Abstract
The relationship between Epstein Barr Virus (EBV) and miR-155 is well established. EBV infection induces miR-155 expression, which is expressed at higher levels in EBV latency type III cells compared to EBV latency type I cells. However, the mechanism by which EBV latency genes activate miR-155 expression is still unclear. Here we present data showing that DNA methylation regulates miR-155 expression. We also provide evidence that the AP1 signaling pathway is involved in EBV-mediated miR-155 activation, and that Bay11 influences signaling of the miR-155 promoter AP1 element. Lastly, we show that LMP2A, LMP1 and EBNAs cannot activate miR-155 expression alone, indicating that the regulation of miR-155 by EBV is dependent on more than one EBV gene or cell signaling pathway. We conclude that the regulation of miR-155 in EBV-positive cells occurs through multiple cell signaling processes involving EBV-mediated chromatin remodeling, cell signaling regulation and transcription factor activation.
Keywords: Epstein Barr Virus, EBV, microRNA, miRNA, miR-155, pri-miR-155, AP1, DNA methylation, LMPs, EBNAs
Introduction
Epstein Barr Virus (EBV) is a ubiquitous DNA tumor virus associated with several hematologic cancers and non-hematological tumors, such as Hodgkin’s lymphoma, Burkitt’s lymphoma, nasopharyngeal carcinoma, and gastric cancer. EBV expresses lytic genes and latent genes at different points in its infection cycle. The EBV immediate early genes (Zta and Rta) and EBV early genes (BMRF1, BGLF4, VCA) are expressed during EBV lytic reactivation. The expression of different sets of latent genes such as the latent membrane proteins (LMPs) and Epstein Barr nuclear antigens (EBNAs) determines the EBV latency stage (latency type I, II or III). Whereas only one EBV latent gene (EBNA1) is expressed on EBV latency type I cells, the full repertoire of latency genes; EBNA1, EBNA3A, 3B, 3C, LP, EBNA2 and LMP2B are expressed in EBV latency type III where many cellular transcription factors are upregulated including AP1. The latent membrane protein 2A (LMP2A) regulates ERK-MAPK (Chen et al., 2002a; Engels et al., 2012), PI3K/Akt (Pan et al., 2008; Portis and Longnecker, 2004), NF-κB (Stewart et al., 2004), STAT (Shair et al., 2012) and the Notch/Wnt pathway (Anderson and Longnecker, 2008; Garuti et al., 2014). Latent membrane protein 1 (LMP1) is similarly involved in multiple cellular signaling pathways, such as NF-κB (Fries et al., 1999), hedgehog (Port et al., 2013), IRF7(Bentz et al., 2012; Ersing et al., 2013; Ning et al., 2003), LKB1-AMPK (Pacchiarotti et al., 2013), PI3K, ERK-MAPK, Wnt/b-catenin, miTOR, p38, JAK/STAT, and EGFR. EBNA3A, 3B and 3C interact with CBF1/RBPJ (kappa) (Maruo et al., 2005; Radkov et al., 1997; Radkov et al., 1999). EBNA2 activates Notch signal transduction (Strobl et al., 2000) through its interaction with CBF1 to regulate cell proliferation and survival.
The EBV EBNA promoters, Wp, Cp and Qp determine the latency type of EBV. Wp and Cp drive expression of the latency replication factor, EBNA1 as well as the EBV latency type III-specific genes EBNA2, EBNA3A-C and EBNA-LP. Qp only drives EBNA1 expression in EBV latency type I. The LMP promoters drive LMP1 and LMP2 expression in EBV latency type II and type III (Schaefer et al., 1991; Zetterberg et al., 1999). Epigenetic mechanisms such as DNA methylation contribute to Wp, Cp and Qp activity and EBV gene expression by blocking the binding of transcription factors to DNA and/or by remodeling chromosome structure. In addition to differences in viral methylation patterns between EBV latency type I and type III cells, latency type differences in DNA methylation also exists in cellular DNA. Low-level methylation of cellular genes in latency type III is associated with high expression of cell transcription factors and the activation of cell signaling. DNA methylation typically causes gene inactivation and silencing (Hutchins et al., 2002; Jones, 2003) and epigenetic DNA methylation-associated gene silencing plays a major role in tumorigenesis. Methylation of tumor suppressor genes generally leads to tumor development and progression (Galm et al., 2005; Herman and Baylin, 2003; House et al., 2003a; House et al., 2003b; Paz et al., 2003) whereas methylation of oncogenes inhibits tumorigenesis.
The 23 nucleotide (nt) non-coding RNA miR-155 is among the most abundant cellular miRNAs expressed in EBV-positive LCLs (Skalsky et al., 2012) and is essential for the growth and survival of LCLs in vitro (Linnstaedt et al., 2010). The basis for most EBV-related cancers is also thought to include the dysregulation of oncogenic miR-155, and there are binding sites for AP1 and NF-κB in its promoter region (Costinean et al., 2006; Eis et al., 2005). Expression of miR-155 can be upregulated via cellular signaling pathways such as the B-cell receptor (BCR) pathway (Yin et al., 2008b), the TGF-beta pathway (Kong et al., 2008) and via NF-κB signaling (Wang et al., 2010). The conjugation of BCR and TGF-beta signaling activates miR-155 expression in EBV-negative cells and induces the EBV lytic cycle in EBV-positive cells (Mutu and Akata). However, these signaling pathways do not appear to induce miR-155 expression in EBV-positive cells. EBV infection can induce miR-155 expression (Cameron et al., 2008; Imig et al., 2011; Jonigk et al., 2013; Yin et al., 2008a; Zhu et al., 2014), and EBV latent genes such as LMP1 and LMP2A have been shown to induce miR-155 expression (Du et al., 2011; Gatto et al., 2008; Rahadiani et al., 2008). Nevertheless, the level of miR-155 activation by LMP1 and LMP2A is not comparable to the level of miR-155 induction by EBV infection. We have reported that the DNA methyltransferase inhibitor, 5-aza-deoxycytidine (5-aza-C) induces expression of both the primary and mature forms of miR-155 in the EBV latency type I cells, Akata and MutuI (Yin et al., 2008a), but the detailed methylation status of the miR-155 promoter in EBV latency type I, II and III cells is not well documented. Albeit a report shows that hypermethylation of miR-155-3p correlates with miR-155-3p repression in Mantle cell lymphoma and other non-Hodgkin’s lymphomas (Yim et al., 2014). The mechanism regarding how EBV activates miR-155 expression is not fully understood; however, it is very clear that EBV latent infection activates AP1 protein expression, and that AP1 proteins mediate BCR-regulated miR-155 in EBV negative cells (Yin et al., 2008b). Therefore, we postulate that EBV-induced miR-155 expression occurs through AP1 proteins under the condition of miR-155 promoter hypomethylation.
The induction of miR-155 expression in EBV positive cells is associated with EBV oncogenesis and pathogenesis. The aim of this study is to analyze and identify the signaling pathways that regulate EBV-mediated miR-155 expression, and to further evaluate which EBV gene(s) play a key role in regulating miR-155 expression.
Results
1. Methylation of the miR-155 CpG island is involved in the regulation of miR-155 expression in EBV positive cells
In line with our previous finding that BCR signaling induces miR-155 expression in EBV-negative B lymphocyte cells (Ramos), we conjectured that BCR signaling could also activate miR-155 expression in EBV-positive latency type I lymphocyte cells (Mutu and Akata). As previously reported, the EBV latent genes, LMP1, LMP2A, EBNAs (3A, 3B and 3C), and the lytic genes, Zta, Rta, BMRF1, were activated in anti-IgG-treated Akata and anti-IgM-treated Mutu cells. However, there was no significant change in miR-155 expression (Supplemental Fig. 1). This inability of BCR signaling in conjunction with induced high expression of EBV latency genes to activate miR-155 may be due to methylation of the miR-155 promoter in EBV-positive latency type I cells. Notably, there is no published literature showing that BCR signaling promotes DNA demethylation.
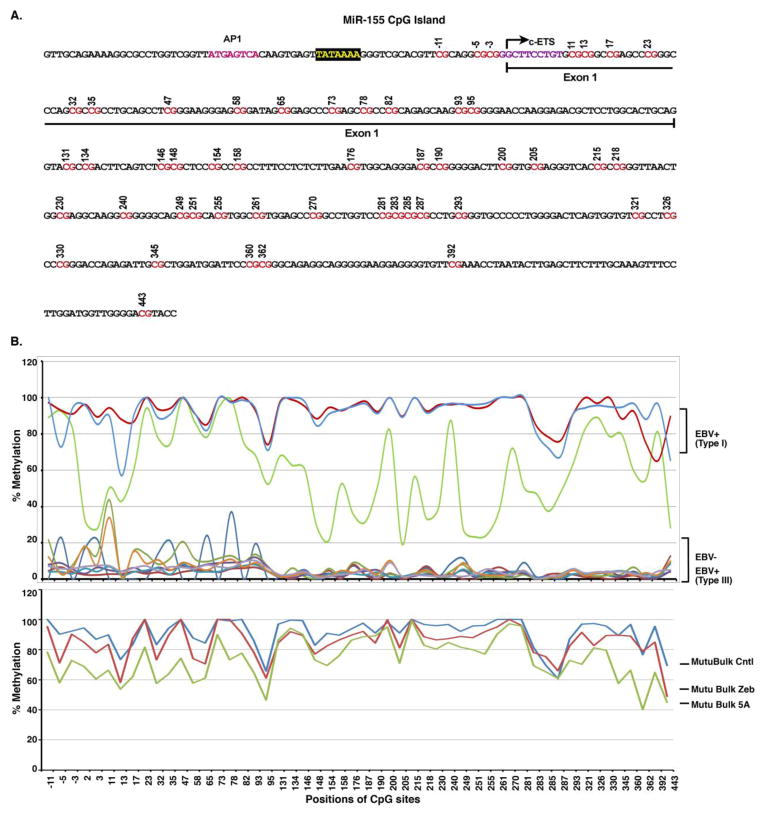
In our previous research, we found that 5′-aza-dc induces both miR-155 and EBV latency gene expression in EBV-positive latency type I cells, but not in EBV-negative cells (Yin et al., 2008a). Moreover, another DNA methylation inhibitor Zebularine, which induces cellular DNA demethylation but not EBV reactivation does not activate miR-155 expression. These results indicate that DNA demethylation alone cannot induce miR-155 expression and that activation of miR-155 expression requires both EBV latency gene expression and DNA demethylation in EBV-positive cells. To investigate this issue further, first, we analyzed the human miR-155 coding sequence (pri-miR-155) from −2000 to +500 relative to the transcription start site (TTS) by the CpG island prediction algorithm, EMBOSS Cpgplot (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/). We found that the miR-155 sequence contained a high confidence CpG island that spans from the 3′ end of the promoter region through the entire first exon and ending 443 bases downstream from the TTS (Fig. 1A). Next, DNA methylation status of the miR-155 island was assessed in EBV-positive and EBV-negative cells (Fig. 1B and Supplemental Table 1). Methylation in EBV latency type I cells (Rael, Akata and Mutu) was higher (52–93%) than that of EBV-negative cells (Ramos, DG75, BL41 and BL30) (3–34%) and EBV latency type III cells (JC5, Dana, Boston, Alwife, JY and P3HR1) (2–5%). Both DNA methylation inhibitors, 5-aza-C and Zebularine, reduced miR-155 CpG island methylation from 97–100% to 46–89% in EBV latency type I cells, Mutu Bulk Cntl (Fig. 1B and Supplemental Table 1). This demonstrates that the failure of Zebularin to activate miR-155 expression is not associated with miR-155 DNA demethylation but instead may be associated with EBV latency type III genes. Furthermore, we have reported that infection of BL41 and BL30 cells with EBV induced miR-155 expression (Yin et al., 2008a). The level of DNA methylation at the miR-155 island in BL41 and BL30 was low (around 6%), and EBV infection did not change the methylation status (Supplemental Table 1). However, a variety of EBV latency genes were highly expressed after EBV infection (Yin et al., 2008a). These data suggest that EBV infection can induce miR-155 expression in cells bearing low-level miR-155 CpG island methylation. Thus, the demethylated miR-155 CpG island is likely critical for inducing miR-155 expression in EBV-positive latency type I cells. Yet DNA demethylation alone cannot activate miR-155 expression without other cell signaling pathways that are activated by EBV genes or other factors (such as the BCR signaling pathway).
Fig. 1. Analysis of the miR-155 CpG island methylation profile in EBV-positive and EBV-negative cells.
DNA methylation was sequenced across part of the promoter and first exon of miR-155 from −11 to 443 relative to the TTS. A. The sequence of the miR-155 CpG island and the sequenced CpG sites are labeled in red. B. MiR-155 CpG island methylation status in EBV-positive and EBV-negative cells. DNA methylation levels at indicated regions were shown as a percentage value in the various cell lines. The analyzed cells include EBV latency type I cells (Mutu, Akata and Rael), EBV latency type III cells (JC5, Dana, Boston, Alwife, JY and P3HR1) and EBV negative cell (DG75). Mutu Bulk Cntl, an EBV-positive latency type I cell line, was treated with DNA demethylating agents Zebularine (Mutu Bulk Zeb) or 5-aza-deoxycytidine (Mutu Bulk 5A).
2. AP1 factors are responsible for miR-155 expression in EBV latency type III cells
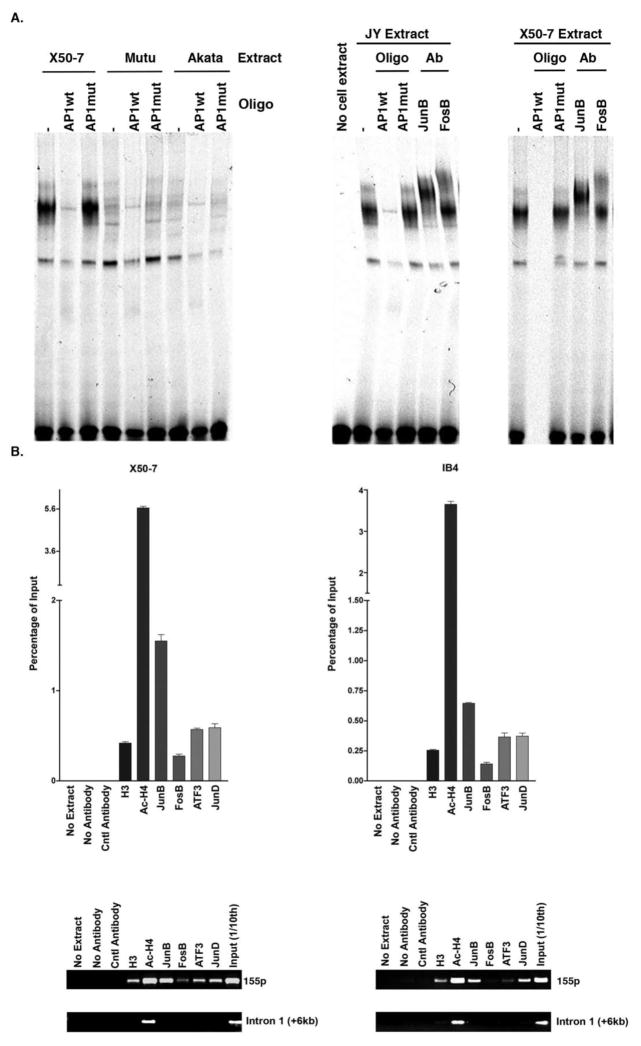
We have previously demonstrated that AP1 proteins play a key role in regulating miR-155 expression in response to BCR activation in the EBV-negative lymphoma cell line Ramos (Yin et al., 2008b). We have also shown that EBV induces miR-155 expression in EBV positive latency type III lymphoma cells, and that miR-155 promoter activity is decreased when the AP1 binding site is mutated (Yin et al., 2008a), implying that EBV genes may induce miR-155 via the activation of AP1 signaling. Yet, it is unclear which AP1 proteins activate miR-155 expression and through which signaling pathways activation of AP1 signaling occurs. To address this issue, we conducted an electrophoretic mobility shift assay (EMSA), which showed that the AP1 proteins JunB and FosB, bind to the miR-155 promoter at high levels in latency type III cells (JY& X50-), but low levels in EBV latency type I cells (Mutu and Akata). These proteins bind to the AP1 element of the miR-155 promoter and supershift the AP1 band (Fig. 2A). Furthermore, an in vivo chromatin immune-precipitation (ChIP) binding assay with antibodies against AP1 proteins confirmed the binding of JunB and FosB and also showed the binding of ATF3 and JunD to the miR-155 promoter region in EBV positive latency type III cells X50-7 and IB4 (Fig. 2B). As a negative control, PCR was performed using primers spanning the miR-155 intron 1 where a lack of AP1 factor signals was observed.
Fig. 2. AP1 proteins bind to the miR-155 promoter in EBV latency type III cells.
A. An electrophoretic motility shift analysis (EMSA) was performed using the miR-155 AP1 elements as the probe; with or without competitor oligos, the wild type miR-155 AP1 (AP1wt) and the mutant miR-155 AP1 (AP1mut); and supershift was conducted with AP1 antibodies, JunB and FosB; in EBV latency type I cells (MutuI or Akata) or type III (X50-7 or JY) cells. − indicates without competitor oligonucleotide (oligo). B. A ChIP assay was conducted with antibodies against Histone 3 (H3), acetyl-Histone 4 (Ac-H4) and AP1 protein, JunB, FosB, ATF3 and JunD, and PCR with specific primers spanning the miR-155 promoter (155p) region and non-specific primers spanning the first intron of the miR-155 gene (Intron 1) in EBV latency type III cells (X50-7 and IB4). Input was 1/10 of each ChIP reaction. ImageJ software was used for quantitation.
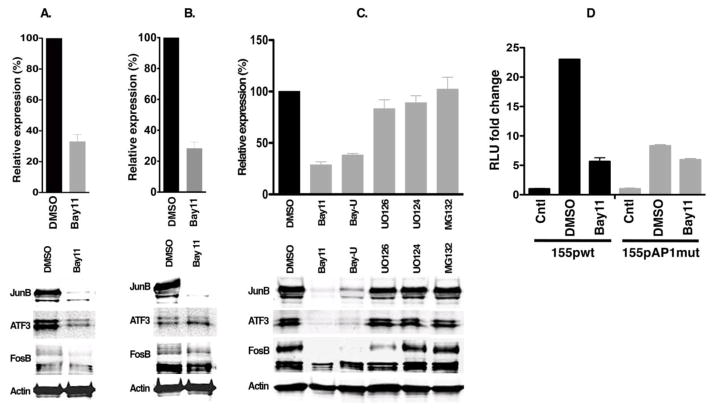
AP1 activity can be induced by several cellular signaling pathways including MAPK (JNKK, p38, PI3, MEKK) and NF-κB pathways that are known to be activated in EBV positive cells. In order to determine which cell signaling pathway known to activate AP1 is involved in miR-155 expression in EBV positive latency type III cells, several cell signaling inhibitors were applied in EBV latency type III cells (X50-7 and IB4); followed by measuring endogenous pri-miR-155 expression using qRT-PCR and analyzing AP1 protein expression via western blotting. As shown in Fig. 3A–C, the MAP kinase inhibitor, UO126 and proteasome inhibitor, MG132 were not able to inhibit AP1 protein expression or pri-miR-155 expression; however, the IκB kinase inhibitor BAY 11-7082 (Bay11) inhibited both pri-miR-155 and AP1 protein expression in these cells. A luciferase reporter assay showed that mutation of the AP1 site of the miR-155 promoter resulted in diminished activity in latency type III cells. Moreover, Bay11 inhibited the promoter activity in the wild type miR-155 promoter (155pwt) but not in the AP1 mutant promoter (155pAP1mut) (Fig. 3D).
Fig. 3. Bay11 inhibits pri-miR-155 and AP1 gene expression in EBV latency type III cells.
A–C. X50-7 (A) and IB4 (B & C) cells were treated with an NF-κB inhibitor and MEK kinase inhibitors for 48 hours and cells were harvested for RT-PCR to detect pri-miR-155 expression (top panel) and for western blot to detect AP1 protein expression (bottom panel). Bay-U indicates treatment with Bay 11 plus UO126. Relative expression in percentage was compared to the DMSO 2−ΔΔCT value. D. A luciferase assay was performed to analyze the activity of miR-155 promoters; wild type (155pwt) vs. AP1 mutant (155pAP1mut), in response to treatment with signaling pathway inhibitor Bay11 in IB4 cells. The relative luminescence units (RLU) fold change value is compared with the RLU measured in control (Cntl), pGL3-basic.
3. EBV genes stimulate miR-155 expression via the AP1 signaling pathway
Several studies including our papers have demonstrated that miR-155 is expressed in EBV latency type III cells but not in EBV latency type I cells (Cameron et al., 2008; Jiang et al., 2006; Kluiver et al., 2006; Yin et al., 2008a), indicating latency type III genes (EBNA-LP, EBNA2, EBNA3A, EBNA3B, EBNA3C, LMP1 and/or LMP2) may play a critical role in miR-155 expression. It has been reported that EBV latency genes are involved in inducing miR-155 expression (Du et al., 2011; Rahadiani et al., 2008) and in regulating cell signaling pathways that are known to activate AP1 activity (Chen et al., 2002b; Johansen et al., 2003; Kieser et al., 1997; O’Neil et al., 2008). Our previous report has shown that the usage of latency type III promoters Cp and LMP1 led to induction of miR-155 expression (Yin et al., 2008a). In line with this finding, infection of BL-41 and Ramos with EBV showed variant EBV latency promoter usage and miR-155 expression. Lower usage EBV latency promoters (Wp, Cp, and LMP) display lower miR-155 expression levels, whereas higher usage of EBV latency promoters is associated with higher miR-155 expression (Yin et al., 2008a). Since Cp/Wp drives EBV latency type III gene expression, these data further verify that EBV latency genes regulate miR-155 expression. However, it is not clear which EBV gene regulates miR-155 expression and through which signaling pathways.
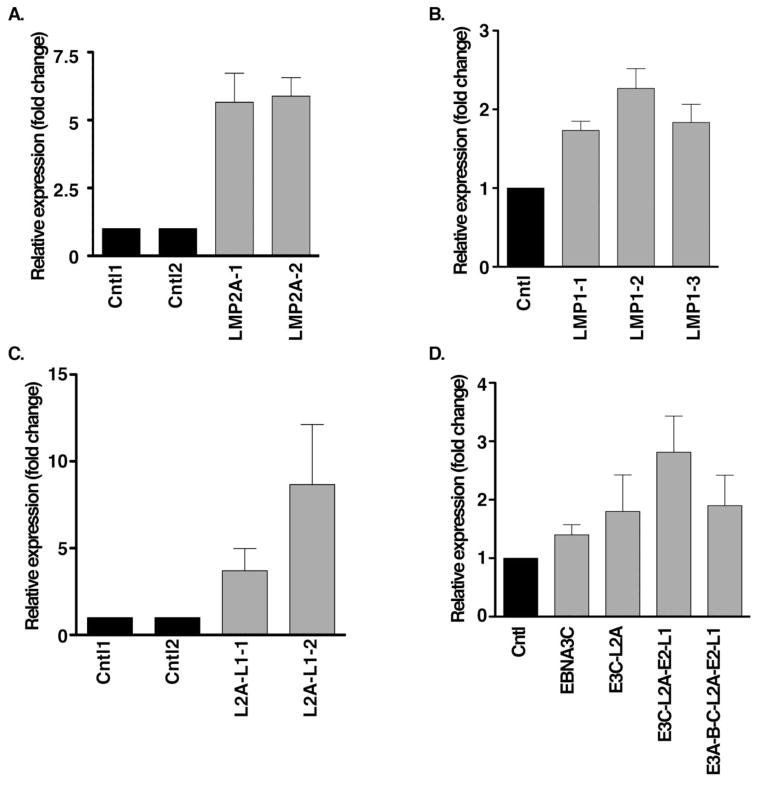
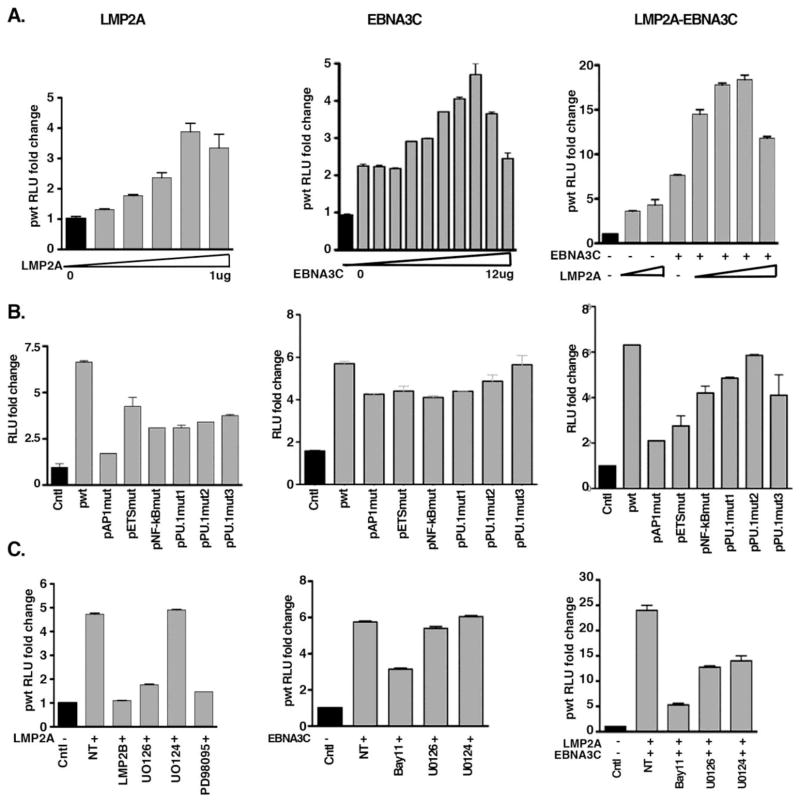
In order to explore which EBV genes play a critical role in miR-155 activation, we evaluated EBV gene function individually in EBV-negative cells BL30 and DG75, in which the level of DNA methylation at the miR-155 CpG island is low. In response to LMP1 and LMP2A, cellular miR-155 expression exhibits induction as reported before (Du et al., 2011; Rahadiani et al., 2008; Yin et al., 2008a) showing that LMP2A and LMP1 slightly induced endogenous pri-miR-155 expression alone or corporately in LMP2A and/or LMP1 stable expression cells, BL30 (Fig. 4A–C). We also found that LMP1, LMP2A, EBNA3C or EBNA3A/3B/3C and EBNA2 can slightly induce endogenous pri-miR-155 expression cooperatively after a 72 hour transient transfection in EBV-negative cells DG75 (Fig. 4D). Furthermore, both LMP2A and EBNA3C promoted miR-155 promoter activity alone or together (Fig. 5A). Mutation of the AP1 site of the miR-155 promoter reduced LMP2A and LMP2A-EBNA3C-mediated miR-155 promoter activation, but not EBNA3C-mediated miR-155 promoter activation (Fig. 5B). In order to further elucidate whether the AP1 signaling pathway plays a key role in miR-155 activation, we transiently expressed LMP2A and/or EBNA3C and treated cells with MEK/ERK and NF-κB inhibitors. A luciferase assay demonstrated that ERK inhibitors UO126 and PD98059 block LMP2A-mediated miR-155 promoter activity. Bay11 inhibited LMP2A and EBNA3C-mediated miR-155 promoter activity (Fig. 5C). These data indicate that LMP2A activates miR-155 expression through the AP1 element and the ERK signaling pathway. However, EBNA3C-mediated miR-155 promoter activation is not dependent on AP1 activity, since neither the miR-155 promoter AP1 mutant nor ERK inhibitors affect EBNA3C’s ability to induce miR-155 promoter activity. Therefore, EBNA3C-mediated miR-155 activation likely occurs through another promoter element. Notably, however, treatment with Bay11 can partially block EBNA3C-mediated miR-155 promoter activation (Fig. 5C). In conclusion, some EBV genes can stimulate miR-155 expression via AP1 elements, though we didn’t find any single or combination of EBV gene/s capable of inducing miR-155 expression dramatically.
Fig. 4. EBV genes activate endogenous pri-miR-155 expression.
A–C. Latency membrane proteins induced pri-miR-155 expression. Stable expression of EBV genes, LMP2A (LMP2A-1 & LMP2A-2, L2A) and/or LMP1 (LMP1-1, LMP1-2 & LMP1-3, L1) was performed in the EBV-negative cell line BL30, and RNA was extracted for real-time RT-PCR to detect endogenous pri-miR-155 expression. D. Assessment of the effect of EBV latent genes on pri-miR-155 expression. Transient transfection was conducted to co-express EBNA3C, LMP2A and EBNA3C (E3C-L2A), LMP2A, EBNA3C, EBNA2 and LMP1 (E3C-L2A-E2-L1), or LMP2A, EBNA3A/3B/3C, EBNA2 and LMP1 (E3A-B-C-L2A-E2-L1) in EBV-negative cells (DG75), and followed by RNA extraction for RT-PCR 72 hours post transfection. The relative expression fold change of pri-miR-155 was calculated based on the 2−ΔΔCT value.
Fig. 5. LMP2A and EBNA3C regulate miR-155 promoter activity.
A. LMP2A and EBNA3C activate miR-155 promoter activity in a dose dependent manner. Co-transfection of wild type miR-155 promoter (pwt) with LMP2A and/or EBNA3C at the indicated concentrations into EBV-negative cells (MutuE1dn) was performed and a luciferase assay was conducted after 48 hours. The RLU fold change was calculated on the basis of the RLU value without LMP2A and/or EBNA3C. B. The miR-155 promoter AP1 mutant (pAP1mut) was activated by 5ug of EBNA3C but not by 0.5ug of LMP2A or 0.5ug of LMP2A plus 5ug of EBNA3C (LMP2A-EBNA3C). LMP2A and/or EBNA3C were co-transfected with miR-155 promoter wild type (pwt) or miR-155 promoter mutants (pAP1mut, pEtsmut, pNF-κBmut and pPU.1muts) into EBV-negative MutuE1dn cells and cells were harvested for a luciferase assay after 48 hours. The RLU fold change was calculated on the basis of the RLU value of control (Cntl), pGL3-basic. C. MEK kinase inhibitors inhibit LMP2A-mediated miR-155 promoter activity but not EBNA3C-mediated miR-155 promoter activity. Wild type miR-155 promoter (pwt) was co-transfected with LMP2A or/and EBNA3C, then cells were treated with the MEK inhibitors U126 or PD98095, or with the NF-κB inhibitor Bay11, and a luciferase assay was carried out after 48 hours. The RLU fold change was calculated on the basis of the RLU value of control (Cntl), no LMP2A and/or EBNA3C.
Discussion
Genetic and epigenetic research has shown that DNA methylation is related to carcinogenesis. Although we have demonstrated that EBV induced the expression of oncogenic miR-155 in EBV-positive latency type III cells and in EBV-negative cells (Ramos, BL41 and BL30) (Yin et al., 2008a), the miR-155 DNA methylation profiles have not been reported in detail. Our results here demonstrate that pri-miR-155 contains a high confidence CpG island spanning the promoter and the first exon. The analysis of methylation status further elucidates lower DNA methylation at the miR-155 promoter correlates with high miR-155 and EBV latency gene expression in EBV-positive latency type III cells; and higher DNA methylation correlates with low miR-155 and EBV latency gene expression in EBV-positive latency type I cells. Since EBV genes interact with DNA methylation pathways (Hino et al., 2009; Tsai et al., 2006), EBV genes may regulate miR-155 promoter methylation to promote miR-155 expression. In order to address this issue, EBV-negative lymphoma cells (Ramos, BL41 and BL30) were infected with EBV (B95-8 or B652) followed by analysis of miR-155 promoter methylation. We observed that there is no significant change in DNA methylation of the miR-155 promoter after EBV infection in Ramos, BL41 and BL30, which have low DNA methylation status in the miR-155 promoter region. This may be because the level of methylation in these cells is already low on miR-155 promoter region or because EBV infection does not change the methylation status of miR-155 promoter. On the other hand, 5-aza-C can decrease DNA methylation of the miR-155 promoter in EBV-positive latency type I cells, Mutu and Akata, which have the highest DNA methylation status in the miR-155 promoter region. Of relevance, we have previously shown that 5-aza-C can activate miR-155 expression in Mutu and Akata cells (Yin et al., 2008a). These data suggest that EBV-induced miR-155 expression is not due to DNA demethylation in EBV-negative cells, and that DNA demethylation is required for miR-155 expression in EBV-positive cells, but may not regulate miR-155 expression alone.
Furthermore, our other studies on the role of EBV latent genes in the regulation of miR-155 didn’t reveal significant findings in latency type I cells exhibiting a hypermethylated miR-155 CpG island. For example, we have studied the role of EBNA1 in regulating miR-155 expression by knocking down EBNA1 in Mutu cells. We have also evaluated the function of EBV latent genes by stable transduction of EBV latent genes such as LMP1 or LMP2A in EBV latency type I cells. Our studies did not reveal induction of miR-155 after diminishing EBNA1 or overexpression of latency genes in EBV latency type I cells (data not shown) We have previously shown that the AP1 proteins FosB and JunB bind to the miR-155 promoter and result in miR-155 activation following BCR cross-linking (Yin et al., 2008b). EBV latent infection is associated with AP1 activation. Here, we demonstrate that the AP1 element is required for miR-155 expression in EBV-positive latency type III cells. Also FosB and JunB, as well as possibly other AP1 proteins, ATF3 and JunD are likely involved in EBV-induced miR-155 expression in EBV-positive latency type III cells through binding to the miR-155 promoter. NF-κB p65 has been previously shown to play a role in LMP1-induced miR-155 expression in EBV positive cells (Gatto et al., 2008; Thompson et al., 2013). In order to investigate the NF-κB function in miR-155 activation, we blocked the NF-κB signaling pathway by using the IkBa phosphorylation inhibitor Bay 11-7082. Interestingly, we did not see that Bay11 acts through the major NF-κB site in the miR-155 promoter. Instead, expression of AP1 proteins JunB, ATF3 and FosB were inhibited dramatically after Bay11 treatment. Moreover, Bay 11-7082 also reduced miR-155 expression. What is more, the MEK/ERK pathway inhibitor UO126 does not prevent the expression of AP1 proteins or miR-155 expression in EBV-positive latency type III cells (Fig. 3C). MG132, a proteasome inhibitor, did not affect AP1 protein expression or miR-155 expression, indicating that Bay 11-mediated AP1 protein inhibition is not dependent on ubiquitination.
LMP2A and EBNA3C alone or cooperatively can activate miR-155 promoter activity, and LMP2A activation of miR-155 is dependent on AP1 function since the reporter assay in Fig. 5B indicated that LMP2A-mediated miR-155 promoter activation decreases when the miR-155 promoter AP1 site is mutated, and in the presence MEK/ERK inhibitors. Therefore, LMP2A-mediated miR-155 expression is regulated by AP1 proteins through the MAPK signaling pathway. Consistent with previous reports, LMP2A is involved in influencing various signaling pathways, such as the B-cell receptor, MAPK and NF-κB pathways. However, EBNA3C-mediated miR-155 activation appears unrelated to either of the AP1 protein or MAPK signaling pathways. We intend to investigate next the role of EBNA3s-regulated cellular signaling in regulating miR-155 expression. Our data demonstrate that multiple EBV genes cooperate with each other in the induction of cellular oncogene miR-155 expression, thereby contributing to various EBV-mediated malignancies.
Conclusions
In this manuscript, we defined a molecular mechanism by which EBV activates miR-155 expression in B-cell lymphoma cells (Fig. 6). Activation of miR-155 by EBV is one factor contributing to lymphocyte oncogenesis. AP1 proteins and DNA hypomethylation are essential elements for EBV-mediated miR-155 activation. Targeting miR-155, AP1 proteins or DNA methylation may be an effective anti-cancer therapy for EBV-related lymphoma.
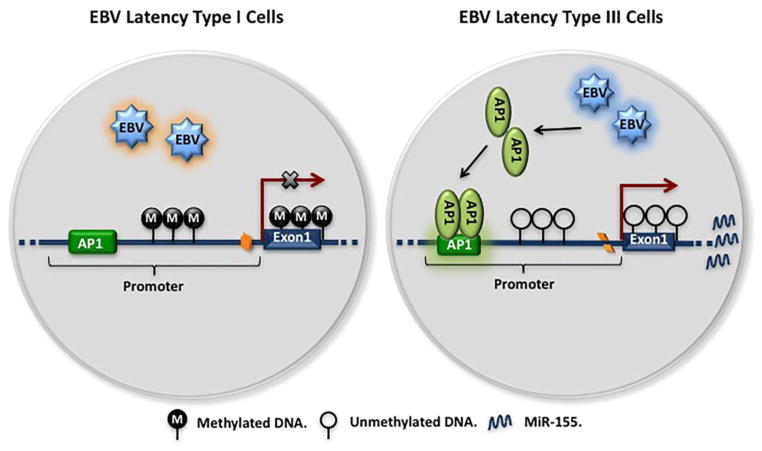
Fig. 6. The regulation of miR-155 expression by DNA methylation and AP1 in EBV-positive cells.
In EBV latency type I cells, hypermethylated DNA at the miR-155 CpG island represses miR-155 expression. In EBV latency type III cells, hypomethylation of the CpG island allows for miR-155 expression. What is more, EBV latency genes such as LMPs induce expression of AP1 genes, which bind to the AP1 site on the miR-155 promoter to activate miR-155 expression.
Materials and methods
Cell culture and treatments
Cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 0.5% penicillin-streptomycin (Invitrogen). 293 cells were cultured in Dulbecco’s modified Eagles Medium (DMEM - Invitrogen) plus 10% FBS and 0.5% penicillin-streptomycin. Retroviral transduced derivatives of cells cultured in the above respective culture media plus 250 μg/ml hygromycin for pEHyg-LMP1transduced cells and/or 1 mg/ml gentamicin for pMSCV-neo-LMP2A transduced cells (Yin et al., 2008a). All cells were maintained at 37°C with 5% CO2 in a tissue culture incubator. All inhibitors including Bay11, UO126, UO124 and MG132 were purchased from Calbiochem.
Methylation analysis of CpG assay
DNA was prepared following the Qiagen protocol and the DNA was bisulfite modification with EZ DNA methylation™ kit (ZYMO research), PCR and pyrosequencing analysis was conducted by EpigenDx.
Transfection and luciferase reporter analysis
Either 1.25 μg of the wild type miR-155 promoter or mutant reporter plasmids (pGL3basic-miR-155p, and pGL3basic-miR-155p-AP1mut, -NF-κBmut, -ETSmut and -PU.1muts) were co-transfected with the indicated concentration of the pSG5-LMP2A and/or pcDNA3-EBNA3C plasmid into 2 × 106 Mutu cells using Lipofectamine (Invitrogen) per the manufacturer’s instructions. Cells were harvested 48 hours post-transfection and analyzed for luciferase reporter activity according to the manufacturer’s protocol (Promega). Co-transfection of pcDNA3-EBNA3A, -3B, -3C, pSG5-LMP2A, -LMP1, and -EBNA2 were performed with Amaxa® Nucleofector™ Kit V in EBV-negative cells DG75 and cells were harvested for RT-PCR after 72 hours.
Real-time RT-PCR analysis
Total RNA was prepared using an RNeasy plus mini kit (Qiagen, cat# 74136) and cDNA was synthesized using the iScript cDNA Synthesis kit (BioRad, Cat# 170-8890). Real-time RT-PCR for pri-miR-155 and G3PDH was performed using the published condition and primers (Yin et al., 2008b). The relative gene expression was calculated by 2−ΔΔCT method, in which ΔΔCT = (ΔCT average in the control cells) − (ΔCT in the EBV gene-transduced cells), and ΔCT = (CT value of pri-miR-155) − (CT value of G3PDH).
Western blot analysis and antibodies
Cells were lysed in 1X SDS page loading buffer, sheared with ultrasound and heated at 95 C for 10 minutes. Protein concentrations were determined using a ND-1000 spectrophotometer (NanoDrop). Twenty-five μg of total protein was loaded in each well and separated in a 4–20% Tri-HCl Criterion Precast gel (BioRad, cat# 345-0033), then transferred to a nitrocellulose transfer membrane (Whatman Schleicher & Schuell, cat# 10 401196). The membranes were incubated overnight at 4°C with primary antibody in Tris-buffered saline (TBS) containing 5% low-fat powdered milk. The membranes were washed 3X (10 minutes each) in TBS buffer, incubated for 1 hour at room temperature with the appropriate secondary antibody in TBS buffer containing 5% low-fat powdered milk. Membranes were washed 3X (15 minutes each) in TBS buffer and subjected to image analysis using an Odyssey infrared imaging system (Li-Cor Biosciences). Actin, JunB, FosB, JunD and ATF3 antibodies were purchased from Santa Cruz Biotechnology.
Generation of stable LMP1/LMP2A expressing cell lines
Cell lines with stable expression of EBV latency genes, pEHyg-LMP1 and/or pMSCV-neo-LMP2A were generated using a retrovirus infections method following the published protocol (Yin et al. 2008a) in the BL30 EBV-negative cell line.
Chromosome Immunoprecipitation (ChIP)
ChIP assay was performed as described before (Yin et al., 2008b). Briefly, 2 × 107 cells were cross-linked with 1% of formaldehyde and lysed in 400ul of RIPA buffer. The chromatin was sonicated to an average size of 600bp. The 400ug of resulting protein was used for IP with antibodies, JunB, FosB, ATF3, JunD, H3 or Acetylate-H4, and 100 ug of protein as input. The resulting immunoprecipitate and inputs were incubated with RNase A for 1 hour at 37°C then digested for 5 hours using proteinase K. Samples were resuspended in 20 μl of H2O for PCR after phenol/chloroform-extraction and ethanol-precipitation. PCR condition and primers were used as described before (Yin et al. 2008b).
Electrophoretic Mobility Shift Analysis (EMSA)
EMSA experiments were carried out following the protocol described as previously (Yin et al., 2008b). The AP1 probe and its competitor oligonucleotides are identical to those previously described in Yin (Yin et al., 2008b).
Supplementary Material
EBV latency type I cells, Mutu (A.) and Akata (B.) were treated with antibodies against IgM (Mutu) or IgG (Akata) for the indicated time point, and cells were harvested for protein extraction and RNA preparation. Expression of latent (LMP1 and LMP2A), immediate-early (Zta and Rta), early (BMRF1 and VCA) proteins shown by immunoblotting (upper panel) and expression of pri-miR-155 shown by CT value of real-time RT-PCR (lower panel).
The numbers on the top row of each table indicate the position of CpG sites. The methylation status was assessed by bisulfite sequencing and the percentage methylation of each CpG site was shown. Neg11, neg5 and neg3 indicate the position of −11, −5 and −3 from TTS. Mutu Bulk Cntl (an EBV-positive latency type I cell line) was treated with DNA demethylating agents Zebularine (Mutu Bulk Zeb) or 5-aza-deoxycytidine (Mutu Bulk 5A). The EBV-negative cell lines (Ramos, BL41 and BL30) were infected with EBV virus (B95-8 or B652) to generate EBV-positive cell lines, Ramos (B95-8), BL41(B652) and BL30 (B652).
Highlights.
The DNA methylation of miR-155 CpG island regulates miR-155 expression in EBV-positive cells.
AP1 signaling pathway is involved in EBV-mediated miR-155 activation.
The regulation of miR-155 by EBV is dependent on more than one EBV gene or multiple cell signaling pathways.
Acknowledgments
We thank Ms. Melody C. Banboo and Dr. Jennifer E. Cameron for their technical assistance in protein and RNA preparation. We thank Drs. Fayong Luo and Mark D. Sides for expert review of the manuscripts. This work was supported by US National Institutes of Health Ruth L. Kirschstein National Research Service Awards 5T32HL007973 (Q.Y.), Ladies Leukemia League (J.A.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Qinyan Yin, Email: qyin@tulane.edu.
Xia Wang, Email: xwang4@tulane.edu.
Claire Roberts, Email: cfewell@tulane.edu.
Erik K. Flemington, Email: erik@tulane.edu.
Joseph A. Lasky, Email: jlasky@tulane.edu.
References
- Anderson LJ, Longnecker R. An auto-regulatory loop for EBV LMP2A involves activation of Notch. Virology. 2008;371:257–266. doi: 10.1016/j.virol.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz GL, Shackelford J, Pagano JS. Epstein-Barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J Virol. 2012;86:12251–12261. doi: 10.1128/JVI.01407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JE, Fewell C, Yin Q, McBride J, Wang X, Lin Z, Flemington EK. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology. 2008;382:257–266. doi: 10.1016/j.virol.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Lu J, Shih YC, Tsai CH. Epstein-Barr virus latent membrane protein 2A regulates c-Jun protein through extracellular signal-regulated kinase. J Virol. 2002a;76:9556–9561. doi: 10.1128/JVI.76.18.9556-9561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Lu J, Shih YC, Tsai CH. Epstein-Barr virus latent membrane protein 2A regulates c-Jun protein through extracellular signal-regulated kinase. J Virol. 2002b;76:9556–9561. doi: 10.1128/JVI.76.18.9556-9561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZM, Hu LF, Wang HY, Yan LX, Zeng YX, Shao JY, Ernberg I. Upregulation of MiR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One. 2011;6:e19137. doi: 10.1371/journal.pone.0019137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels N, Yigit G, Emmerich CH, Czesnik D, Schild D, Wienands J. Epstein-Barr virus LMP2A signaling in statu nascendi mimics a B cell antigen receptor-like activation signal. Cell Commun Signal. 2012;10:9. doi: 10.1186/1478-811X-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersing I, Bernhardt K, Gewurz BE. NF-kappaB and IRF7 pathway activation by Epstein-Barr virus Latent Membrane Protein 1. Viruses. 2013;5:1587–1606. doi: 10.3390/v5061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries KL, Miller WE, Raab-Traub N. The A20 protein interacts with the Epstein-Barr virus latent membrane protein 1 (LMP1) and alters the LMP1/TRAF1/TRADD complex. Virology. 1999;264:159–166. doi: 10.1006/viro.1999.9980. [DOI] [PubMed] [Google Scholar]
- Galm O, Wilop S, Luders C, Jost E, Gehbauer G, Herman JG, Osieka R. Clinical implications of aberrant DNA methylation patterns in acute myelogenous leukemia. Annals of Hematology. 2005;84:39–46. doi: 10.1007/s00277-005-0005-0. [DOI] [PubMed] [Google Scholar]
- Garuti A, Rocco I, Cirmena G, Chiaramondia M, Baccini P, Calabrese M, Palermo C, Friedman D, Zoppoli G, Ballestrero A. Quantitative Real Time PCR assessment of hormonal receptors and HER2 status on fine-needle aspiration pre-operatory specimens from a prospectively accrued cohort of women with suspect breast malignant lesions. Gynecol Oncol. 2014;132:389–396. doi: 10.1016/j.ygyno.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 2008;36:6608–6619. doi: 10.1093/nar/gkn666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada K, Fukayama M. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- House MG, Guo M, Efron DT, Lillemoe KD, Cameron JL, Syphard JE, Hooker CM, Abraham SC, Montgomery EA, Herman JG, Brock MV. Tumor suppressor gene hypermethylation as a predictor of gastric stromal tumor behavior. J Gastrointest Surg. 2003a;7:1004–1014. doi: 10.1016/j.gassur.2003.08.002. discussion 1014. [DOI] [PubMed] [Google Scholar]
- House MG, Wistuba II, Argani P, Guo M, Schulick RD, Hruban RH, Herman JG, Maitra A. Progression of gene hypermethylation in gallstone disease leading to gallbladder cancer. Ann Surg Oncol. 2003b;10:882–889. doi: 10.1245/aso.2003.02.014. [DOI] [PubMed] [Google Scholar]
- Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. Gene silencing quantitatively controls the function of a developmental trans-activator. Molecular Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, Tinguely M, Faggioni A, Trivedi P, Meister G, Renner C, Grasser FA. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011;39:1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Lee EJ, Schmittgen TD. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- Johansen LM, Deppmann CD, Erickson KD, Coffin WF, Thornton TM, Humphrey SE, Martin JM, Taparowsky EJ. EBNA2 and activated notch induce expression of BATF. J Virol. 2003;77:6029–6040. doi: 10.1128/JVI.77.10.6029-6040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Epigenetics in carcinogenesis and cancer prevention. Epigenetics in Cancer Prevention: Early Detection and Risk Assessment. 2003;983:213–219. doi: 10.1111/j.1749-6632.2003.tb05976.x. [DOI] [PubMed] [Google Scholar]
- Jonigk D, Izykowski N, Maegel L, Schormann E, Maecker-Kolhoff B, Laenger F, Kreipe H, Hussein K. MicroRNA expression in Epstein-Barr virus-associated post-transplant smooth muscle tumours is related to leiomyomatous phenotype. Clin Sarcoma Res. 2013;3:9. doi: 10.1186/2045-3329-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser A, Kilger E, Gires O, Ueffing M, Kolch W, Hammerschmidt W. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 1997;16:6478–6485. doi: 10.1093/emboj/16.21.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J, Haralambieva E, de Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, van den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–153. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, Cheng JQ. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–6784. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J Virol. 2010;84:11670–11678. doi: 10.1128/JVI.01248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Johannsen E, Illanes D, Cooper A, Zhao B, Kieff E. Epstein-Barr virus nuclear protein 3A domains essential for growth of lymphoblasts: transcriptional regulation through RBP-Jkappa/CBF1 is critical. J Virol. 2005;79:10171–10179. doi: 10.1128/JVI.79.16.10171-10179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning S, Hahn AM, Huye LE, Pagano JS. Interferon regulatory factor 7 regulates expression of Epstein-Barr virus latent membrane protein 1: a regulatory circuit. J Virol. 2003;77:9359–9368. doi: 10.1128/JVI.77.17.9359-9368.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil JD, Owen TJ, Wood VH, Date KL, Valentine R, Chukwuma MB, Arrand JR, Dawson CW, Young LS. Epstein-Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J Gen Virol. 2008;89:2833–2842. doi: 10.1099/vir.0.2008/003392-0. [DOI] [PubMed] [Google Scholar]
- Pacchiarotti I, Bond DJ, Baldessarini RJ, Nolen WA, Grunze H, Licht RW, Post RM, Berk M, Goodwin GM, Sachs GS, Tondo L, Findling RL, Youngstrom EA, Tohen M, Undurraga J, Gonzalez-Pinto A, Goldberg JF, Yildiz A, Altshuler LL, Calabrese JR, Mitchell PB, Thase ME, Koukopoulos A, Colom F, Frye MA, Malhi GS, Fountoulakis KN, Vazquez G, Perlis RH, Ketter TA, Cassidy F, Akiskal H, Azorin JM, Valenti M, Mazzei DH, Lafer B, Kato T, Mazzarini L, Martinez-Aran A, Parker G, Souery D, Ozerdem A, McElroy SL, Girardi P, Bauer M, Yatham LN, Zarate CA, Nierenberg AA, Birmaher B, Kanba S, El-Mallakh RS, Serretti A, Rihmer Z, Young AH, Kotzalidis GD, MacQueen GM, Bowden CL, Ghaemi SN, Lopez-Jaramillo C, Rybakowski J, Ha K, Perugi G, Kasper S, Amsterdam JD, Hirschfeld RM, Kapczinski F, Vieta E. The International Society for Bipolar Disorders (ISBD) task force report on antidepressant use in bipolar disorders. Am J Psychiatry. 2013;170:1249–1262. doi: 10.1176/appi.ajp.2013.13020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YR, Vatsyayan J, Chang YS, Chang HY. Epstein-Barr virus latent membrane protein 2A upregulates UDP-glucose dehydrogenase gene expression via ERK and PI3K/Akt pathway. Cell Microbiol. 2008;10:2447–2460. doi: 10.1111/j.1462-5822.2008.01221.x. [DOI] [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Port RJ, Pinheiro-Maia S, Hu C, Arrand JR, Wei W, Young LS, Dawson CW. Epstein-Barr virus induction of the Hedgehog signalling pathway imposes a stem cell phenotype on human epithelial cells. J Pathol. 2013;231:367–377. doi: 10.1002/path.4245. [DOI] [PubMed] [Google Scholar]
- Portis T, Longnecker R. Epstein-Barr virus (EBV) LMP2A mediates B-lymphocyte survival through constitutive activation of the Ras/PI3K/Akt pathway. Oncogene. 2004;23:8619–8628. doi: 10.1038/sj.onc.1207905. [DOI] [PubMed] [Google Scholar]
- Radkov SA, Bain M, Farrell PJ, West M, Rowe M, Allday MJ. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkov SA, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday MJ. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahadiani N, Takakuwa T, Tresnasari K, Morii E, Aozasa K. Latent membrane protein-1 of Epstein-Barr virus induces the expression of B-cell integration cluster, a precursor form of microRNA-155, in B lymphoma cell lines. Biochemical and biophysical research communications. 2008;377:579–583. doi: 10.1016/j.bbrc.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Schaefer BC, Woisetschlaeger M, Strominger JL, Speck SH. Exclusive Expression of Epstein-Barr-Virus Nuclear Antigen-1 in Burkitt-Lymphoma Arises from a 3rd Promoter, Distinct from the Promoters Used in Latently Infected Lymphocytes. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:6550–6554. doi: 10.1073/pnas.88.15.6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shair KH, Bendt KM, Edwards RH, Nielsen JN, Moore DT, Raab-Traub N. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) and LMP2A function cooperatively to promote carcinoma development in a mouse carcinogenesis model. J Virol. 2012;86:5352–5365. doi: 10.1128/JVI.07035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS pathogens. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Dawson CW, Takada K, Curnow J, Moody CA, Sixbey JW, Young LS. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc Natl Acad Sci U S A. 2004;101:15730–15735. doi: 10.1073/pnas.0402135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobl LJ, Hofelmayr H, Marschall G, Brielmeier M, Bornkamm GW, Zimber-Strobl U. Activated Notch1 modulates gene expression in B cells similarly to Epstein-Barr viral nuclear antigen 2. J Virol. 2000;74:1727–1735. doi: 10.1128/jvi.74.4.1727-1735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Vardinogiannis I, Gilmore TD. Identification of an NF-kappaB p50/p65-responsive site in the human MIR155HG promoter. BMC Mol Biol. 2013;14:24. doi: 10.1186/1471-2199-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Li HP, Lu YJ, Hsueh C, Liang Y, Chen CL, Tsao SW, Tse KP, Yu JS, Chang YS. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668–11676. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- Wang P, Hou J, Lin L, Wang C, Liu X, Li D, Ma F, Wang Z, Cao X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- Yim RL, Wong KY, Kwong YL, Loong F, Leung CY, Chu R, Lam WW, Hui PK, Lai R, Chim CS. Methylation of miR-155-3p in mantle cell lymphoma and other non-Hodgkin’s lymphomas. Oncotarget. 2014;5:9770–9782. doi: 10.18632/oncotarget.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, McBride J, Fewell C, Lacey M, Wang X, Lin Z, Cameron J, Flemington EK. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008a;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. J Biol Chem. 2008b;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg H, Stenglein M, Jansson A, Ricksten A, Rymo L. Relative levels of EBNA1 gene transcripts from the C/W, F and Q promoters in Epstein-Barr virus-transformed lymphoid cells in latent and lytic stages of infection. J Gen Virol. 1999;80(Pt 2):457–466. doi: 10.1099/0022-1317-80-2-457. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang Y, Sun Y, Zheng J, Zhu D. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Otorhinolaryngol. 2014;271:1939–1945. doi: 10.1007/s00405-013-2818-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EBV latency type I cells, Mutu (A.) and Akata (B.) were treated with antibodies against IgM (Mutu) or IgG (Akata) for the indicated time point, and cells were harvested for protein extraction and RNA preparation. Expression of latent (LMP1 and LMP2A), immediate-early (Zta and Rta), early (BMRF1 and VCA) proteins shown by immunoblotting (upper panel) and expression of pri-miR-155 shown by CT value of real-time RT-PCR (lower panel).
The numbers on the top row of each table indicate the position of CpG sites. The methylation status was assessed by bisulfite sequencing and the percentage methylation of each CpG site was shown. Neg11, neg5 and neg3 indicate the position of −11, −5 and −3 from TTS. Mutu Bulk Cntl (an EBV-positive latency type I cell line) was treated with DNA demethylating agents Zebularine (Mutu Bulk Zeb) or 5-aza-deoxycytidine (Mutu Bulk 5A). The EBV-negative cell lines (Ramos, BL41 and BL30) were infected with EBV virus (B95-8 or B652) to generate EBV-positive cell lines, Ramos (B95-8), BL41(B652) and BL30 (B652).