Abstract
Studying phospholipases A2 (PLA2s) is a challenging task since they act on membrane-like aggregated substrates and not on monomeric phospholipids. Multidisciplinary approaches that include hydrogen/deuterium exchange mass spectrometry (DXMS) and computational techniques have been employed with great success in order to address important questions about the mode of interactions of PLA2 enzymes with membranes, phospholipid substrates and inhibitors. Understanding the interactions of PLA2s is crucial since these enzymes are the upstream regulators of the eicosanoid pathway liberating free arachidonic acid (AA) and other polyunsaturated fatty acids (PUFA). The liberation of AA by PLA2 enzymes sets off a cascade of molecular events that involves downstream regulators such as cyclooxygenase (COX) and lipoxygenase (LOX) metabolites leading to inflammation. Aspirin and other nonsteroidal anti-inflammatory drugs (NSAIDs) work by inhibiting COX, while Zileuton inhibits LOX and both rely on PLA2 enzymes to provide them with AA. That means PLA2 enzymes can potentially also be targeted to diminish inflammation at an earlier point in the process. In this review we describe extensive efforts reported in the past to define the interactions of PLA2 enzymes with membranes, substrate phospholipids and inhibitors using DXMS, molecular docking, and molecular dynamics (MD) simulations.
Keywords: Phospholipase A2, PAPC, catalytic cycle, allosteric regulation, eicosanoid pathway, molecular dynamics simulations, DXMS
Introduction
Through their catalytic action, phospholipases A2 (PLA2) de-esterify and release free fatty acids from the sn-2 position of membrane phospholipids including arachidonic acid (AA) and related polyunsaturated fatty acids (PUFA) (Dennis et al. , 2011). Eicosanoids, which are associated with inflammation, are produced by the downstream enzymes of the eicosanoid pathway such as cyclooxygenases (COX), lipoxygenases (LOX), and cytochrome P450s (CYP) that rely on PLA2s to provide them with AA and related PUFA (Buczynski et al. , 2009, Dennis and Norris, 2015, Funk, 2001). Existing nonsteroidal anti-inflammatory drugs (NSAID) target the downstream enzymes of the prostaglandin pathway. As the upstream regulators of the eicosanoid pathway, PLA2 enzymes can be targeted to diminish inflammation at an earlier stage in the process. Besides their implication in the inflammatory process, PLA2 enzymes are also implicated in various diseases including atherosclerosis,(Karabina et al. , 2010, Rosenson and Hurt-Camejo, 2012) diabetes,(Ayilavarapu et al. , 2010, Bone et al. , 2015) arthritis(Hegen et al. , 2003, Masuda et al. , 2005) and cancer(Nakanishi and Rosenberg, 2006, Park et al. , 2012, Scott et al. , 2010) and inhibition of the enzymatic activity of specific PLA2s should be beneficial for the treatment of these diseases (Bone et al. , 2015, Karabina et al. , 2010). Therefore, there has been great interest in understanding the interactions of PLA2 enzymes with membranes, substrates and inhibitors in order to develop effective new therapeutic agents.
The PLA2 superfamily consists of 16 groups and many subgroups and each has been classified into one of the six main types based on their characteristics of sequence and structure. The six main types are secreted (sPLA2), cytosolic (cPLA2), calcium-independent (iPLA2), platelet-activating factor acetylhydrolase (PAF-AH) also known as lipoprotein-associated (LpPLA2), lysosomal (LPLA2), and adipose (AdPLA) (Dennis et al. , 2011). Even though the PLA2 superfamily constitutes a diverse set of enzymes, they share the same catalytic activity because of convergent evolution. Nevertheless, divergent PLA2 enzymes have a distinct 3D-structure, binding pocket and catalytic residues as well as a unique, in terms of topology and function, interfacial surface through which they associate with the membrane to extract and bind their phospholipid substrate. Since PLA2 enzymes have to associate with biological membranes to complete their catalytic cycle, they evolved to act on membrane-like phospholipids such as micelles, membranes and liposomes rather than on monomeric phospholipids. Thus, PLA2s follow a special kinetic model which we named “surface dilution kinetics” that was successfully employed to interpret the kinetic characteristics of these enzymes (Carman et al. , 1995).
This short review focuses on the application of hydrogen/deuterium exchange mass spectrometry (DXMS)(Cao et al. , 2013) and computational techniques(Durrant and McCammon, 2011, Sliwoski et al. , 2014) to understanding the mechanism of action and inhibition of Group IVA cytosolic PLA2 (GIVA cPLA2)(Clark et al. , 1991, Sharp et al. , 1991) and Group VIA calcium-independent PLA2 (GVIA iPLA2) (Larsson et al. , 1998). Molecular dynamics (MD) simulations, molecular docking, and other computer-aided design techniques guided by DXMS experimental data have been utilized with great success in understanding the association and interactions of these two intracellular enzymes with membranes in a biphasic water-lipid system as well as with specific phospholipid substrates and inhibitors (Mouchlis et al. , 2015).
Structure and function of GIVA cPLA2 and GVIA iPLA2
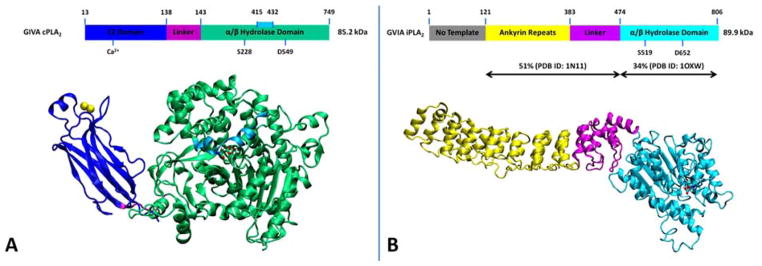
The 85 kDa human GIVA cPLA2 (cPLA2α, calcium-dependent) contains 749 amino acids and it was cloned and sequenced in 1991 from U937 cells (Clark et al. , 1991, Kramer et al. , 1991). The crystal structure of the enzyme revealed an N-terminal C2 domain (residues: 13–137), a linker region (residues: 138–143) and a C-terminal α/β hydrolase catalytic domain (residues: 144–749, Fig. 1A) (Dessen et al. , 1999). GIVA cPLA2 has a deep channel-like binding site that is located in the α/β hydrolase catalytic domain and contains the catalytic dyad of Ser/Asp (Mouchlis et al. , 2015). A peptide region consisting of residues 415–432 is on the top of the binding pocket and is defined as the lid (Dessen et al. , 1999). The lid region is amphipathic showing increased on-exchanged rates in DXMS experimental data upon binding with phospholipid vesicles and inhibitors (Burke et al. , 2009, Burke et al. , 2008). Our recent studies using MD simulations guided by DXMS data showed high flexibility of the lid region during the extraction of a phospholipid molecule from the membrane into the binding pocket (Mouchlis et al. , 2015). GIVA cPLA2 is regulated by intracellular calcium through a calcium binding site located at the C2 domain that activates the translocation of the enzyme to the phospholipid membrane (Fig. 1A). Using DXMS experiments, we showed that calcium causes conformational changes in the C2 domain to stabilize the C2 domain and facilitate association of the enzyme with the membrane as shown by decreased hydrogen/deuterium (H/D) exchange rates (Hsu et al. , 2008). GIVA cPLA2 activity is enhanced by binding to phosphatidylinositol-4,5-bisphosphate (PIP2) and this activation is calcium-independent (Balsinde et al. , 2000, Leslie and Channon, 1990, Six and Dennis, 2003). Four lysine residues Lys488, Lys541, Lys543, and Lys544, have been shown to constitute a PIP2 binding site and to be critical for PIP2-mediated activation of GVIA cPLA2 (Das and Cho, 2002, Six and Dennis, 2003). Ceramide 1-phosphate (C1P) which is a phosphorylated sphingolipid has also been suggested to activate GIVA cPLA2 (Chalfant and Spiegel, 2005, Pettus et al. , 2004). This enzyme exhibits specificity for phospholipids containing AA esterified at the sn-2 position (Clark et al. , 1991, Ghosh et al. , 2006).
Figure 1.
3D structure of the enzymes. (A) The X-ray crystal structure of GIVA cPLA2 (PDB ID 1CJY). (B) The homology model of GVIA iPLA2. Reprinted from reference Mouchlis et al. , 2015.
The human GVIA iPLA2 (iPLA2β, PNPLA9, calcium-independent) has a molecular weight of 89.9 kDa and it has longest sequence among the multiple splice variants expressed by the human gene (Larsson Forsell et al. , 1999, Larsson et al. , 1998). The X-ray crystal structure of the enzyme has not been solved yet but homology modeling studies showed that region 121–474 has 51% homology with human ankyrinR (PDB ID: 1N11)(Michaely et al. , 2002) and region 475–806 has 34% homology with patatin (PDB ID: 1OXW) (Rydel et al. , 2003). According to the homology models the structure of GVIA iPLA2 consists of the ankyrin repeats (residues: 121–383), a linker region (residues 384–474), and an α/β hydrolase catalytic domain (residues: 475–806) (Dennis et al. , 2011, Mouchlis et al. , 2015). No homology template was found for region 1–120 (Mouchlis et al. , 2015). ATP was found to enhance the activity of GVIA iPLA2, but it is not a substrate or cofactor for this enzyme (Hazen and Gross, 1991, Lio and Dennis, 1998). GVIA iPLA2 also contains a catalytic dyad of Ser/Asp and has low specificity for the free fatty acid esterified at the sn-2 position (Lio and Dennis, 1998). Besides its phospholipase A2 activity, GVIA iPLA2 also exhibits lysophospholipase, transacylase and acyl-CoA thioesterase activity (Jenkins et al. , 2006, Winstead et al. , 2000).
GIVA cPLA2 and GVIA iPLA2 in eicosanoid biosynthesis
Eicosanoid biosynthesis requires free AA which is provided by PLA2 enzymes under physiological or inflammatory cell activation. GIVA cPLA2 and GVIA iPLA2 have been implicated in cellular eicosanoid production. GIVA cPLA2 is activated by Toll-like receptors (LTR), purinergic receptors and other receptors and it is translocated to the perinuclear and endoplasmic reticulum membranes during inflammatory responses (Dennis and Norris, 2015). After its translocation, it releases AA from membrane phospholipids containing esterified AA at the sn-2 position leading to the production of pro-inflammatory eicosanoids (Buczynski et al. , 2009, Buczynski et al. , 2007). Our group showed that in macrophages GIVA cPLA2 may also hydrolyze phospholipids that have other PUFAs such as EPA, DPA and DHA esterified at the sn-2 position, which are precursors for anti-inflammatory resolvins (Keyes et al. , 2010, Norris and Dennis, 2012, Norris et al. , 2014). GVIA iPLA2 constitutively releases a low level of free fatty acids and it is primarily involved in membrane homeostasis and remodeling(Balsinde et al. , 1997, Dennis et al. , 2011) The specificity of GVIA iPLA2 for the free fatty acid at the sn-2 position is not great; thus, its activity may include hydrolysis of AA that is esterified at this position.
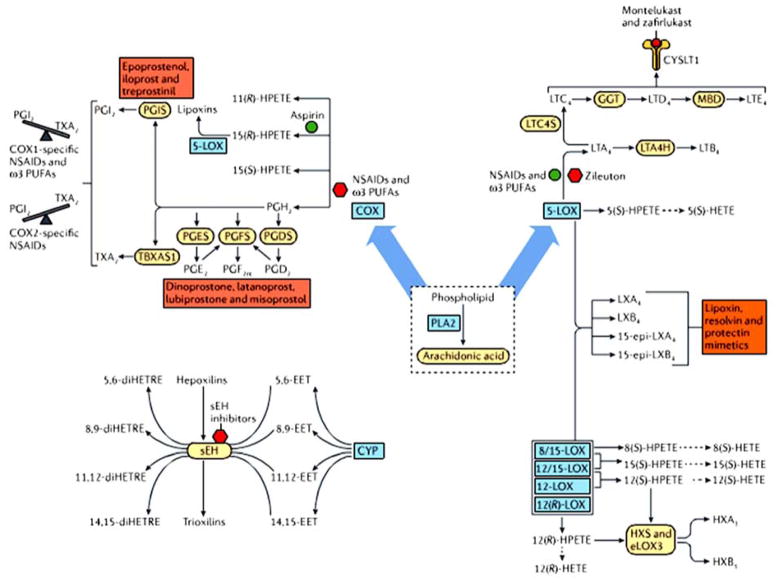
Downstream enzymes in the cyclooxygenase (COX) pathway generate prostaglandins (PG), thromboxanes (TX) and lipoxins (LX, Fig.2). In the lipoxygenase (LOX) pathway, the downstream enzymes generate leukotrienes (LTs), hydroxyeicosatetraenoic acids (HETE) and hepoxilins (HX, Fig. 2). Finally, in the cytochrome P450 (CYP) epoxy hydrolase pathway, the downstream enzymes generate epoxides and dihydroxy polyunsaturated fatty acids (PUFA, Fig. 2) (Buczynski et al. , 2009, Dennis and Norris, 2015). Pharmaceutical agents that target the prostaglandin pathway have been widely used for decades. Aspirin is the best example among the numerous NSAID that have been commercially available. NSAID inhibit COX-1 and COX-2 enzymes and as a result inhibit prostaglandin biosynthesis (Fig. 2) (Vane, 2002). Zileuton is a 5-LOX inhibitor, which is also a major therapeutic target for the treatment of allergic and asthmatic conditions (Fig. 2) (Rossi et al. , 2010). Other pathways including CYP and sEH have also been considered as pharmaceutical targets for inflammation and inflammatory diseases. Since PLA2 enzymes are the upstream regulators of the eicosanoid pathway, they should be potential therapeutic targets for inflammation and inflammation-related diseases such as cancer, arthritis and diabetes (Fig. 2).
Figure 2.
Eicosanoid pathway and its therapeutic targets. Downstream regulators such as COX, LOX and CYP rely on PLA2 enzymes to provide them with AA. Reprinted from Dennis and Norris, 2015.
Catalytic cycle of GIVA cPLA2 and GVIA iPLA2
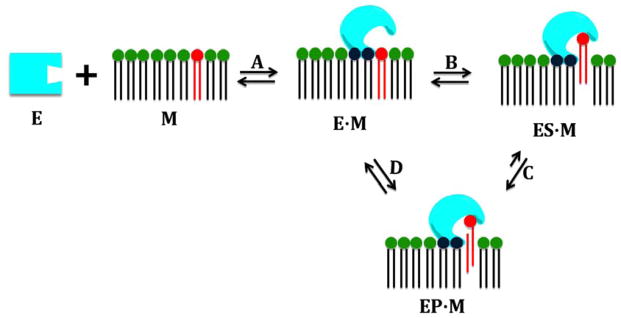
PLA2 enzymes are often water-soluble, but act as membrane associated enzymes showing a significant increase in activity when their phospholipid substrate is in a membrane-like form such as micelles, vesicles or liposomes rather than in a monomeric form. Most of the PLA2 enzymes have to associate with a lipid-water interface through their interfacial surface, and access their phospholipid substrate in order to complete their catalytic cycle which is the hydrolysis of the ester bond at the sn-2 position of the phospholipid molecule (Fig. 3). Thus, PLA2 enzymes can best be described with a special kinetic model called “surface dilution kinetics” (Carman et al. , 1995). This model includes four steps: A) association of the enzyme with the membrane, B) extraction and binding of a phospholipid molecule in the catalytic site, C) hydrolysis of the phospholipid molecule, and D) diffusion of the products in the membrane, followed by repetition of the cycle (Fig. 3). All of these steps exist in equilibrium. The membrane is a very important factor in the catalytic cycle of these enzymes, and recently we have introduced the idea that the membrane acts as an allosteric ligand shifting the conformation of PLA2 enzymes from the inactive (in water) to the active (on the membrane surface) form (Mouchlis et al. , 2015). These four steps are described in detail below.
Figure 3.
The catalytic cycle of GIVA cPLA2 and GVIA iPLA2. Reprinted from reference Mouchlis et al. , 2015.
A. Association of the enzyme with the membrane
GIVA cPLA2 and GVIA iPLA2 associate with membranes to extract and bind their phospholipid substrates even though they are in principal water-soluble enzymes. Therefore, membranes are an important factor for these enzymes. Elucidating their interactions with and conformational changes during the association with membrane is vital in understanding the mechanism of action of these two enzymes.
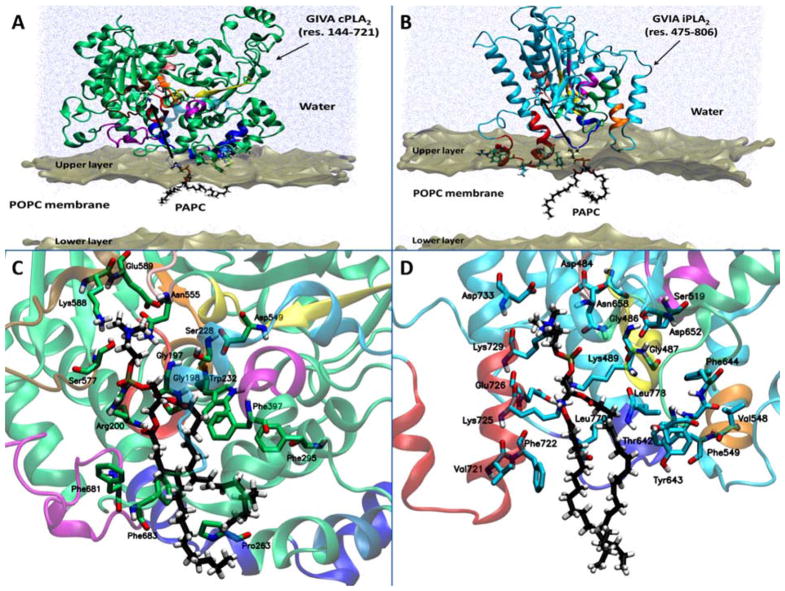
According to DXMS experimental data, five peptide regions of GIVA cPLA2 (28– 35, 36–39, 258–265, 268–279, and 466–470) exhibited decreased levels of deuteration in the presence of phospholipid vesicles (Burke et al. , 2008). Regions 28–35 and 36–39 are located on the C2 domain and their association with the phospholipid vesicles was found to be calcium-dependent because they exhibited a larger change in exchange in the presence of calcium (Burke et al. , 2008, Hsu et al. , 2008). Region 96–98 of the C2 domain was reported to play a significant role in the association of GIVA cPLA2 with the membrane (Málková et al. , 2005, Perisic et al. , 1999). Unfortunately, no peptides covering region 96–98 were identified during the digestion of GIVA cPLA2, so no DXMS data are available for interactions of that region with phospholipid vesicles (Burke et al. , 2008). Peptide regions 258–265, 268–279, and 466–470 (blue and light blue region in Fig. 4A) are located on the catalytic domain and were used to generated a three-dimensional model of the catalytic domain of GIVA cPLA2 on the surface of the membrane (Fig. 4A) (Mouchlis et al. , 2015). It is worth mentioning that the C2 domain penetrates the membrane approximately 15 Å, leading to the catalytic domain being near the surface of the membrane. Residues like Trp464 and Met468 penetrate the membrane surface interacting with the hydrophobic tails of the phospholipids, while residues such as Lys273, Lys274, and Arg467 interact with the head groups of the membrane phospholipids (Fig. 4A) (Mouchlis et al. , 2015).
Figure 4.
Association of the catalytic domain with the membrane bilayer for (A) GIVA cPLA2 and (B) GVIA iPLA2. Substrate binding of a PAPC phospholipid molecule in the catalytic site of (C) GIVA cPLA2 and (D) GVIA iPLA2 is shown. Reprinted from reference Mouchlis et al. , 2015.
The association of GVIA iPLA2 with the membrane has also been studied by our group using coarse-grained and atomistic simulations guided by DXMS experimental data (Bucher et al. , 2013, Hsu et al. , 2009). The ankyrin repeats (Fig. 1B) showed no interactions with phospholipid vesicles (Hsu et al. , 2009). MD simulations of the catalytic domain showed that region 708–730 (red region in Fig. 4B) is forming an amphipathic helix after penetrating the membrane with its hydrophilic residues such as Arg710 and Lys719 (Fig. 4B) to interact with the phospholipid head groups, and its hydrophobic residues like Pro711, Pro714, Trp715, Leu717, Val721, and Phe722 to interact with the fatty acid tails of the phospholipids (Bucher et al. , 2013). Our previously published DXMS data and MD simulations indicated that region 708–730 is likely the anchor region from which the enzyme associates with the membrane in contrast to GIVA cPLA2 that contains the C2 domain which assists membrane association of the enzyme (Bucher et al. , 2013, Burke et al. , 2008, Hsu et al. , 2009).
B. Extraction and binding of a phospholipid molecule in the catalytic site
Steered molecular dynamics (SMD) simulations guided by DXMS results gave insight into the extraction pathway of a PAPC phospholipid molecule from the membrane into the catalytic site of both GIVA cPLA2 and GVIA iPLA2(Mouchlis et al. , 2015). The direction that PAPC was forced into the catalytic site is indicated by the thick black arrow in Figures 4A and 4B. Reference Mouchlis et al. , 2015 includes animations in the supporting information that show the detailed extraction of PAPC from the membrane into the catalytic site of both enzymes (Mouchlis et al. , 2015).
During the extraction of the PAPC into the catalytic site of GIVA cPLA2, three regions that are located near the entrance of the site (261–268, 405–415, and 670–686) were found to play a significant role in the process. Residues like Asn268, Ser408, Gln411, Asn682, and Gln684 were found to participate in hydrogen-bonding with the head group of the PAPC molecule while is extracted into the catalytic site (see Movie S1 in reference Mouchlis et al. , 2015). Arg200 is stabilized by Ser680 and Glu418 when PAPC is not in the catalytic site, while PAPC enters the site approximately two-thirds of its length; Arg200 interacts with the phosphate group stabilizing its binding. The lid region (cyan region in Fig 4A and 4C) exhibits high flexibility whereas PAPC enters the binding pocket (Mouchlis et al. , 2015). Once PAPC completely enters the active site and during an MD simulation on the GIVA cPLA2-PAPC complex, Arg200 and Asn555 interact with the phosphate group while Glu418 and Glu589 interact with the positively charged choline group of the phospholipid head group (Fig. 4C). The oxygen atom of the sn-2 carbonyl group is located near the oxyanion hole (Gly197/Gly198) participating in hydrogen-bonding, while its carbon atom is located near catalytic Ser228. Finally the fatty acid chains are placed in the hydrophobic region of the catalytic site with the four double bonds of the sn-2 arachidonyl chain participating in π-π stacking with the aromatic residues Trp232, Phe295, Phe397, Phe681, and Phe683.
Simulations of the extraction of the PAPC molecule into the catalytic site of GVIA iPLA2 also gave useful information for its extraction pathway (see Movie S2 in reference Mouchlis et al. , 2015). The peptide regions 720–730 (red region in Fig. 4B and 4D) and 640–648 (green region in Fig. 4B and 4D) were found to be involved in the extraction process of PAPC. Residues like Thr642 and Tyr643 serve as gate keepers controlling the volume of the GIVA iPLA2 catalytic site and along with Lys725 assist also the extraction of the PAPC through hydrogen-bonding with its phosphate group (Mouchlis et al. , 2015). MD simulations of the GVIA iPLA2-PAPC complex showed the phosphate group interacts with Lys489, Asn658 and Lys729 while the choline group interacts with Asp484 and Asp733. The oxygen atom of the sn-2 carbonyl group is located near the oxyanion hole (Gly486/Gly487) participating in hydrogen-bonding, while its carbon atom is located near catalytic Ser519. An interesting separation of the sn-2 from the sn-1 fatty acid chain occurs in the case of GVIA iPLA2 when the substrate binds in the hydrophobic region of its catalytic site. In the hydrophobic region, the fatty acyl chains interact with residues like Val548, Phe549, Phe643, Phe644, Val721, Phe722, Leu770, and Leu778 located on regions that showed a decreased deuteration level in DXMS experimental data.
C. Hydrolysis of the phospholipid molecule and D. diffusion of the products
Since GIVA cPLA2 and GVIA iPLA2 share a common catalytic dyad of Ser/Asp, they also share a common hydrolytic mechanism (see Fig. S9 in reference Mouchlis et al. , 2015). After the extraction of a phospholipid molecule its binding is stabilized through interactions of its phosphate group with an asparagine and an arginine (GIVA cPLA2) or a lysine (GVIA iPLA2). Hydrolysis starts with the abstraction of a proton from the catalytic serine by the aspartic acid. The negatively charged serine attacks the carbon atom of the sn-2 carbonyl group forming a tetrahedral intermediate which is stabilized by two glycine residues referred to as the oxyanion hole. The tetrahedral intermediate or transition state is then cleaved leading to a serine-acyl enzyme intermediate while the aspartic acid is transferring a proton atom to the lysophospholipid product. In a similar manner, the serine-acyl enzyme intermediate breaks down releasing the free fatty acid (Dessen et al. , 1999, Mouchlis et al. , 2015). The products of the hydrolysis are then diffused back into the membrane and the hydrolytic cycle starts over again. The details of the products diffusion mechanism have not been elucidated yet, but we are planning to study it in future projects.
Allosteric regulation of GIVA cPLA2 and GVIA iPLA2 by membrane
The association of GIVA cPLA2 and GVIA iPLA2 through their interfacial surface (surface dilution kinetics)(Carman et al. , 1995, Deems et al. , 1975) is the most interesting part of their catalytic cycle (Fig. 3). Our previously published MD simulation supported the notion that the interfacial surface of these two enzymes contains allosteric sites for binding to the membrane (Fig. S8 in reference Mouchlis et al. , 2015). During the association of these enzymes with the membrane, the membrane surface acts as an allosteric ligand shifting the conformational state of the enzyme from the inactive to the active (step A in Fig. 3) in an analogous manner to that described by Changeux (Changeux, 2012, Changeux, 2009, Monod et al. , 1965). In contrast to Changeux’s original model, our PLA2-membrane models suggest that PLA2 enzymes have a very large allosteric site or surface interacting with the membrane (Mouchlis et al. , 2015).
Binding mode of GIVA cPLA2 and GVIA iPLA2 inhibitors
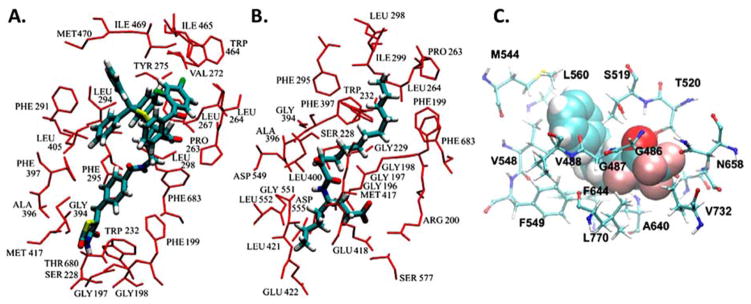
Experimental combined with computational techniques have successfully been used to develop potent and selective inhibitors for PLA2 enzymes (Mouchlis et al. , 2011a, Mouchlis et al. , 2011b, Mouchlis et al. , 2010a, b, Mouchlis et al. , 2012). MD simulations guided by DXMS experimental data were also successfully used to help in understanding the binding mode of pyrrophenone and a 2-oxoamide in the catalytic site of GIVA cPLA2,(Burke et al. , 2009) and a trifluoromethyl ketone in the catalytic site of GVIA iPLA2 (Hsu et al. , 2013). Pyrrophenone contains a thiazolidinedione ring designed to target Arg200 and a carbonyl group bridging the two benzoyl groups to target the GIVA cPLA2 catalytic Ser228 and was synthesized in 2001 (Seno et al. , 2001). A 2-oxoamide AX007 contains a carboxylic acid designed to also target Arg200 and a 2-oxoamide to target catalytic Ser228 of GIVA cPLA2 and was synthesized in 2002 (Kokotos et al. , 2002). The peptide regions 256–265, 268–279, 466–470, 473–478, and 684–689 exhibited decreased deuteration levels upon binding of pyrrophenone and AX007. MD simulations led to enzyme-inhibitor complexes (Fig. 5A and B) that showed interactions of the inhibitors with residues such as Pro263, Leu264, Leu267, Val272, Tyr275, Trp464, Ile465, Ile469, Met470, and Phe683 (Burke et al. , 2009). Aromatic trifluoromethylketone inhibitors such as PHFK were synthesized by Kokotos’ group to target the catalytic Ser519 of GVIA iPLA2 (Baskakis et al. , 2008, Kokotos et al. , 2010). DXMS experiments showed that peptide regions 483–493, 516–525, 544–549, 631–655, and 773–778 demonstrated a significant decrease in deuteration levels upon binding of PHFK. Docking of PHFK in the catalytic site of GVIA iPLA2 showed interactions with residues like Thr520, Phe549, Met544, Val548, Phe549, and Leu560 (Fig. 5C).
Figure 5.
Binding mode of GIVA cPLA2 and GVIA iPLA2 inhibitors elucidated using MD simulations guided by DXMS data. (A) Pyrrophenone in GIVA cPLA2. (B) 2-Oxoamide in GIVA cPLA2. (C) PHFK in GVIA iPLA2. Adopted from references 25 and 61.
Acknowledgments
We would like to thank the National Institutes of Health RO1-GM20501-40 (E.A.D.) for supporting our investigations of phospholipase A2 over the years.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayilavarapu S, Kantarci A, Fredman G, Turkoglu O, Omori K, Liu H, et al. Diabetes-induced oxidative stress is mediated by Ca2+-independent phospholipase A2 in neutrophils. J Immunol. 2010;184:1507–15. doi: 10.4049/jimmunol.0901219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsinde J, Balboa MA, Dennis EA. Antisense inhibition of group VI Ca2+-independent phospholipase A2 blocks phospholipid fatty acid remodeling in murine P388D1 macrophages. J Biol Chem. 1997;272:29317–21. doi: 10.1074/jbc.272.46.29317. [DOI] [PubMed] [Google Scholar]
- Balsinde J, Balboa MA, Li WH, Llopis J. Cellular regulation of cytosolic group IV phospholipase A2 by phosphatidylinositol bisphosphate levels. J Immunol. 2000 doi: 10.4049/jimmunol.164.10.5398. [DOI] [PubMed] [Google Scholar]
- Baskakis C, Magrioti V, Cotton N, Stephens D, Constantinou-Kokotou V, Dennis EA, et al. Synthesis of polyfluoro ketones for selective inhibition of human phospholipase A2 enzymes. J Med Chem. 2008;51:8027–37. doi: 10.1021/jm800649q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone RN, Gai Y, Magrioti V, Kokotou MG, Ali T, Lei X, et al. Inhibition of Ca2+-independent phospholipase A2β (iPLA2β) ameliorates islet infiltration and incidence of diabetes in NOD mice. Diabetes. 2015;64:541–54. doi: 10.2337/db14-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Hsu YH, Mouchlis VD, Dennis EA, McCammon JA. Insertion of the Ca2+-independent phospholipase A2 into a phospholipid bilayer via coarse-grained and atomistic molecular dynamics simulations. PLoS Comput Biol. 2013;9:e1003156. doi: 10.1371/journal.pcbi.1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski M, Dumlao D, Dennis E. Thematic Review Series: Proteomics. An integrated omics analysis of eicosanoid biology. J Lipid Res. 2009;50:1015–38. doi: 10.1194/jlr.R900004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczynski MW, Stephens DL, Bowers-Gentry RC, Grkovich A, Deems RA, Dennis EA. TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264. 7 cells. J Biol Chem. 2007;282:22834–47. doi: 10.1074/jbc.M701831200. [DOI] [PubMed] [Google Scholar]
- Burke J, Babakhani A, Gorfe A, Kokotos G, Li S, Woods V, et al. Location of inhibitors bound to group IVA phospholipase A2 determined by molecular dynamics and deuterium exchange mass spectrometry. J Am Chem Soc. 2009;131:8083–91. doi: 10.1021/ja900098y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J, Hsu Y-H, Deems R, Li S, Woods V, Dennis E. A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2. J Biol Chem. 2008;283:31227–36. doi: 10.1074/jbc.M804492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Burke J, Dennis E. Using hydrogen/deuterium exchange mass spectrometry to define the specific interactions of the phospholipase A2 superfamily with lipid substrates, inhibitors, and membranes. J Biol Chem. 2013;288:1806–13. doi: 10.1074/jbc.R112.421909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G, Deems R, Dennis E. Lipid signaling enzymes and surface dilution kinetics. J Biol Chem. 1995;270:18711–4. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- Chalfant CE, Spiegel S. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J Cell Sci. 2005;118:4605–12. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- Changeux J-P. Allostery and the Monod-Wyman-Changeux model after 50 years. Annu Rev Biophys. 2012;41:103–33. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- Changeux J-PP. Allosteric receptors: from electric organ to cognition. Annu Rev Pharmacol Toxicol. 2009;50:1–38. doi: 10.1146/annurev.pharmtox.010909.105741. [DOI] [PubMed] [Google Scholar]
- Clark JD, Lin LL, Kriz RW, Ramesha CS, Sultzman LA, Lin AY, et al. A novel arachidonic acid-selective cytosolic PLA2 contains a Ca2+-dependent translocation domain with homology to PKC and GAP. Cell. 1991;65:1043–51. doi: 10.1016/0092-8674(91)90556-e. [DOI] [PubMed] [Google Scholar]
- Das S, Cho W. Roles of catalytic domain residues in interfacial binding and activation of group IV cytosolic phospholipase A2. J Biol Chem. 2002;277:23838–46. doi: 10.1074/jbc.M202322200. [DOI] [PubMed] [Google Scholar]
- Deems RA, Eaton BR, Dennis EA. Kinetic analysis of phospholipase A2 activity toward mixed micelles and its implications for the study of lipolytic enzymes. J Biol Chem. 1975;250:9013–20. [PubMed] [Google Scholar]
- Dennis E, Cao J, Hsu Y-H, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111:6130–85. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–23. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen A, Tang J, Schmidt H, Stahl M, Clark J, Seehra J, et al. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–60. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- Durrant JD, McCammon JA. Molecular dynamics simulations and drug discovery. BMC Biol. 2011 doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–5. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the Group IV phospholipase A2 family. Prog Lipid Res. 2006;45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Hazen SL, Gross RW. Human myocardial cytosolic Ca2+-independent phospholipase A2 is modulated by ATP. Concordant ATP-induced alterations in enzyme kinetics and mechanism-based inhibition. Biochem J. 1991;280(Pt 3):581–7. doi: 10.1042/bj2800581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegen M, Sun L, Uozumi N, Kume K, Goad ME, Nickerson-Nutter CL, et al. Cytosolic phospholipase A2α-deficient mice are resistant to collagen-induced arthritis. J Exp Med. 2003;197:1297–302. doi: 10.1084/jem.20030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-H, Bucher D, Cao J, Li S, Yang S-W, Kokotos G, et al. Fluoroketone Inhibition of Ca2+-Independent Phospholipase A2 through Binding Pocket Association Defined by Hydrogen/Deuterium Exchange and Molecular Dynamics. J Am Chem Soc. 2013;135:1330–7. doi: 10.1021/ja306490g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-H, Burke J, Li S, Woods V, Dennis E. Localizing the membrane binding region of Group VIA Ca2+- independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J Biol Chem. 2009;284:23652–61. doi: 10.1074/jbc.M109.021857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-HH, Burke JE, Stephens DL, Deems RA, Li S, Asmus KM, et al. Calcium binding rigidifies the C2 domain and the intradomain interaction of GIVA phospholipase A2 as revealed by hydrogen/deuterium exchange mass spectrometry. J Biol Chem. 2008;283:9820–7. doi: 10.1074/jbc.M708143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CM, Yan W, Mancuso DJ, Gross RW. Highly selective hydrolysis of fatty acyl-CoAs by calciumindependent phospholipase A2beta. Enzyme autoacylation and acyl-CoA-mediated reversal of calmodulin inhibition of phospholipase A2 activity. J Biol Chem. 2006;281:15615–24. doi: 10.1074/jbc.M511623200. [DOI] [PubMed] [Google Scholar]
- Karabina SA, Gora S, Atout R, Ninio E. Extracellular phospholipases in atherosclerosis. Biochimie. 2010;92:594–600. doi: 10.1016/j.biochi.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Keyes KT, Ye Y, Lin Y, Zhang C. Resolvin E1 protects the rat heart against reperfusion injury. Am J Physiol Heart Circ Physiol. 2010 doi: 10.1152/ajpheart.01057.2009. [DOI] [PubMed] [Google Scholar]
- Kokotos G, Hsu Y-H, Burke J, Baskakis C, Kokotos C, Magrioti V, et al. Potent and selective fluoroketone inhibitors of group VIA calcium-independent phospholipase A2. J Med Chem. 2010;53:3602–10. doi: 10.1021/jm901872v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokotos G, Kotsovolou S, Six DA, Constantinou-Kokotou V, Beltzner CC, Dennis EA. Novel 2-Oxoamide Inhibitors of Human Group IVA Phospholipase A2. J Med Chem. 2002;45:28912893. doi: 10.1021/jm025538p. [DOI] [PubMed] [Google Scholar]
- Kramer RM, Roberts EF, Manetta J, Putnam JE. The Ca2+-sensitive cytosolic phospholipase A2 is a 100-kDa protein in human monoblast U937 cells. J Biol Chem. 1991;266:5268–72. [PubMed] [Google Scholar]
- Larsson Forsell PK, Kennedy BP, Claesson HE. The human calcium-independent phospholipase A2 gene multiple enzymes with distinct properties from a single gene. Eur J Biochem. 1999;262:575–85. doi: 10.1046/j.1432-1327.1999.00418.x. [DOI] [PubMed] [Google Scholar]
- Larsson PK, Claesson HE, Kennedy BP. Multiple splice variants of the human calcium-independent phospholipase A2 and their effect on enzyme activity. J Biol Chem. 1998;273:207–14. doi: 10.1074/jbc.273.1.207. [DOI] [PubMed] [Google Scholar]
- Leslie CC, Channon JY. Anionic phospholipids stimulate an arachidonoyl-hydrolyzing phospholipase A2 from macrophages and reduce the calcium requirement for activity. Biochim Biophys Acta. 1990;1045:261–70. doi: 10.1016/0005-2760(90)90129-l. [DOI] [PubMed] [Google Scholar]
- Lio YC, Dennis EA. Interfacial activation, lysophospholipase and transacylase activity of group VI Ca2+- independent phospholipase A2. Biochim Biophys Acta. 1998;1392:320–32. doi: 10.1016/s0005-2760(98)00049-6. [DOI] [PubMed] [Google Scholar]
- Malkova S, Long F, Stahelin RV, Pingali SV, Murray D, Cho W, et al. X-ray reflectivity studies of cPLA2α-C2 domains adsorbed onto Langmuir monolayers of SOPC. Biophysical journal. 2005;89:1861–73. doi: 10.1529/biophysj.105.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Murakami M, Komiyama K, Ishihara M, Ishikawa Y, Ishii T, et al. Various secretory phospholipase A2 enzymes are expressed in rheumatoid arthritis and augment prostaglandin production in cultured synovial cells. FEBS J. 2005;272:655–72. doi: 10.1111/j.1742-4658.2004.04489.x. [DOI] [PubMed] [Google Scholar]
- Michaely P, Tomchick D, Machius M, Anderson R. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–483. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J, Wyman J, Changeux JP. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Mouchlis VD, Barbayianni E, Mavromoustakos TM, Kokotos G. The application of rational design on phospholipase A2 inhibitors. Curr Med Chem. 2011a;18:2566–82. doi: 10.2174/092986711795933678. [DOI] [PubMed] [Google Scholar]
- Mouchlis VD, Bucher D, McCammon JA, Dennis EA. Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. PNAS. 2015;112:E516–E25. doi: 10.1073/pnas.1424651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis VD, Magrioti V, Barbayianni E, Cermak N, Oslund RC, Mavromoustakos TM, et al. Inhibition of secreted phospholipases A2 by 2-oxoamides based on alpha-amino acids: Synthesis, in vitro evaluation and molecular docking calculations. Bioorg Med Chem. 2011b;19:735–43. doi: 10.1016/j.bmc.2010.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchlis VD, Mavromoustakos TM, Kokotos G. Design of new secreted phospholipase A2 inhibitors based on docking calculations by modifying the pharmacophore segments of the FPL67047XX inhibitor. J Comput-Aided Mol Des. 2010a;24:107–15. doi: 10.1007/s10822-010-9319-7. [DOI] [PubMed] [Google Scholar]
- Mouchlis VD, Mavromoustakos TM, Kokotos G. Molecular docking and 3D-QSAR CoMFA studies on indole inhibitors of GIIA secreted phospholipase A2. J Chem Inf Model. 2010b;50:1589–601. doi: 10.1021/ci100217k. [DOI] [PubMed] [Google Scholar]
- Mouchlis VD, Michopoulou V, Constantinou-Kokotou V, Mavromoustakos T, Dennis EA, Kokotos G. Binding conformation of 2-oxoamide inhibitors to group IVA cytosolic phospholipase A2 determined by molecular docking combined with molecular dynamics. J Chem Inf Model. 2012;52:243–54. doi: 10.1021/ci2005093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Rosenberg DW. Roles of cPLA2α and arachidonic acid in cancer. Biochim Biophys Acta. 2006;1761:1335–43. doi: 10.1016/j.bbalip.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PC, Dennis EA. Omega-3 fatty acids cause dramatic changes in TLR4 and purinergic eicosanoid signaling. PNAS. 2012;109:8517–22. doi: 10.1073/pnas.1200189109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris PC, Gosselin D, Reichart D, Glass CK, Dennis EA. Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. PNAS. 2014;111:12746–51. doi: 10.1073/pnas.1404372111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee C, Jang J-H, Ghim J, Kim Y-J, You S, et al. Phospholipase signalling networks in cancer. Nat Rev Cancer. 2012;12:782–92. doi: 10.1038/nrc3379. [DOI] [PubMed] [Google Scholar]
- Perisic O, Paterson HF, Mosedale G, Lara-Gonzalez S, Williams RL. Mapping the phospholipid-binding surface and translocation determinants of the C2 domain from cytosolic phospholipase A2. J Biol Chem. 1999;274:14979–87. doi: 10.1074/jbc.274.21.14979. [DOI] [PubMed] [Google Scholar]
- Pettus BJ, Bielawska A, Subramanian P, Wijesinghe DS, Maceyka M, Leslie CC, et al. Ceramide 1- phosphate is a direct activator of cytosolic phospholipase A2. J Biol Chem. 2004;279:11320–6. doi: 10.1074/jbc.M309262200. [DOI] [PubMed] [Google Scholar]
- Rosenson RS, Hurt-Camejo E. Phospholipase A2 enzymes and the risk of atherosclerosis. Eur Heart J. 2012;33:2899–909. doi: 10.1093/eurheartj/ehs148. [DOI] [PubMed] [Google Scholar]
- Rossi A, Pergola C, Koeberle A. The 5-lipoxygenase inhibitor, zileuton, suppresses prostaglandin biosynthesis by inhibition of arachidonic acid release in macrophages. Br J Pharmacol. 2010 doi: 10.1111/j.1476-5381.2010.00930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- Scott KF, Sajinovic M, Hein J, Nixdorf S, Galettis P, Liauw W, et al. Emerging roles for phospholipase A2 enzymes in cancer. Biochimie. 2010;92:601–10. doi: 10.1016/j.biochi.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Seno K, Okuno T, Nishi K, Murakami Y, Yamada K, Nakamoto S, et al. Pyrrolidine inhibitors of human cytosolic phospholipase A2. Part 2: synthesis of potent and crystallized 4-triphenylmethylthio derivative 'pyrrophenone'. Bioorg Med Chem Lett. 2001;11:587–90. doi: 10.1016/s0960-894x(01)00003-8. [DOI] [PubMed] [Google Scholar]
- Sharp JD, White DL, Chiou XG, Goodson T, Gamboa GC, McClure D, et al. Molecular cloning and expression of human Ca2+-sensitive cytosolic phospholipase A2. J Biol Chem. 1991;266:14850–3. [PubMed] [Google Scholar]
- Six DA, Dennis EA. Essential Ca2+-independent Role of the Group IVA Cytosolic Phospholipase A2 C2 Domain for Interfacial Activity. J Biol Chem. 2003;278:23842–50. doi: 10.1074/jbc.M301386200. [DOI] [PubMed] [Google Scholar]
- Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014;66:334–95. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane JR. Biomedicine. Back to an aspirin a day? Science. 2002;296:474–5. doi: 10.1126/science.1071702. [DOI] [PubMed] [Google Scholar]
- Winstead MV, Balsinde J, Dennis EA. Calcium-independent phospholipase A2: structure and function. Biochim Biophys Acta. 2000;1488:28–39. doi: 10.1016/s1388-1981(00)00107-4. [DOI] [PubMed] [Google Scholar]