filtration and extraction step, an E. coli cell standard carrying a diagnostic gene fragment for qPCR detection at the chromosome was applied. The so called defined genetic target number cell standard (DeTaCs) was directly spiked into 50% of the collected wastewater samples as previously described (Kaiblinger, 2008) at concentrations of log10 7.0 (details on the design, production and detection of the DeTaCs are given in detail in the supplementary material section). The low variability of the concentrations obtained from the DeTaCs spikes by qPCR further proved the reliability and reproducibility of the filtration and extraction process. The multiplicative standard deviation for the compiled data set resulted in a value of s* = 2.1.
2.5. Cultivation-based enumeration of standard faecal indicator bacteria (SFIB)
Cultivation-based enumeration of SFIB (i.e., E. coli, Enterococci and Clostridium perfringens spores) was performed in the frame of our ISO 17025 accreditation. Before analysis, the samples were homogenized in an ultrasonic bath for 5 min (Bandelin Sonorex, RK 100, 35 kHz; Berlin, Germany). For membrane filtration, appropriate dilutions were performed (Farnleitner et al., 2010; Vierheilig et al., 2013). Enumeration of presumptive E. coli was based on the ISO standard 16649-1 (ISO, 2001a), using the chromogenic TBX agar (Oxoid, Thermo Fisher Scientific Inc., United Kingdom) and incubation at 44 ± 0.5 °C for 24 ± 0.5 h. Enumeration of Enterococci was based on the ISO standard 7899-2 (ISO, 2000), using Slanetz–Bartley medium (Oxoid) and incubation at 36 ± 2 °C for 44 ± 4 h. For quantification of C. perfringens spores, 5 ml influent and 15 ml effluent were pasteurized at 60 ± 2 °C for 15 min. C. perfringens was analysed according to the established ISO method 14189 (ISO, 2013), based on selective cultivation using TSC agar (Scharlau, Spain) at 44 ± 0.5 °C for 21 ± 3 h and subsequent identification of colonies by acid phosphatase reaction (Ryzinska-Paier et al., 2011). For quality assurance (media control as well as performance control of the methods), the reference strains E. coli NCTC 9001, Enterococcus faecalis NCTC 775 and Clostridium perfringens NCTC 8237 were used.
2.6. Quantification of human-specific viral faecal indicators by qPCR
To concentrate the desired viral DNA, 50 ml of influent and 500 ml of effluent were used. For membrane bioreactor 5000–10,000 ml of effluent grab samples, were used for the skimmed milk flocculation process as established by Calgua (Calgua et al., 2013a, 2008). Viral concentrates were resuspended in 1 ml of phosphate buffer (1:2, v/v of Na2HPO4 0.2 M and NaH2PO4 0.2 M) at pH 7.5. A control spike (adenovirus type 35) was also added as a process control to aliquoted composite influent and aliquoted composite effluent samples. The HAdV35 spiking stock was prepared in A549 cells and quantified by plaque assays and quantitative PCR (cf. for more details on plaque assay see supplemental material). A GC number of 105 HAdV35 were used for spiking. The qPCR assay used to quantify the spiked HAdV in the samples showed a limit of detection of 11 GC per reaction. According to the protocol of concentration applied, numbers lower than 102–103 GC/L were under the sample limit of detection. All process controls tested in this study were above the given sample limit of detection and well within the range of recovery rates, as previously reported for this specific protocol of concentration (Calgua et al., 2013b). The process of treatment for all samples investigated was thus considered acceptable (for more information see supplemental material) Ultrapure water (ISO 3696 grade 1) produced from tap water was used as negative control of the process. The water treatment system consists of a pretreatment cartridge followed by a reverse osmosis unit, UV irradiation chamber, a deionizing polishing cartridge and a 0.2 µm membrane filter (ELGA Medica-R 7, Veolia, Water Systems, Austria). Viral DNA was extracted from all samples using the QIAamp Viral RNA kit (Qiagen, Inc.). Nucleic acid eluates (holding time < 48 h at 21 °C). were sent at room temperature to the laboratory in Barcelona for quantification by qPCR. Specific real-time qPCR assays were used to quantify HAdV and JCPyV as previously described in detail (Bofill-Mas et al., 2006; Hernroth et al., 2002; Pal et al., 2006). Amplifications were performed in a 25-µl reaction mixture containing 10 µl of DNA and 15 µl of TaqMan Environmental Master Mix (Life Technologies) and 0.9 µM of each primer (AdF and AdR) and 0,225 µM of fluorogenic probe (AdP1) for HAdV detection. After activation of the AmpliTaq Gold for 10 min at 95 °C, 40 cycles (15 s at 95 °C and 1 min at 60 °C) were performed in a Stratagene Mx3000P detection system. DNA suspensions were analysed in duplicates using un-diluted and ten-fold dilutions to analyse environmental samples, whereas each dilution of standard DNA suspensions (cf. supplemental material) from 100 to 107 gene copies was run in triplicates. In all of the qPCRs carried out, the amount of DNA was defined as the mean of the data obtained. A non-template control (NTC) and a non-amplification control (NAC, DNA polymerase was omitted from the reaction to monitor background signal and probe stability) were added to each run. Tests for inhibition were performed according to the procedure described in section 2.5. To verify negative results known amounts of targeted viral DNA was added to negative samples and analysis was repeated.
2.7. Enumeration of human-associated bacteriophages infecting B. thetaiotaomicron
Bacteriophages infecting B. thetaiotaomicron were enumerated according to the standard method ISO 10705-4 (ISO, 2001b) as described for Bacteroides fragilis RYC2056 or HSP40 infecting phages. The host strain applied for the phage analyses was B. thetaiotaomicron (GA17), kindly provided by Prof. Maite Muniesa, University of Barcelona, Spain. To reduce the high concentration of background flora, samples were filtered through a low protein-binding membrane (0.2 µm; Minisart 16534, Sartorius). For enumeration of the phages, 1 ml of the host strain (inoculum culture) was added to tubes containing 2.5 ml of semisolid agar and aliquots of the samples to be tested, gently mixed, and poured onto solid agar plates. The plates were incubated under anaerobic conditions (AnaeroGen AN0025A, Oxoid) at 36 ± 2 °C for 21 ± 3 h. The results are expressed as the number of plaque-forming units (pfu) per sample volume.
2.8. Data analysis and statistics
All microbial data are expressed as log10 (x+1), after having performed all the needed calculations on the untransformed data. Reductions were calculated as log10 (effluent) minus log10 (influent). Microbial loads were calculated as numbers per inhabitant and day. To achieve this, the respective microbiological concentrations were multiplied by the amount of discharge and divided by the number of connected people. Visual and statistical data were analysed with Visplore 2.0 (Piringer et al., 2010) (VRVis GmbH, Austria, Vienna) and Sigma Plot 11.0 (SPSS Inc., Chicago, USA). To account for multiple testing, statistical significance levels were corrected according to Bonferroni. All graphs were prepared using Sigma Plot 13.0, Visplore 2.0 and CorelDraw X5 (Corel, Canada). To support correct comparisons of the variability of log-normal distributed variables, the multiplicative standard deviation s* was calculated for the recovered results according to Limpert et al. (2001). The multiplicative standard deviations s* ranged from 1.5 to 6.4 and from 1.5 to 1.7 for microbiological and chemical parameters, respectively. A s* > 1 is considered indicative to apply multiplicative standard deviation statistics (further details see Limpert et al., 2001).
3. Results
3.1. Characteristics of selected municipal WWTPs and chemical wastewater quality
Five municipal activated sludge WWTPs (WWTP2-6) in the metropolitan area of Vienna, Austria, with design capacities ranging from 20000 to 140000 p.e. and average loadings of 6600–78400 p.e. were selected. The number of connected inhabitants ranged from approx. 2000 up to approx. 31000 (Table 1). The catchments could be described as a mix of rural and urbanized areas. Sewers were constructed as combined systems, with a pressure pipe as inflowing sewer in case of WWTP4. Industrial influence in the catchment was evaluated as low to moderate. A potential impact on raw wastewater quality due to seasonal events relating to crop harvest and processing (i.e., wine production in the catchments of WWTPs 2, 4, 5, 6) and tourism (i.e., summer tourism in the catchment of WWTP5) could not be excluded. The average flow at the influent varied from 1600 m3 per day at WWTP5 to 18700 m3 per day at WWTP4 (Table 1).
Table 1.
WWTP: wastewater treatment plant; PE: population equivalent; M: primary treatment: mechanical treatment step; C secondary treatment: biological carbon (C) removal; N,D,P: tertiary treatment: nutrient removal including nitrification (N): denitrification (D) and phosphorus removal (P); COD: chemical oxygen demand; TP: total phosphorus; TN: total nitrogen; NO3–N: nitrate nitrogen; NH4–N: ammonium nitrogen; infl.: influent; effl.: effluent of WWTP; * Annual mean COD load (kg a−1) divided by a COD load per person of 110 g COD d−1;** during summer.
| WWTP | Design capacity [PE] | Actual average loading (PE)* | Inhabitants connected | Type | Advanced treatment/specifics | COD [mg L−1] median |
TP [mg L−1] median |
TN [mg L−1] median |
NO3–N [mg L−1]
median |
NH4–N [mg L−1]
median |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| infl | effl | infl | effl | infl | effl | effl | effl | ||||||||
| 2 | 40,000 | 49,000 | 23,500 | M, C, N, P | overloaded | 466 | 36 | 9.3 | 0.7 | 58 | 25 | 21 | 0.3 | ||
| 3 | 23,000 | 16,300 | 10,800 | M, C, N, D, P | – | 553 | 16 | 8.8 | 0.3 | 55 | 9 | 4 | 0.1 | ||
| 4 | 140,500 | 78,400 | 30,800 | M, C, N, D, P | – | 441 | 16 | 5 | 0.3 | 43 | 9 | 4 | 0.1 | ||
| 5 | 20,000 | 6,600 | 2,100 | M, C, N, D, P | UV disinfection** | 443 | 16 | 5.5 | 0.2 | 38 | 2 | 0.4 | 0.2 | ||
| 6 | 45,000 | 20,000 | 15,000 | M, C, N, D, P | 555 | 24 | 8.5 | 0.7 | 63 | 14 | 11 | 0.4 | |||
| 7 | 21,000 | 17,200 | 5,900 | M, C, N, D, P | membrane system | 905 | 19 | 9 | 0.5 | 47 | 5 | 3 | 0.1 | ||
All WWTPs were using an activated sludge process with mechanical treatment and carbon removal. WWTPs 3–6 also performed nitrification and denitrification, while WWTP2 was overloaded and not designed for denitrification, resulting in lower nitrogen removal rates compared to other WWTPs (Table 1). The average sludge age ranged from 8 to 57 days (Table S1, additional material). P removal was achieved at all WWTPs by chemical precipitation with iron and/or aluminium salts, which is required for sensitive areas in the European Union (EC, 1991). WWTP5 was also equipped with additional UV-disinfection (UV-2000, Trojan Technologies, Canada; 48 UV lamps, operational parameters: UV Transmission UVT 10 mm: 65%, max discharge: 135 m3 h−1) at the effluent, which was operated only during summer. One additional activated sludge plant (WWTP7) was selected in the rural area of Bavaria, Germany. WWTP7 is a membrane bioreactor with a system of ultra-filtration membranes submerged in the aeration tank. The three vacuum rotation membrane units, each having a membrane surface of 2264 m2 and a pore size of approximately 38 nm, are used to separate the activated sludge flocs from the treated wastewater by means of a pressure difference. (physical solid-liquid separation process, Table 1).
In raw wastewater of WWTPs 2–7, median COD, TN, and TP showed values ranging from 440 to 900 mg L−1, from 40 to 60 mg L−1, and from 5 to 9 mg L−1, respectively (Table 1). Observed elimination rates were 94–98%, 77–93% (except WWTP2 with ~50%), and 90–96% for COD, TN, and TP, respectively, (Table S1, additional material). The median water temperature at the wastewater effluents was 12.5 °C–15.5 °C (further details in Table 1 and Table S1).
3.2. Characteristics of selected domestic WWTPs and chemical wastewater quality
Eight small dWWTPs (numbers 8–15) with a design capacity of 6–130 p.e., were selected in the metropolitan area of Vienna, Austria (Table S1, additional material). Two types of dWWTPs were encountered, including Dr. Renner® technology (also known as Gallé wastewater technique®) and Putox® technology (also known as Purator®). Two dWWTPs were linked to little taverns in the mountainous area of Vienna. Up to 50 persons were contributing their excreta to these sewer systems. Faecal load were strongly fluctuating. One dWWTP was localized at a horse barn with approximately 25 persons as permanent faecal sources. The rest of the dWWTPs was connected to individual households, with up to 5 contributing persons. Determined COD in the treated wastewater of dWWTPs 8–14 ranged from 15 to 56 mg L−1; the settle-able solids were generally less than 0.1 mg L−1. Ammonium and pH yielded values of 0.2–1.7 mg L−1 and 6.9–7.6, respectively, in the treated wastewater (see Table S1 additional material for more details).
3.3. Does wastewater from different municipal WWTPs show differences in GeBaM and SFIB concentrations?
One of the aims was to evaluate whether human-associated genetic Bacteroidetes faecal markers (GeBaM) and SFIB concentrations in raw and treated wastewater show significant differences with regard to the investigated municipal disposal systems or background conditions. The GeBaM BacHUM, HF183 TaqMan and AllBac as well as the cultivation-based parameter E. coli, enterococci and C. perfringens spores were selected as test parameters (for details see Table S3, additional material, n = 6–10 per WWTP). Statistical comparisons were performed between all the individually investigated municipal WWTPs, covering all WWTP2 to WWTP6 combinations (type 1 comparisons). Comparisons were also performed for the concentrations of microorganisms in wastewater from cool vs. warm seasons for the lumped results from WWTP 2–6 (type 2 comparisons). The results of type 1 and type 2 comparisons revealed no significant differences (Mann-Whitney Rank Sum Test, p < 0.05, Bonferroni-corrected). Hence, the results from WWTP2 to WWTP6 were pooled for further analysis (section 3.4). The results from WWTP7 (membrane filtration) and WWTP5 (UV-disinfection) are shown separately in sections 3.4.4. and 3.6.
3.4. Occurrence of microbial indicators in raw and biological treated wastewater from municipal systems
3.4.1. Prevalence and abundance of GeBaM and comparison with SFIB and human viral faecal markers
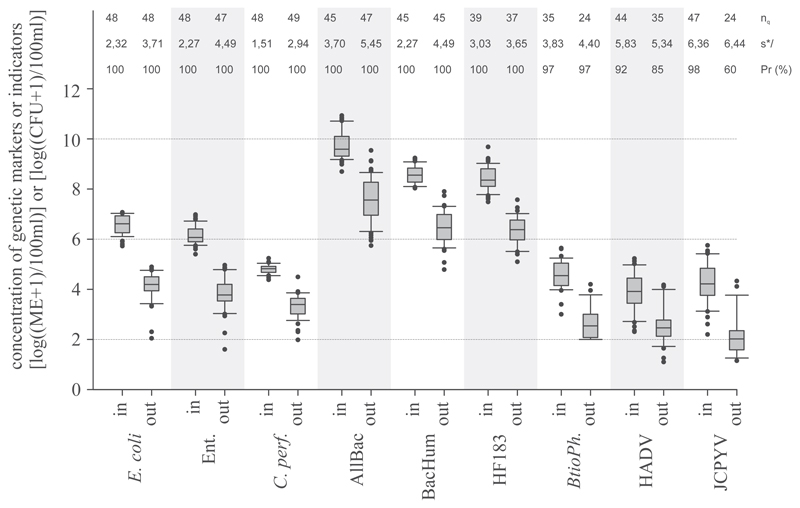
All investigated GeBaM markers (n = 37–45 per parameter and influent or effluent samples) showed 100% occurrence in raw and biologically treated wastewater with primary, secondary and tertiary treatments. The human-associated faecal markers BacHUM and HF183 TaqMan revealed remarkably similar concentrations, with medians of log10 8.6, log10 8.6 (raw) and log10 6.5, log10 6.4 (treated) ME per 100 ml wastewater, respectively (Fig. 1). The AllBac maker showed concentrations one order of magnitude higher, with medians of log10 9.6 (raw) and log10 7.8 (treated) ME per 100 ml wastewater.
Fig. 1.
Concentration of standard faecal indicators and genetic microbial source tracking markers in raw (in) and treated (out) wastewater in pooled data from WWTP 2-6. Data shown are a pooled set. AllBac: genetic faecal marker for the total Bacteroidetes populations; BacHum, HF183: genetic faecal marker for human-associated Bacteroidetes populations; C. perf: Clostridium perfringens spores, Ent: enterococci, BtioPh: bacteriophages infecting Bacteroides thetaiotaomicron, HAdV: human adenovirus, JCPyV: JC polyomavirus, nq: number of quantifiable samples, s*: estimated multiplicative standard deviation, PR (%): Prevalence of investigated markers. Boxes cover the 25th to 75th percentile, line within the boxes, median; whiskers the 10th to 90th percentile.
SFIB (n = 47–49 per parameter and influent or effluent samples) also proved 100% prevalent in the investigated raw and treated wastewater, but with concentrations 2 to 3 orders of magnitude lower compared to GeBaM. The median concentrations for E. coli, Enterococci, and C. perfringens spores were log10 6.6, log10 6.1, log10 4.8 (raw) and log10 4.2, log10 3.8, and log10 3.4 (treated) CFU per 100 ml of wastewater, respectively (Fig. 1). The analysed bacteriophages and human viruses could not be detected in all samples. The prevalence of the human faecal-associated bacteriophage BtioPh (n = 24, 35 influent or effluent samples) was 97% in raw and treated wastewater. Prevalence rates for the human viruses HAdV and JCPyV (n = 24–47 per parameter and influent or effluent samples) were 92%, 98% (raw) and 85%, 60% (treated), respectively. Median concentrations for BtioPh, HAdV, and JCPyV were log10 4.5, log10 3.9, log10 4.2 (raw) and log10 2.5, log10 2.5, and log10 2.1 (treated) PFU or GC per 100 ml of wastewater (Fig. 1).
3.4.2. Variability of GeBaM concentrations in wastewater and comparisons with SFIB and human viral markers
In raw wastewater, the s* were remarkably low for the genetic marker concentrations of the BacHUM and HF183 TaqMan assay and were comparable with the variability of E. coli and enterococci, ranging from s* = 2.3 to s* = 3.0 (Fig. 1). C. perfringens spores proved to be the most constantly occurring indicator in raw wastewater (s* = 1.5).
A general increase in the variability of indicator concentrations between influent and effluent samples, irrespective of the considered parameter and WWTP, was obvious (p < 0.05, n = 9, Kruskal-Wallis). The multiplicative standard deviation s* increased by an average factor of 1.5 (range 0.9–2.0) during wastewater treatment (Fig. 1). In treated wastewater the variability of GeBaM concentrations matched the variability of SFIB concentrations as well (s* = 3.6 to 5.5).
The concentration variability of viral markers was higher, ranging from s* = 3.8 to 6.4 in raw wastewater and s* = 4.4 to 6.4 in treated wastewater. The investigated chemical parameters showed a statistical variability in the range of C. perfringens spores in raw wastewater (s* = 1.5, 1.8, 1.6 and 1.7 for COD, BOD, TN and TP, respectively). In treated wastewater the chemical parameters revealed lower variability compared to the microbiological parameter (s* = 1.6, 2.4, 3.1, 2.4 for COD, BOD, TN, and TP, respectively).
3.4.3. Establishing GeBaM loads per connected person and day
Medians for the calculated faecal marker loads AllBac, BacHUM, and HF183 TaqMan resulted in log10 13.2, log10 12.2, and log10 12.2 (raw wastewater) and log10 11.5, log1010.1, log10 9.9 (treated wastewater) ME per connected persons and day, respectively (Fig. S2, supplementary material). Quantitative relationships and statistical variability between GeBaM, SFIB and human viral faecal marker loads were similar to the obtained relationships regarding the concentrations (details are shown in Fig. S2, supplementary material).
3.4.4. Achieved microbiological reductions by wastewater treatment
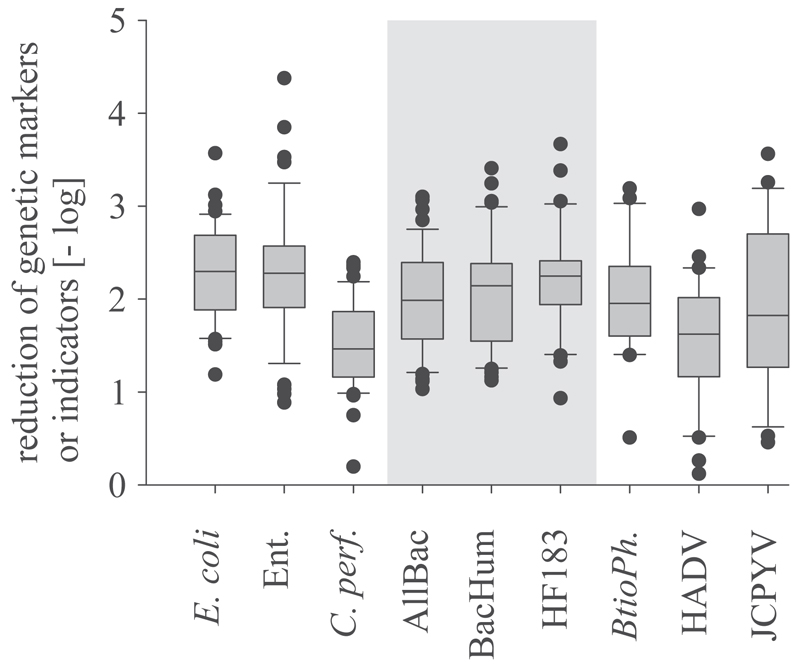
C. perfringens revealed significantly lower reductions compared with the other SFIB and GeBaM. HAdV achieved lower reductions compared with E. coli (Kruskall Wallis, p < 0.001) during wastewater treatment (including primary, secondary and tertiary treatment in WWTPs 2–6). Some basic trends were obvious. E. coli achieved the highest reduction and showed a 10th-percentile value of -log101.6. GeBaM, enterococci, and bacteriophage BtioPh revealed very similar 10th-percentile values that ranged from -log101.0 to -log101.2 (Fig. 3). The lowest 10th-percentile reduction values were achieved by the human-specific viral faecal indicator HAdV and the bacterial faecal indicator C. perfringens spores, at -log10 0.5 and -log10 0.9 reductions, respectively (Fig. 3).
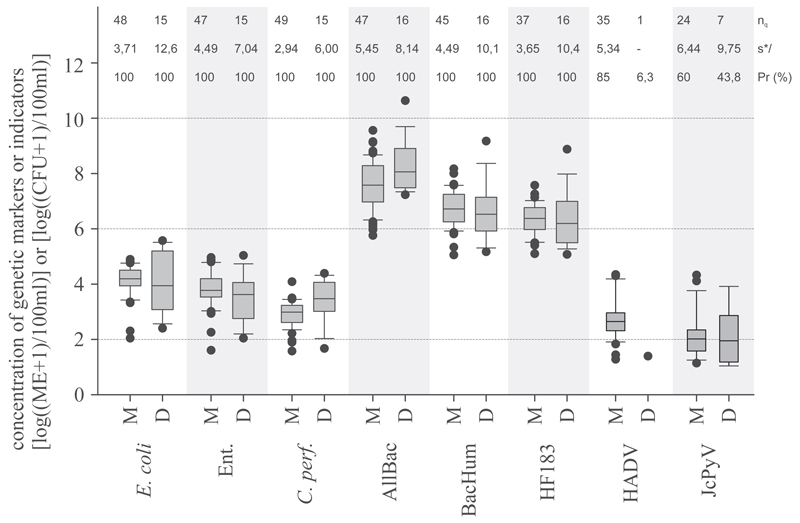
Fig. 3.
Concentration of standard faecal indicators and genetic microbial source tracking markers in pooled data of municipal (M) WWTPs 2-6 effluents versus pooled data from domestic (D) dWWTPs 8-14 effluents. AllBac: genetic faecal marker for the total Bacteroidetes populations; BacHum, HF183: genetic faecal markers for human-associated Bacteroidetes populations; C. perf: Clostridium perfringens spores, Ent: Enterococci, nq: number of quantifiable samples, s*: estimated multiplicative standard deviation, PR (%): Prevalence of investigated markers. Boxes cover the 25th to 75th percentile, line within the boxes, median; whiskers the 10th to 90th percentile.
The reduction of GeBaM at the activated sludge membrane bioreactor (WWTP7) revealed a median 2.8 to 3.6 orders of magnitude increase in treatment efficacy of GeBaM compared to the conventional activated sludge treatment plants WWTPs 2–6 (Fig. S3, supplementary material).
3.4.5. Elucidating the relationships amongst GeBaM and other microbial/chemical variables
Except for total suspended solids (SS), statistical analysis of the pooled data set from WWTPs 2–6, including information from raw and treated wastewater, resulted in significant relationships amongst all parameters (correlation coefficients rho = 0.73–0.95, p < 0.0045; Table 2). Due to the inhomogeneous distribution of the raw vs. the treated wastewater data, such correlation analysis with pooled data led to a statistically biased relationship. Separate analysis was thus performed for the raw and treated wastewater data. Viral data were not included in this detailed correlation analysis because the replicate number was considered too low. Results for viral data are thus only given exemplarily for the raw waster data set, where appropriate.
Table 2.
Correlation table showing the Spearman rank coefficient for the pooled influent and effluent (in & out), the influent (in) and the effluent (out) data of WWTP 2-6. AllBac: genetic faecal marker for the total Bacteroidetes populations; BacHum, HF183: genetic faecal markers for human-associated Bacteroidetes populations; C. perf: Clostridium perfringens spores, Ent: Enterococci; COD: chemical oxygen demand; TN: total nitrogen; NH4–N: ammonium nitrogen; TP: total phosphorus; TSS: total suspended solids.
| AllBac | * = significant | Bonfer roni P = 0,0045 | ||||||||||||||||||||
| 0.85* |
BacHum | |||||||||||||||||||||
| 0.25 | 0.62* | |||||||||||||||||||||
| 0.81* |
0.95* |
HF183 | in & out |
|||||||||||||||||||
| 0.19 | 0.45 | 0.80* | 0.84* | in | out | |||||||||||||||||
| 0.77* |
0.83* |
0.78* |
E. coli | |||||||||||||||||||
| 0.11 | 0.42* | 0.06 | 0.59* | 0.08 | 0.58* | |||||||||||||||||
| 0.76* |
0.83* |
0.78* |
0.86* |
Ent | ||||||||||||||||||
| 0.24 | 0.17 | 0.14 | 0.49* | 0.04 | 0.46 | 0.39 | 0.52* | |||||||||||||||
| 0.73* |
0.82* |
0.77* |
0.80* |
0.82* |
C.perf. | |||||||||||||||||
| 0.26 | -0.06 | 0.41* | 0.15 | 0.22 | 0.18 | 0.39 | -0.01 | 0.39 | 0.22 | |||||||||||||
| 0.75* |
0.81* |
0.76* |
0.80* |
0.82* |
0.89* |
COD | ||||||||||||||||
| 0.29 | 0.03 | 0.36 | 0.17 | 0.19 | 0.23 | 0.43* | -0.03 | 0.39 | 0.13 | 0.65* | 0.47* | |||||||||||
| 0.60* |
0.71* |
0.67* |
0.63* |
0.65* |
0.77* |
0.80* |
TN | |||||||||||||||
| 0.27 | 0.01 | 0.51* | 0.14 | 0.47 | 0.01 | 0.38 | 0.07 | 0.46* | 0.08 | 0.62* | 0.36 | 0.62* | 0.66* | |||||||||
| 0.74* |
0.83* |
0.79* |
0.77* |
0.82* |
0.81* |
0.85* |
0.71* |
NH4 | ||||||||||||||
| 0.29 | -0.04 | 0.48* | 0.22 | 0.32 | 0.21 | 0.01 | 0.15 | 0.43 | 0.20 | 0.60* | -0.04 | 0.49* | 0.32 | 0.73* | 0.37 | |||||||
| 0.73* |
0.80* |
0.77* |
0.78* |
0.78* |
0.84* |
0.90* |
0.82* |
0.87* |
TP | |||||||||||||
| 0.10 | 0.04 | 0.44 | 0.03 | 0.37 | 0.04 | 0.28 | 0.04 | 0.24 | 0.07 | 0.49* | 0.28 | 0.58* | 0.59* | 0.76* | 0.83* | 0.61* | 0.42 | |||||
| 0.06 |
0.23 |
0.24 |
-0.03 |
0.20 |
0.52* |
0.34 |
0.22 |
-0.03 |
0.13 |
TSS | ||||||||||||
| n.a | 0.06 | n.a | 0.23 | n.a | 0.24 | n.a | -0.03 | n.a | 0.20 | n.a | 0.52* | n.a | 0.34 | n.a | 0.22 | n.a | -0.03 | n.a | 0.13 | |||
Correlation analysis for the raw wastewater data from catchments WWTPs 2–6 indicated a tight relationship between the human-associated faecal markers BacHUM and the HF183 TaqMan (rho = 0.80, p < 0.0045). A significant relationship became also obvious between the concentrations of the faecal marker HF183 TaqMan and the human-specific JC polyomavirus (rho = 0.45, p < 0.0045). In sharp contrast, the AllBac marker did not show a discernible relationship with the human-associated GeBaM or SFIB (Table 2). Interesting but non-significant correlation coefficients were obtained amongst the SFIB (rho = 0.39, p > 0.0045). Remarkably, a relationship between the human-associated GeBaM and the SFIB with the biological oxygen demand, the nitrogen content, and the phosphorus content in raw wastewater became evident. This was indicated by a range of significant correlations, including BacHUM, E. coli and enterococci with one or several components of the chemical parameters (rho = 0.46–0.51, p < 0.0045). Amongst the microbiological parameters, C. perfringens spores had the most pronounced relationship with the chemical quality characteristics of raw wastewater (rho = 0.49–0.65, p < 0.0045). Except for SS, a general interrelationship between all investigated chemical variables was obvious (rho = 0.49–0.76, p < 0.0045).
A different situation could be found for the data set of the treated wastewater. Except for C. perfringens spores, an increased tendency in associations amongst the microbiological variables during the treatment process was observed (i.e. 7 out of the 10 pairwise comparisons became significant after treatment, Table 2). In this respect, a correlation between the genetic AllBac and the human-associated BacHUM marker was discernible after the treatment (rho = 0.62, p < 0.0045). In contrast, relationships between the human-associated GeBaM and SFIB with the chemical quality characteristics of the wastewater disappeared during the treatment process (i.e. no significant correlation out of 25 pair-wise comparisons, Table 2). Again, the exception were C. perfringens spores, showing significant relationships with the COD and SS (Table 2).
3.5. Occurrence of GeBaM and SFIB in treated wastewater of small domestic WWTPs and comparison to municipal WWTPs
GeBaM in treated wastewater from dWWTPs showed 100% prevalence. The human-associated faecal markers BacHUM and HF183 TaqMan revealed similar concentrations, with medians of log10 6.3 and log10 6.2 ME per 100 ml wastewater (Fig. 3). The AllBac maker showed concentrations two orders of magnitude higher, with a median of log10 8.0 ME per 100 ml wastewater. FIB markers in treated domestic wastewater also resulted in 100% prevalence. The medians for E. coli, enterococci, and C. perfringens spores were log10 3.9, log10 3.6 and log10 3.9 CFU per 100 ml treated wastewater, respectively (Fig. 3). JCPyV were detected in 3 of 6 dWWTPs (log10 2.0-log10 3.0 ME per 100 ml of wastewater) evaluated that treated wastewater from 6 to 130 PE.
GeBaM and SFIB in treated wastewater had very similar concentrations for both the small domestic and the municipal WWTPs (Fig. 3). No differences in concentration could be detected (Man Whitney, p < 0.008, n from WWTPs randomly adjusted to number of dWWTPs). Information on the variability of results is reflected by the boxplots and the multiplicative standard deviations statistics (s*) directly given at Fig. 3.
3.6. Observed reductions due to UV irradiation (254 nm) (WWTP5)
After UV irradiation, the observed reductions for enterococci, E. coli, and somatic coliphages, given as 5th-percentile values (i.e., only 5% of the values showed a lower reduction), were log10 3.4, log10 3.0, and log 2.7, respectively (Fig. S4). C. perfringens spores were only slightly inactivated, resulting in a 5th-percentile reduction of log10 0.69. In contrast, no statistically significant reduction of GeBaM was detectable (one-way ANOVA, p < 0.05).
4. Discussion
Human-associated genetic Bacteroidetes faecal markers (GeBaM) were consistently detected in high concentrations in the investigated samples from raw and biological treated wastewater. The size of the studied wastewater systems varied over 4 orders of magnitude, with populations ranging from as few as 3 individuals up to 49000 inhabitants connected. Statistical analysis also demonstrated that GeBaM concentrations did not reveal differences regarding the type of the wastewater system or the time of the season investigated. Our results thus provide strong empirical evidence of the ubiquitous and abundant occurrence of GeBaM in raw and biologically treated wastewater, regardless whether the wastewater is derived from single households, larger settlements, or towns. Information on the quantitative occurrence and dynamics of GeBaM in wastewater along the wastewater and sanitation pathway has been limiting so far. The few studies available have focussed on individual samples or single systems (Ervin et al., 2013; Silkie and Nelson, 2009; Srinivasan et al., 2011; Stapleton et al., 2009). To our knowledge, our results provide the first comprehensive information on the occurrence and dynamics of GeBaM in raw and biologically treated wastewater from several well-characterized wastewater systems and treatment plants. Characterisation was facilitated by standard physicochemical and chemical analysis of raw and treated wastewater.
The selected systems were predominantly influenced by wastewater from households. No signs of significant influence from agriculture or industry could be found, and chemical analysis did not show any deviations from quality characteristics as expected for raw wastewater of municipal or domestic origin (Gujer, 2002). Data on the discharge dynamics also indicated that large rain events did not happen during the seasonal sampling campaigns (Table S1). A relevant influence on the wastewater quality due to surface runoff in the catchment area, potentially leading to strong dilution effects or to the input of faecal material from non-human sources, was thus not expected.
This study further supports the fact that faecal pollution based on GeBaM qPCR quantification can be performed with at least equal precision compared with traditional ISO-based cultivation techniques (Stapleton et al., 2009). The determined concentrations of GeBaM and SFIB (E.coli and enterococci) indicated equal statistical variability in raw and treated wastewater. This finding is of special interest regarding the current evaluation of GeBaM as a potentially new means to complement routine water quality testing (Betancourt and Fujioka, 2006; McQuaig et al., 2012; Molina et al., 2014). The statistical variability of GeBaM and SFIB concentrations was lowest for raw wastewater and, interestingly, increased during biological wastewater treatment. It can be assumed, that the raw wastewater (apparently) underlies sufficient mixing in the sewer channel to balance potential differences of input concentrations from the connected households. Diurnal variations were accounted for by the volume-proportional 24 h automated sampling. Interestingly, C. perfringens showed a statistical variability in the range of the measured chemical parameters, which was far lower than that of the rest of the microbiological parameters. A very low variability of C. perfringens in water has been reported previously (Byamukama et al., 2005).
It has to be mentioned that the statistical comparison of variability was supported by the implementation of two methodical innovations. To obtain representative samples from the influent and effluent of WWTPs, an automated 24-h volume-proportional and cooled sample was taken (Mayer et al., 2015). In contrast to this study, most studies dealing with microbiological investigations rely on randomly collected grab samples. In addition, the multiplicative standard deviation s* was introduced to the field of pollution microbiology to obtain an appropriate measure of statistical variability for log-normal distributed parameters (Limpert et al., 2001).
Unlike chemical load calculations, load calculations for microbial source tracking markers are very rare (Wilkes et al., 2014, 2013). The presented results suggest the future use of human-associated GeBaM loads as a valuable metric to estimate the impact of municipal and domestic wastewater input into the environment. The established median loads for raw and biological treated wastewater of approximately 1012 and 1010 molecule equivalents of human-associated GeBaM per person and day, respectively, demonstrate the sensitivity of GeBaM as a general measure of faecal pollution from municipal wastewater. Assuming a defecation rate of 100 g–1000 g of faecal excrement per person and day (Cummings et al., 1992; Geldreich, 1978), the estimated median load for raw wastewater can be converted back to a concentration range of 109 to 1010 human-associated GeBaM per g of faeces. This estimated range of GeBaM concentration compares well with concentrations in human faeces measured by qPCR (Haugland et al., 2010; Kildare et al., 2007).
The recovered GeBaM concentrations were in good agreement with previously reported levels (Reischer et al., 2013; Silkie and Nelson, 2009; Stapleton et al., 2009). GeBaM had concentrations at least two orders of magnitude higher than SFIB (cf. Fig. 1). Given the reported occurrence of intestinal microbiota in intestinal systems and human faeces, the dominance of GeBaM over SFIB is well known and expected (Ley et al., 2008; Reischer et al., 2007). This quantitative dominance of GeBaM in raw and biological treated wastewater is of high practical importance, regarding the sensitivity of molecular faecal pollution detection in comparison with cultivation-based standards. SFIB enumeration in water requires only minimal processing efforts. Samples are either directly analysed (MPN procedures) or subjected to membrane filtration before cultivation is started (ISO, 2000, 2005). PCR analysis involves several additional manipulation steps, including nucleic acid extraction, purification, and partial analysis of the extracted volumes (Ervin et al., 2013). Molecular detection methods thus have to apply higher sampling volumes or have to focus on more abundant targets to achieve comparable sample limits of detection (SLOD). The highly abundant nature of GeBaM in wastewater supports equal to superior sensitivity in comparison to SFIB methods, without the need for largely increased sampling volumes. This fact is the basis for the generation of large comparative sampling sets to appropriately cover pollution dynamics in aquatic systems (Ervin et al., 2013; Reischer et al., 2008, 2011; Riedel et al., 2014).
A high statistical association between the concentrations of the two human-associated GeBaM assays, the HF183 Taqman and the BacHUM, was observed for raw and treated wastewater (cf. Table 2). A possible explanation for this tight relationship can be found in the nature of the targeted human-associated Bacteroidetes populations. The most widely used human-associated GeBaM assays (including the above-mentioned ones) still focus on the same or similar phylogenetic sequence targets originally described by Bernhard and Field (Bernhard and Field, 2000). Recent research indicates that the HF183 Taqman and the BacHUM assay target populations within the species of Bacteroides dorei (McLellan and Eren, 2014). Although these assays revealed quite different specificity and sensitivity characteristics in a recent multi-laboratory study (Layton et al., 2013), our data elucidate the redundant nature of the HF183 Taqman and BacHUM assays for the detection of human-associated faecal pollution along the pathway of wastewater disposal. However, the tight association between these two independently performed assays proved the analytical precision of the recovered results within our study.
Statistically significant correlations between concentrations of GeBaM and SFIB could not be detected, although significant associations between the concentrations of faecal indicators and chemical parameters became obvious. A remarkable exception for raw wastewater was the slight but significant relationship between the human-associated BacHUM and the human-specific JCPyV (rho = 0.45, p < 0.02), pointing to the human-associated faecal pollution indication capacity of these molecular targets. Additionally, C. perfringens showed a slight correlation with BacHUM (rho = 0.41, p < 0.02). This is in line with a recent study, where C. perfringens revealed to be associated with human and carnivorous fecal sources, rather than with faecal emissions form herbivorous animals (Vierheilig et al., 2013).
The process of biological wastewater treatment had an equalising effect on the relationship amongst the microbiological variables, whilst the correlation of microbiological to chemical parameters, with the exception of a few cases, totally disappeared. To our knowledge, such a shift of correlation between microbiological parameters during wastewater treatment has not been reported so far and needs further verification.
Although the prevalence and abundance of the human-specific viruses HAdV and JCPyV found in the Austrian WWTP are lower than the range previously reported (Bofill-Mas et al., 2006; Rusinol et al., 2014), the nonetheless high prevalence of HAdV (92%) and JCPyV (98%) in the raw wastewater of the investigated municipal disposal systems in all seasons, and the detection of JCPyV in small dWWTPs, supports the usefulness of these specific tools as markers to trace human faecal pollution from WWTPs. The low abundance and prevalence were probably due to differences in the applied protocol (for instance, 500 ml of effluent municipal and domestic wastewater were concentrated instead of the 10 L tested in other studies). JCPyV and HAdV have been described as highly stable in the environment and present in nearly 100% of raw wastewater samples with concentrations up to log10 7.0 ME 100 ml−1 (Bofill-Mas et al., 2013). JCPyV is a highly specific human marker excreted in urine, and the detection of HAdV has been recently described as particularly useful for the prediction of risk in bathing waters (Marion et al., 2014). These parameters seem to be suitable tools to complement GeBaM/SFIB-based surface water monitoring for selected sampling locations or situations when higher sampling volumes can be processed. HAdV and JCPyV have been successfully applied for the identification of the source of contamination in river catchments covering various geographical areas (Rusinol et al., 2014). These viruses can also support verification of MST results in situations when the specificity level from human-associated GeBaM is deemed insufficient (Reischer et al., 2013).
GeBaM, E. coli and enterococci revealed similar reductions rates in the representatively chosen municipal activated sludge WWTPs (Fig. 2). Only C. perfringens spores demonstrated a lower reduction, most likely due to its resistant nature (Vierheilig et al., 2013). These results clearly demonstrate that GeBaM emission from municipal WWTP can be expected at fairly constant concentrations, and treatment just eliminates 2 log orders of magnitude from raw wastewater. The activated sludge membrane bioreactor removed approximately 5.4 and 5.0 log orders (10th percentile) of magnitude of BacHUM and HF183 TaqMan from wastewater, which is in line with previously reported data on bacterial removal in a membrane bioreactor (van den Akker et al., 2014). Data on UV irradiation (254 nm) indicated no discernible effect on the PCR detectable concentrations of GeBaM in wastewater, which is in agreement with a recently published study (Chern et al., 2014). It should be mentioned that the effect of chlorination was not investigated. This type of disinfection is not applied in European WWTPs. This study did not focus on other important factors, such as mobility, persistence, or specificity of the investigated indicators and markers in the aquatic environment. which also have to be considered for the appropriate interpretation of monitoring results.
Fig. 2.
Reduction of standard faecal indicators and genetic microbial source tracking markers in municipal WWTPs 2-6 during wastewater treatment (pooled data). AllBac: genetic faecal marker for the total Bacteroidetes populations; BacHum, HF183: genetic faecal marker for human-associated Bacteroidetes populations; Ent: enterococci; C. perf: Clostridium perfringens spores,; BtioPh: bacteriophages infecting Bacteroides thetaiotaomicron, HAdV: human adenovirus, JCPyV: JC polyomavirus. Boxes cover the 25th to 75th percentile, line within the boxes, median; whiskers the 10th to 90th percentile.
5. Conclusions
In our study the occurrence and the dynamics of GeBaM in point sources along the human wastewater pathway as expected in the Central European Region have been shown.
A comparison to bacterial, viral, and process parameters at treatment (including membrane bioreactor and UV irradiation) was performed and volume-proportional 24-h auto-sampling and multiplicative standard deviation statistics were introduced.
The results strongly support the application of human-associated GeBaM to complement faecal pollution monitoring programs in water resources based on E. coli and enterococci.
Human-associated GeBaM were consistently detected in high concentrations in raw and biologically treated wastewater samples, irrespective of plant size, type and time of the season.
Human-specific JC polyomaviruses and adenoviruses seem to be a valuable support if highly specific markers are needed for MST.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.watres.2015.12.031.
Acknowledgements
This paper was supported by the Austrian Science Fund (FWF) as part of the research projects P22309 and P23900, the “Vienna Doctoral Programme on Water Resource Systems” (W1219-N22) and the research project “Groundwater Resource Systems Vienna” in cooperation with the Vienna Waterworks as part of the “(New) Danube–Lower Lobau Network Project” (Gewässervernetzung (Neue) Donau–Untere Lobau (Nationalpark Donau-Auen)) funded by the Government of Austria (Federal Ministry of Agriculture, Forestry, Environment & Water Management), the Government of Vienna, and the European Agricultural Fund for Rural Development (project LE 07-13). We thank Prof. Maite Muniesa, University of Barcelona, Spain for providing us the host strain GA17. We thank also Dr. Annika Allard from the University of Umeå, for providing us HAdV35. This is a joint investigation of the Interuniversity Cooperation Centre for Water & Health (www.waterandhealth.at). We kindly acknowledge the support by the laboratory assistance of Sonja Knetsch and Andrea Lettl.
References
- AEV. 1. AEV für kommunales Abwasser. Verordnung des Bundesministers für Land- und Forstwirtschaft über die Begrenzung von Abwasseremissionen aus Abwasserreinigungsanlagen für Siedlungsgebiete (StF: BGBl. Nr. 210/1996) 1996 Letzte Änderung: BGBl II Nr 392/2000. [Google Scholar]
- Ahmed W, Sritharan T, Palmer A, Sidhu JPS, Toze S. Evaluation of bovine feces-associated microbial source tracking markers and their correlations with fecal indicators and Zoonotic pathogens in a Brisbane, Australia, reservoir. Appl Environ Microbiol. 2013;79(8):2682–2691. doi: 10.1128/AEM.03234-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Field KG. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol. 2000;66(10):4571–4574. doi: 10.1128/aem.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt WQ, Fujioka RS. Bacteroides spp. as reliable marker of sewage contamination in Hawaii's environmental waters using molecular techniques. Water Sci Technol. 2006;54(3):101–107. doi: 10.2166/wst.2006.455. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Van De Werfhorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, Wang D, Weisberg SB. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res. 2013;47(18):6812–6828. doi: 10.1016/j.watres.2012.12.046. [DOI] [PubMed] [Google Scholar]
- Boehm AB, Yamahara KM, Love DC, Peterson BM, McNeill K, Nelson KL. Covariation and photoinactivation of traditional and novel indicator organisms and human viruses at a sewage-impacted Marine Beach. Environ Sci Technol. 2009;43(21):8046–8052. doi: 10.1021/es9015124. [DOI] [PubMed] [Google Scholar]
- Bofill-Mas S, Albinana-Gimenez N, Clemente-Casares P, Hundesa A, Rodriguez-Manzano J, Allard A, Calvo M, Girones R. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl Environ Microbiol. 2006;72(12):7894–7896. doi: 10.1128/AEM.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66(1):238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S, Rusinol M, Fernandez-Cassi X, Girones R. Potential risk of MCPyV infection through water. J Neurovirol. 2013;19(3):297–297. [Google Scholar]
- Byamukama D, Mach RL, Kansiime F, Manafi M, Farnleitner AH. Discrimination efficacy of fecal pollution detection in different aquatic habitats of a high-altitude tropical country, using presumptive coliforms, Escherichia coli, and Clostridium perfringens spores. Appl Environ Microbiol. 2005;71(1):65–71. doi: 10.1128/AEM.71.1.65-71.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. Enterococci in the environment. Microbiol Mol Biol Rev. 2012;76(4):685–706. doi: 10.1128/MMBR.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgua B, Fumian T, Rusinol M, Rodriguez-Manzano J, Mbayed VA, Bofill-Mas S, Miagostovich M, Girones R. Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res. 2013a;47(8):2797–2810. doi: 10.1016/j.watres.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Calgua B, Mengewein A, Grunert A, Bofill-Mas S, Clemente-Casares P, Hundesa A, Wyn-Jones AP, Lopez-Pila JM, Girones R. Development and application of a one-step low cost procedure to concentrate viruses from seawater samples. J Virol Methods. 2008;153(2):79–83. doi: 10.1016/j.jviromet.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Calgua B, Rodriguez-Manzano J, Hundesa A, Sunen E, Calvo M, Bofill-Mas S, Girones R. New methods for the concentration of viruses from urban sewage using quantitative PCR. J Virol Methods. 2013b;187(2):215–221. doi: 10.1016/j.jviromet.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Cankar K, Stebih D, Dreo T, Zel J, Gruden K. Critical points of DNA quantification by real-time PCR - effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. Bmc Biotechnol. 2006;6:15. doi: 10.1186/1472-6750-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern EC, Brenner K, Wymer L, Haugland RA. Influence of wastewater disinfection on densities of culturable fecal indicator bacteria and genetic markers. J Water Health. 2014;12(3):410–417. doi: 10.2166/wh.2013.179. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Bingham SA, Heaton KW, Eastwood MA. Fecal weight, colon cancer risk, and dietary-intake of nonstarch polysaccharides (dietary fiber) Gastroenterology. 1992;103(6):1783–1789. doi: 10.1016/0016-5085(92)91435-7. [DOI] [PubMed] [Google Scholar]
- Desmarais TR, Solo-Gabriele HM, Palmer C. An investigation of the regrowth potential of three indicator microbes. Abstr General Meet Am Soc Microbiol. 2001;101:650. [Google Scholar]
- Domingo JWS, Bambic DG, Edge TA, Wuertz S. Quo vadis source tracking? towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 2007;41(16):3539–3552. doi: 10.1016/j.watres.2007.06.001. [DOI] [PubMed] [Google Scholar]
- EC. European Commission. Council Directive of 21. Mai 1991 Concerning “urban Waste Water Treatment” (91/271/EEC) (No. L 135/40) Amendment: Directive 98/15/EC. 1991 [Google Scholar]
- Ervin JS, Russell TL, Layton BA, Yamahara KM, Wang D, Sassoubre LM, Cao YP, Kelty CA, Sivaganesan M, Boehm AB, Holden PA, et al. Characterization of fecal concentrations in human and other animal sources by physical, culture-based, and quantitative real-time PCR methods. Water Res. 2013;47(18):6873–6882. doi: 10.1016/j.watres.2013.02.060. [DOI] [PubMed] [Google Scholar]
- Farnleitner AH, Ryzinska-Paier G, Reischer GH, Burtscher MM, Knetsch S, Kirschner AKT, Dirnboeck T, Kuschnig G, Mach RL, Sommer R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J Appl Microbiol. 2010;109(5):1599–1608. doi: 10.1111/j.1365-2672.2010.04788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldreich EE. Bacterial populations and indicator concepts in feces sewage storm water and solid wastes. In: Berg Gerald., editor. Indicators of Viruses in Water and Food. Illus. Ann Arbor Science Publishers Inc, Ann Arbor; Mich., USA: 1978. pp. 51–97. ISBN 0-250-40055-3. Viii+424pp. [Google Scholar]
- Griffiths RI, Whiteley AS, O'Donnell AG, Bailey MJ. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol. 2000;66(12):5488–5491. doi: 10.1128/aem.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujer W. Siedlungswasserwirtschaft. 2. Springer Verlag, Auflage; 2002. [Google Scholar]
- Hagedorn C, Harwood VJ, Blanch A. Microbial Source Tracking: Methods, Applications, and Case Studies. Springer; New York, USA: 2011. [Google Scholar]
- Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. Fems Microbiol Rev. 2014;38(1):1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected bacteroidales species and human fecal waste by qPCR. Syst Appl Microbiol. 2010;33(6):348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Hernroth BE, Conden-Hansson AC, Rehnstam-Holm AS, Girones R, Allard AK. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first scandinavian report. Appl Environ Microbiol. 2002;68(9):4523–4533. doi: 10.1128/AEM.68.9.4523-4533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S, Sadowsky MJ. Escherichia coli in the environment: Implications for water quality and human health. Microbes Environ. 2008;23(2):101–108. doi: 10.1264/jsme2.23.101. [DOI] [PubMed] [Google Scholar]
- ISO. Water Quality – Detection and Enumeration of Intestinal Enterococci – Part 2: Membrane Filtration Method (ISO 7899–2: 2000) International Organization of Standardization; Geneva, Switzerland: 2000. [Google Scholar]
- ISO. Microbiology of Food and Animal Feeding Stuffs – Horizontal Method for the Enumeration of Beta-glucuronidase-positive Escherichia coli Colony-count Technique at 44 Degrees C Using Membranes and 5-bromo-4 chloro-3-indolyl Beta-D-glucoronide (ISO 16649–1:2001 04 15) International Organisation of Standardisation; Geneva, Switzerland: 2001a. [Google Scholar]
- ISO. Water Quality – Detection and Enumeration of Bacteriophages – Part 4: Enumeration of Bacteriophages Infecting Bacteroides Fragilis (ISO 10705–4:2001) International Organisation of Standardisation; Geneva, Switzerland: 2001b. [Google Scholar]
- ISO. Water Quality – General Guidance on the Enumeration of Microorganisms by Culture (ISO 8199:2005) International Organisation of Standardisation; Geneva, Switzerland: 2005. [Google Scholar]
- ISO. Water Quality – Enumeration of Clostridium perfringens – Method Using Membrane Filtration (ISO 14189) International Organisation of Standardisation; Geneva, Switzerland: 2013. [Google Scholar]
- Kaiblinger K. Standardisation and Marker Sequence Evaluation of Bacteroidetes Based Quantitative Microbial Source Tracking Methods for Humans and Ruminant Animals. Master Thesis; University of Technology Vienna: 2008. [Google Scholar]
- Karlen Y, McNair A, Perseguers S, Mazza C, Mermod N. Statistical significance of quantitative PCR. Bmc Bioinforma. 2007;8:16. doi: 10.1186/1471-2105-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keity CA, Varma M, Sivaganesan M, Haugland RA, Shanks OC. Distribution of genetic marker concentrations for fecal indicator bacteria in sewage and animal feces. Appl Environ Microbiol. 2012;78(12):4225–4232. doi: 10.1128/AEM.07819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 2007;41(16):3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol. 2006;72(6):4214–4224. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton BA, Cao YP, Ebentier DL, Hanley K, Balleste E, Brandao J, Byappanahalli M, Converse R, Farnleitner AH, Gentry-Shields J, Gidley ML, et al. Performance of human fecal anaerobe-associated PCR-based assays in a multilaboratory method evaluation study. Water Res. 2013;47(18):6897–6908. doi: 10.1016/j.watres.2013.05.060. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert E, Stahel WA, Abbt M. Log-normal distributions across the sciences: keys and clues. Bioscience. 2001;51(5):341–352. [Google Scholar]
- Marion JW, Lee C, Lee CS, Wang QH, Lemeshow S, Buckley TJ, Saif LJ, Lee J. Integrating bacterial and viral water quality assessment to predict swimming-associated illness at a freshwater Beach: a cohort study. Plos One. 2014;9(11):10. doi: 10.1371/journal.pone.0112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RE, Vierheilig J, Egle L, Reischer GH, Saracevic E, Mach RL, Kirschner AK, Zessner M, Sommer R, Farnleitner AH. Automated sampling procedures supported by high persistence of bacterial fecal indicators and bacteroidetes genetic microbial source tracking markers in municipal wastewater during short-term storage at 5°C. Appl Environ Microbiol. 2015;81(15):5134–5143. doi: 10.1128/AEM.00998-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan SL, Eren AM. Discovering new indicators of fecal pollution. Trends Microbiol. 2014;22(12):697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaig S, Griffith J, Harwood VJ. Association of fecal indicator bacteria with human viruses and microbial source tracking markers at coastal beaches impacted by nonpoint source pollution. Appl Environ Microbiol. 2012;78(18):6423–6432. doi: 10.1128/AEM.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina M, Hunter S, Cyterski M, Peed LA, Kelty CA, Sivaganesan M, Mooney T, Prieto L, Shanks OC. Factors affecting the presence of human-associated and fecal indicator real-time quantitative PCR genetic markers in urban-impacted recreational beaches. Water Res. 2014;64:196–208. doi: 10.1016/j.watres.2014.06.036. [DOI] [PubMed] [Google Scholar]
- Pal A, Sirota L, Maudru T, Peden K, Lewis AM. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyornaviruses. J Virological Methods. 2006;135(1):32–42. doi: 10.1016/j.jviromet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Pina S, Puig M, Lucena F, Jofre J, Girones R. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol. 1998;64(9):3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piringer H, Berger W, Krasser J. HyperMoVal: Interactive visual validation of Regression models for real-time simulation. Comput Graph Forum. 2010;29(3):983–992. [Google Scholar]
- Reischer GH, Ebdon JE, Bauer JM, Schuster N, Ahmed W, Astrom J, Blanch AR, Bloschl G, Byamukama D, Coakley T, Ferguson C, et al. Performance characteristics of qPCR assays targeting human- and ruminant-associated bacteroidetes for microbial source tracking across sixteen countries on six continents. Environ Sci Technol. 2013;47(15):8548–8556. doi: 10.1021/es304367t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Haider JM, Sommer R, Stadler H, Keiblinger KM, Hornek R, Zerobin W, Mach RL, Farnleitner AH. Quantitative microbial faecal source tracking with sampling guided by hydrological catchment dynamics. Environ Microbiol. 2008;10(10):2598–2608. doi: 10.1111/j.1462-2920.2008.01682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Kasper DC, Steinborn R, Farnleitner AH, Mach RL. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett Appl Microbiol. 2007;44(4):351–356. doi: 10.1111/j.1472-765X.2006.02094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Kasper DC, Steinborn R, Mach RL, Farnleitner AH. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl Environ Microbiol. 2006;72(8):5610–5614. doi: 10.1128/AEM.00364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer GH, Kollanur D, Vierheilig J, Wehrspaun C, Mach RL, Sommer R, Stadler H, Farnleitner AH. Hypothesis-driven approach for the identification of fecal pollution sources in water resources. Environ Sci Technol. 2011;45(9):4038–4045. doi: 10.1021/es103659s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley CM, Jamieson RC, Hansen LT, Yost CK, Bezanson GS. Baseline and storm event monitoring of bacteroidales marker concentrations and enteric pathogen presence in a rural Canadian watershed. Water Res. 2014;60:278–288. doi: 10.1016/j.watres.2014.04.039. [DOI] [PubMed] [Google Scholar]
- Riedel TE, Zimmer-Faust AG, Thulsiraj V, Madi T, Hanley KT, Ebentier DL, Byappanahalli M, Layton B, Raith M, Boehm AB, Griffith JF, et al. Detection limits and cost comparisons of human- and gull-associated conventional and quantitative PCR assays in artificial and environmental waters. J Environ Manag. 2014;136:112–120. doi: 10.1016/j.jenvman.2014.01.029. [DOI] [PubMed] [Google Scholar]
- Rusinol M, Fernandez-Cassi X, Hundesa A, Vieira C, Kern A, Eriksson I, Ziros P, Kay D, Miagostovich M, Vargha M, Allard A, et al. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 2014;59:119–129. doi: 10.1016/j.watres.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Ryzinska-Paier G, Sommer R, Haider JM, Knetsch S, Frick C, Kirschner AK, Farnleitner AH. Acid phosphatase test proves superior to standard phenotypic identification procedure for Clostridium perfringens strains isolated from water. J Microbiol Methods. 2011;87(2):189–194. doi: 10.1016/j.mimet.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer EP, VandeWalle JL, Bootsma MJ, McLellan SL. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res. 2011;45(14):4081–4091. doi: 10.1016/j.watres.2011.04.049. [DOI] [PubMed] [Google Scholar]
- Shanks OC, Kelty CA, Sivaganesan M, Varma M, Haugland RA. Quantitative PCR for genetic markers of human fecal pollution. Appl Eniron Microbiol. 2009;75(17):5507–5513. doi: 10.1128/AEM.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks OC, Sivaganesan M, Peed L, Kelty CA, Blackwood AD, Greene MR, Noble RT, Bushon RN, Stelzer EA, Kinzelman J, Anan'eva T, et al. Interlaboratory comparison of real-time PCR protocols for quantification of general fecal indicator bacteria. Environ Sci Technol. 2012;46(2):945–953. doi: 10.1021/es2031455. [DOI] [PubMed] [Google Scholar]
- Shibata T, Kojima K, Lee SA, Furumai H. Model evaluation of faecal contamination in coastal areas affected by urban rivers receiving combined sewer overflows. Water Sci Technol. 2014;70(3):430–436. doi: 10.2166/wst.2014.225. [DOI] [PubMed] [Google Scholar]
- Siefring S, Varma M, Atikovic E, Wymer L, Haugland RA. Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems. J Water Health. 2008;6(2):225–237. doi: 10.2166/wh.2008.022. [DOI] [PubMed] [Google Scholar]
- Silkie SS, Nelson KL. Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res. 2009;43(19):4860–4871. doi: 10.1016/j.watres.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Aslan A, Xagoraraki I, Alocilja E, Rose JB. Escherichia coli, enterococci, and Bacteroides thetaiotaomicron qPCR signals through wastewater and septage treatment. Water Res. 2011;45(8):2561–2572. doi: 10.1016/j.watres.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Stapleton CM, Kay D, Wyer MD, Davies C, Watkins J, Kay C, McDonald AT, Porter J, Gawler A. Evaluating the operational utility of a bacteroidales quantitative PCR-based MST approach in determining the source of faecal indicator organisms at a UK bathing water. Water Res. 2009;43(19):4888–4899. doi: 10.1016/j.watres.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Stevens G, Mascarenhas M, Mathers C. Global health risks: progress and challenges. Bull World Health Organ. 2009;87:646–646. doi: 10.2471/BLT.09.070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckel DM, Stelzer EA, Dick LK. Evaluation of two spike-and-recovery controls for assessment of extraction efficiency in microbial source tracking studies. Water Res. 2009;43(19):4820–4827. doi: 10.1016/j.watres.2009.06.028. [DOI] [PubMed] [Google Scholar]
- Tallon P, Magajna B, Lofranco C, Leung KT. Microbial indicators of faecal contamination in water: a current perspective. Water Air Soil Pollut. 2005;166(1–4):139–166. [Google Scholar]
- Tambalo DD, Fremaux B, Boa T, Yost CK. Persistence of host-associated bacteroidales gene markers and their quantitative detection in an urban and agricultural mixed prairie watershed. Water Res. 2012;46(9):2891–2904. doi: 10.1016/j.watres.2012.02.048. [DOI] [PubMed] [Google Scholar]
- Tryland I, Myrmel M, Ostensvik O, Wennberg AC, Robertson LJ. Impact of rainfall on the hygienic quality of blue mussels and water in urban areas in the inner Oslofjord, Norway. Mar Pollut Bull. 2014;85(1):42–49. doi: 10.1016/j.marpolbul.2014.06.028. [DOI] [PubMed] [Google Scholar]
- van den Akker B, Trinh T, Coleman HM, Stuetz RM, Le-Clech P, Khan SJ. Validation of a full-scale membrane bioreactor and the impact of membrane cleaning on the removal of microbial indicators. Bioresour Technol. 2014;155:432–437. doi: 10.1016/j.biortech.2013.12.123. [DOI] [PubMed] [Google Scholar]
- Vierheilig J, Frick C, Mayer RE, Kirschner AKT, Reischer GH, Derx J, Mach RL, Sommer R, Farnleitner AH. Clostridium perfringens is not suitable for the indication of fecal pollution from ruminant wildlife but is associated with excreta from nonherbivorous animals and human sewage. Appl Environ Microbiol. 2013;79(16):5089–5092. doi: 10.1128/AEM.01396-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman RL, Shively DA, Pawlik H, Nevers MB, Byappanahalli MN. Occurrence of Escherichia coli and enterococci in cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl Environ Microbiol. 2003;69(8):4714–4719. doi: 10.1128/AEM.69.8.4714-4719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes G, Brassard J, Edge TA, Gannon V, Gottschall N, Jokinen CC, Jones TH, Khan IUH, Marti R, Sunohara MD, Topp E, et al. Long-term monitoring of waterborne pathogens and microbial source tracking markers in paired agricultural watersheds under controlled and conventional tile drainage management. Appl Environ Microbiol. 2014;80(12):3708–3720. doi: 10.1128/AEM.00254-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes G, Brassard J, Edge TA, Gannon V, Jokinen CC, Jones TH, Marti R, Neumann NF, Ruecker NJ, Sunohara M, Topp E, et al. Coherence among different microbial source tracking markers in a small agricultural stream with or without livestock exclusion practices. Appl Environ Microbiol. 2013;79(20):6207–6219. doi: 10.1128/AEM.01626-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuertz S, Wang D, Reischer GH, Farnleitner AH. In: Microbial source tracking: methods, applications, and case studies. Hagedorn C, Blanch AR, Harwood VJ, editors. Springer; New York, USA: 2011. pp. 61–113. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.