Abstract
The prototype coronavirus mouse hepatitis virus (MHV) exhibits highly selective packaging of its genomic positive-stranded RNA into assembled virions, despite the presence in infected cells of a large excess of subgenomic viral mRNAs. One component of this selectivity is the MHV packaging signal (PS), an RNA structure found only in genomic RNA and not in subgenomic RNAs. It was previously shown that a major determinant of PS recognition is the second of the two RNA-binding domains of the viral nucleocapsid (N) protein. We have now found that PS recognition additionally depends upon a segment of the carboxy-terminal tail (domain N3) of the N protein. Since domain N3 is also the region of N protein that interacts with the membrane (M) protein, this finding suggests a mechanism by which selective genome packaging is accomplished, through the coupling of genome encapsidation to virion assembly.
Keywords: Viral genome packaging, Packaging signal, RNA virus, Murine coronavirus, Mouse hepatitis virus, Nucleocapsid protein
Graphical abstract
Highlights
-
•
Coronavirus MHV genomic RNA packaging depends on the second RNA-binding domain (CTD) of the nucleocapsid N protein.
-
•
MHV genomic RNA packaging also depends on the carboxy-terminal tail (domain N3) of N protein.
-
•
These findings suggest a mechanism for selective genome packaging, through coupling of genome encapsidation to virion assembly.
1. Introduction
The selective packaging of genomic RNA (gRNA) into assembling virions is a fundamental problem faced by many RNA viruses. Coronaviruses occupy a unique niche among positive-strand RNA viruses in two respects. First, they have helically symmetric nucleocapsids and, consequently, are not subject to the architectural constraints that govern packaging for viruses with icosahedral capsids (Prevelige, 2016). Second, although they synthesize a large molar excess of multiple subgenomic RNA (sgRNA) species during infection, coronaviruses very selectively incorporate gRNA into virions (Makino et al., 1990, Escors et al., 2003).
Coronavirus gRNA packaging has been most intensively studied in two betacoronaviruses, mouse hepatitis virus (MHV) and bovine coronavirus (BCoV), and in the alphacoronavirus transmissible gastroenteritis virus (TGEV). For MHV, a genomic packaging signal (PS) was originally identified through analyses of packaged defective-interfering (DI) RNAs, which are extensively deleted genomic remnants that replicate by appropriating the RNA synthesis machinery of a helper virus (Makino et al., 1990, van der Most et al., 1991). The MHV PS is situated roughly 20.3 kb from the 5′ end of the genome, embedded in the segment of gene 1 that encodes the nonstructural protein 15 (nsp15) subunit of the replicase-transcriptase (Fosmire et al., 1992, Cologna and Hogue, 2000, Narayanan and Makino, 2001). This localization places the PS solely in gRNA and not in any of the six species forming the 3′-nested set of sgRNAs of MHV.
A structure recently proposed for the MHV PS (Chen et al., 2007b) models it as a 95-nt bulged stem-loop containing four repeat units, each with an AA (or GA) bulge; an internal loop separates the PS into quasi-symmetric upper and lower halves ( Fig. 1A). This structure is consistent with chemical and enzymatic probing profiles and is highly conserved among lineage A betacoronaviruses (such as MHV and BCoV). We previously manipulated the MHV PS in the intact virus, rather than in DI RNAs, with the goal of obtaining insights into the mechanism of packaging (Kuo and Masters, 2013). Extensive disruption of the PS structure with 20 coding-silent mutations (in a mutant designated silPS) or outright deletion of the PS resulted in the packaging of abundant amounts of sgRNA in addition to gRNA in highly purified virions. We found that the PS was not essential for MHV viability, but it conferred a distinct selective advantage to genomes that harbored it. Additionally, the PS remained functional when transposed to an ectopic genomic site, a noncoding region created downstream of the replicase-transcriptase gene. This work showed that the PS indeed governs the selective incorporation of gRNA into virions.
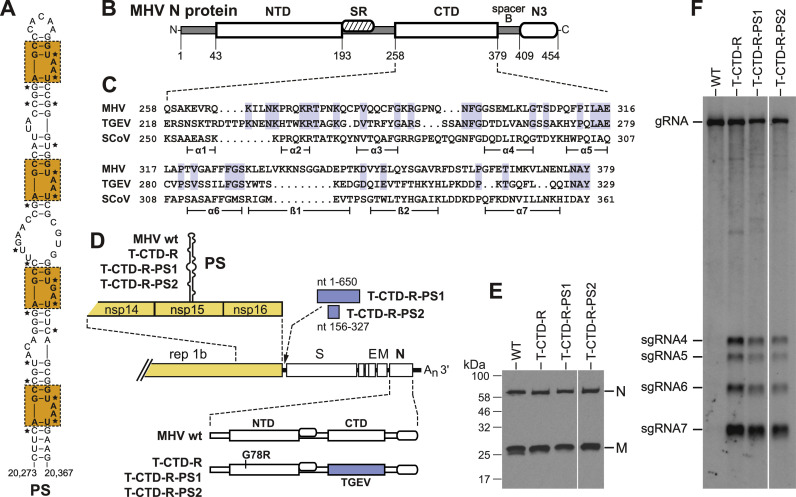
Fig. 1.
Effect of substitution of the TGEV N protein CTD in the MHV N protein. (A) The MHV packaging signal as modeled by the Olsthoorn laboratory (Chen et al., 2007b). The four repeat units are boxed. Stars mark 20 nucleotides that were mutated in the silPS mutant to disrupt the PS structure without altering the encoded segment of nsp15 amino-acid sequence (Kuo and Masters, 2013). (B) Domain structure of the MHV N protein: NTD, amino-terminal RNA-binding domain; SR, serine- and arginine-rich region; CTD, carboxy-terminal RNA-binding domain; spacer B, variable spacer region; N3, carboxy-terminal M-interacting domain. Amino-acid residue numbers are shown beneath the schematic. (C) Alignment of the CTDs of the MHV, TGEV, and SARS-CoV N proteins. GenBank accession numbers for the sequences shown are: MHV-A59, AY700211; TGEV, AJ271965; SARS-CoV, AY278741. Residues that are identical in MHV and TGEV are highlighted. Shown below the alignment are α-helices and β-strands determined for the SARS-CoV CTD crystal structure (Chen et al., 2007a). (D) Compositions of the TGEV CTD chimeras. The N proteins of all three TGEV mutants contain a substitution of the TGEV CTD replacing that of MHV, plus the reverting mutation G78R in the NTD. All mutants (as well as the isogenic wild-type MHV) contain the wild-type MHV PS within the nsp15 coding region and a short linker segment replacing nonessential genes between gene 1 and the S gene, as described previously (Kuo and Masters, 2013). T-CTD-R-PS1 and T-CTD-R-PS2, additionally contain nt 1-650 or nt 156-327 of the TGEV genome, respectively, inserted into the linker between gene 1 and the S gene. (E) Western blots of purified virions probed with anti-N and anti-M monoclonal antibodies; note that the anti-N antibody recognizes an epitope in MHV domain N3. (F) Northern blots of RNA isolated from purified virions. MHV RNA was detected with a probe specific for the 5′ half of the N gene.
To begin to identify interacting partners of the PS, we modified the nucleocapsid (N) protein, the molecule that coats the gRNA and winds it into a helically symmetric filament within the virion (Masters, 2006). N is a mostly basic phosphoprotein containing two structurally separate RNA-binding domains, the amino-terminal domain (NTD) and the carboxy-terminal domain (CTD) (Fig. 1B). (For a comprehensive review of coronavirus N protein structure, see Chang et al., 2014.) The CTD additionally mediates N-N dimerization and higher-order interactions in the nucleocapsid (Chang et al., 2013). This nomenclature can obscure the point that the actual carboxy terminus of N protein consists of a linker (spacer B) joining the CTD to N3, an acidic domain that binds to the endodomain of the membrane (M) protein during virion assembly (Kuo and Masters, 2002; Hurst et al., 2005; Verma et al., 2006, Verma et al., 2007; Kuo et al., 2016). Another linker, between the NTD and CTD, encompasses a serine- and arginine-rich region that associates with replicase subunit nsp3 in a crucial early event of infection (Hurst et al., 2010).
Although the MHV PS is conserved among lineage A betacoronaviruses, it is clear that severe acute respiratory syndrome coronavirus (SARS-CoV), a lineage B betacoronavirus, does not contain a homolog of the MHV PS (Joseph et al., 2007, Chen and Olsthoorn, 2010). We therefore turned to SARS-CoV chimeras for evidence of interacting partners of this RNA element, constructing MHV N protein mutants in which either of the two RNA-binding domains of N was substituted by its SARS-CoV counterpart (Kuo et al., 2014). Strikingly, we found that the SARS-CoV CTD chimera recapitulated the defective packaging phenotype of the silPS mutant and the PS deletion mutant. Furthermore, the region affecting PS recognition was localized in a partial CTD chimera to a central segment of 30 amino acids, which corresponds to helices α4-α6 of the SARS-CoV CTD (Chen et al., 2007a). Conversely, gRNA packaging selectivity in the SARS-CoV NTD chimera was identical to that of wild-type MHV. This demonstrated that the second RNA-binding domain of N, the CTD, is the major protein determinant of MHV PS recognition. In the work reported here, we show that PS recognition additionally depends upon a segment of the carboxy-terminal tail of the N protein, domain N3. This finding suggests a mechanism by which selective genome packaging is carried out.
2. Results
2.1. Substitution of the TGEV CTD in MHV N abolishes recognition of the MHV PS
We found previously that selective packaging of MHV genomic RNA was abrogated by the substitution of the SARS-CoV CTD, but not the NTD, into the MHV N protein. To extend this result to a still more phylogenetically distant N protein, we chose TGEV, an alphacoronavirus for which selective genomic packaging has been clearly demonstrated (Escors et al., 2003, Morales et al., 2013). As is the case with SARS-CoV, the TGEV genome does not contain a counterpart of the MHV PS in the nsp15 coding region (Chen and Olsthoorn, 2010). We would consequently expect that the TGEV N protein did not evolve the capacity to recognize the MHV PS.
To construct TGEV N protein substitution mutants, we used the reverse-genetic technique of targeted RNA recombination (Kuo et al., 2000, Goebel et al., 2004). Because no structures are available for the NTD or CTD of any alphacoronavirus N protein, it was not straightforward to clearly define the boundaries of these domains in TGEV N. In the case of the TGEV NTD, multiple attempts made with each of five different sets of boundaries were all unsuccessful in producing a viable chimeric virus. This possibly indicates that the NTD mediates some sequence-specific RNA-binding function (other than packaging) which cannot be exchanged between alpha- and betacoronaviruses. For the CTD, we generated an alignment of the corresponding domains of MHV and TGEV (Fig. 1C), guided by the MHV and SARS-CoV CTD alignment that was already known to produce a functional substitution (Kuo et al., 2014). The MHV CTD retains only 25% amino-acid identity with the TGEV CTD, an even greater divergence than the 35% identity between the MHV and SARS-CoV CTDs.
Three independent isolates of the TGEV-CTD mutant were obtained. All three produced small plaques and grew slowly compared to the wild type. However, after two to three passages, cultures of the original mutants rapidly became overrun by revertants that produced nearly wild-type-sized plaques at 37 °C. From these we isolated three independent revertants and found that the two most robust growers each contained the mutation G78R in the (MHV) NTD of N protein. Thus, to facilitate better growth for virion purification, we reconstructed the G78R mutation into the TGEV chimeric mutant, now designated T-CTD-R (Fig. 1D). We subsequently learned that the G78R mutation had no impact on the packaging phenotype (data not shown). In this respect, the G78R mutation was similar to a second-site mutation of the previously studied SARS-CoV CTD chimera that enhanced growth of that mutant but had no effect on packaging (Kuo et al., 2014).
RNA was isolated from equal amounts of purified wild-type and T-CTD-R virions that had been normalized by Western blotting of the two most abundant structural proteins, N and M (Fig. 1E). Northern blot analysis of virion RNA with a probe specific for the upstream half of the N gene revealed that, as seen previously, wild-type virions exhibited almost complete stringency in packaging gRNA and excluding sgRNA (Fig. 1F). By contrast, the T-CTD-R mutant packaged plentiful amounts of sgRNAs, proportional to the relative levels of these species found in infected cells. This was the same phenotype we had observed previously for other packaging-defective MHV mutants (Kuo and Masters, 2013, Kuo et al., 2014). It should be noted that in the T-CTD-R mutant only the N gene and not the MHV PS had been altered. Therefore, this result confirmed our previous finding that the CTD is a major determinant of PS recognition.
To determine whether the T-CTD-R mutant could be used to capture the TGEV PS, we constructed two additional mutants in which TGEV sequence was inserted at a site immediately downstream of MHV gene 1 (Fig. 1D). We had previously shown that the MHV PS remains fully functional when it is inserted at this ectopic position and deleted from its native site (Kuo and Masters, 2013). The first mutant, T-CTD-R-PS1, incorporated nt 1-650 of the TGEV genome, which has been shown to harbor the TGEV PS, through analyses of packaged DI RNAs (Escors et al., 2003, Morales et al., 2013). The second mutant, T-CTD-R-PS2, incorporated a subregion, nt 156-327, which contains a triply-repeated hairpin-loop motif that has been proposed to constitute the alphacoronavirus PS (Chen and Olsthoorn, 2010). However, both mutants displayed the same packaging-negative phenotype as the T-CTD-R mutant (Fig. 1E and F); that is, incorporation of the putative TGEV PS did not restore gRNA packaging selectivity to T-CTD-R. Although we could not eliminate other potential explanations for this negative result, this outcome suggested that there exists some viral factor, in addition to the N protein CTD, that is required for PS recognition.
2.2. MHV chimeras containing the SARS-CoV M protein are packaging-defective
One candidate for an additional viral factor involved in genome packaging is the membrane (M) protein (Narayanan and Makino, 2001, Narayanan et al., 2003). We recently constructed MHV chimeras containing whole or partial substitutions of the SARS-CoV M protein for the purpose of learning more about intermolecular interactions of M in virion assembly (Kuo et al., 2016). MHV and SARS-CoV, which fall into different lineages of the betacoronavirus genus, have M proteins that share only 38% amino-acid sequence identity. We reasoned that, as with previously studied N gene chimeras (Hurst et al., 2010, Kuo et al., 2014), this degree of divergence might impair particular viral functions. One conclusion of this work was that the carboxy-terminal tail of N protein, N3, is both necessary and sufficient for virion assembly interactions between the coronavirus N and M proteins, a result that is supported by numerous prior studies (Kuo and Masters, 2002, Hurst et al., 2005, Verma et al., 2006, 2007; Luo et al., 2006). Thus, the constructed SARS-CoV M chimeric mutants necessarily also incorporated SARS-CoV domain N3, which is extremely divergent from that of MHV, retaining little to no amino-acid sequence homology.
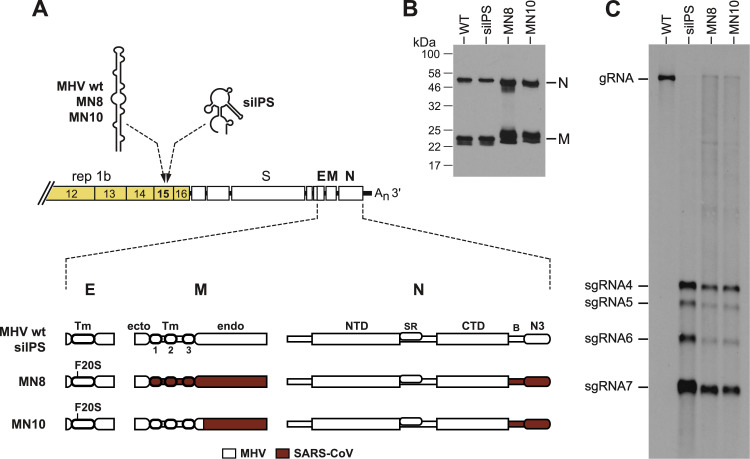
To examine the potential effect of M protein on the selective packaging of MHV gRNA, we examined two chimeric mutants that were sufficiently robust to allow extensive purification of virions. One of these, MN8, contained the three transmembrane domains plus the endodomain of SARS-CoV M protein ( Fig. 2A). The other mutant, MN10, contained the endodomain of SARS-CoV M. Both mutants were engineered to also contain an envelope (E) protein point mutation, F20S, which had been found to enhance the growth of the M chimeras (Kuo et al., 2016). Viruses of the MN8 and MN10 mutants were grown and purified side-by-side with the wild type and the silPS mutant (Kuo and Masters, 2013), the latter two as positive and negative packaging controls, respectively. Purified virions were normalized by Western blots probed with antibodies specific for the N and M proteins (Fig. 2B). It should be noted that the mobilities of the MN8 and MN10 N and M proteins differ from those of the wild type because of the altered molecular masses and amino-acid compositions of the chimeric proteins (Kuo et al., 2016). Analysis of isolated virion RNA by Northern blotting showed that both the MN8 and the MN10 mutant were markedly impaired in their ability to selectively package gRNA (Fig. 2C). Although the M chimeras did not appear to be as thoroughly defective as the silPS mutant, they nonetheless incorporated substantial amounts of sgRNA into virions, compared to the wild type. There also appeared to be considerably less gRNA than we had observed in previous Northern blots of silPS virion RNA (Kuo and Masters, 2013, Kuo et al., 2014) and in dot blots of MN8 virion gRNA (Kuo et al., 2016). We believe that this was due to sample-to-sample variation in the recovery and blotting transfer efficiency of this unusually large (31-kb) RNA molecule.
Fig. 2.
Packaging defect of MHV mutants containing the SARS-CoV M protein. (A) Compositions of the SARS-CoV M chimeras (Kuo et al., 2016). Mutant MN8 contains the three transmembrane (Tm) domains and the endodomain of SARS-CoV M protein. Mutant MN10 contains the endodomain of SARS-CoV M. Both mutants also contain the SARS-CoV N protein spacer B and domain N3, as well as a growth-enhancing mutation (F20S) in the transmembrane domain of E protein. N protein domains are labeled as in Fig. 1. Both MN8 and MN10 (as well as the isogenic wild-type MHV) have the wild-type MHV PS within the nsp15 coding region. The silPS mutant has the wild-type MHV N, M, and E proteins, but carries 20 coding-silent mutations in nsp15 that disrupt the structure of the PS (Kuo and Masters, 2013). (B) Western blots of purified virions probed with anti-N polyclonal and anti-M monoclonal antibodies; note that the anti-M antibody recognizes an epitope in the MHV M ectodomain. (C) Northern blots of RNA isolated from immunopurified virions. MHV RNA was detected with a probe specific for the 5′ half of the N gene.
Unlike, the silPS mutant, MN8 and MN10 each carry the wild-type MHV PS (Fig. 2A). Thus, their packaging defect had to be a consequence of one or more of their structural protein mutations. One of these alterations, the E protein F20S mutation, assists viral growth through aiding incorporation of the spike (S) protein into virions (Kuo et al., 2016). However, this mutation is not relevant to the packaging phenotype, since we observed the same packaging defect in two other M chimeras that contain the wild-type E protein, MN3 and MN9 (data not shown). MN3 has the entire SARS-CoV M protein, while MN9 is identical to MN10, except that it lacks the F20S mutation. Therefore, our data indicate that in addition to the N protein CTD, selective packaging of MHV gRNA must entail a role for the endodomain of M protein or else domain N3 and spacer B of the N protein.
2.3. Domain N3 of the N protein plays a role in PS recognition
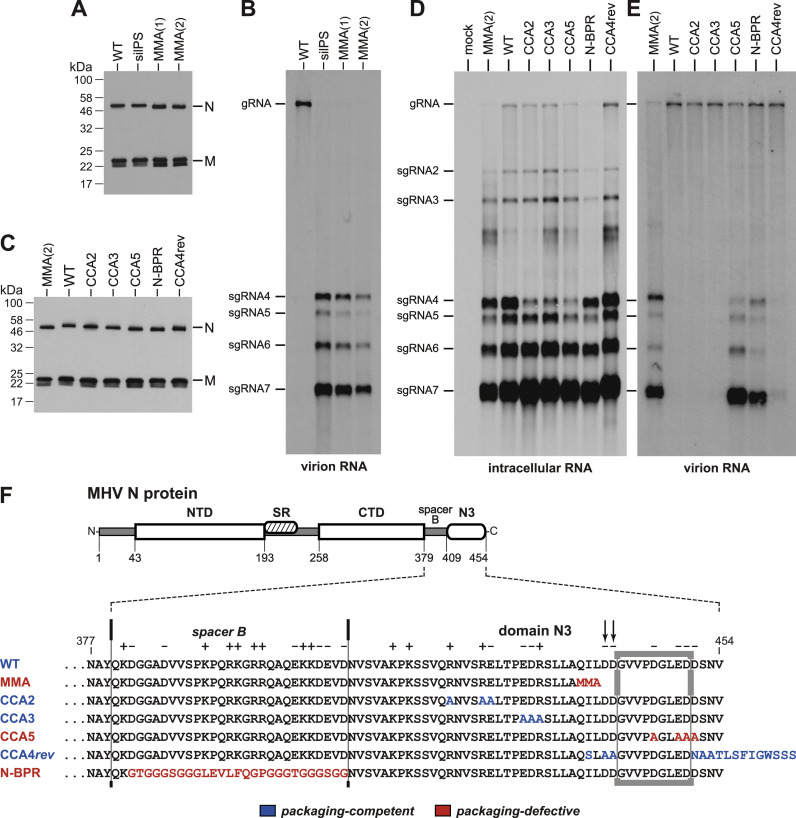
Owing to the essential virion assembly interaction between the M protein endodomain and N protein domain N3, it was not possible to separate their potential roles in packaging through further dissection of the SARS-CoV M chimeras. We had previously found that a chimeric construct that paired the MHV M protein with SARS-CoV domain N3 was lethal; conversely, a chimeric construct that paired the SARS-CoV M protein with MHV domain N3 was lethal (Hurst et al., 2010, Kuo et al., 2016). However, an alternative route to exploring the functions of domain N3 was afforded by a collection of domain N3 mutants that had been constructed for other purposes or had arisen in reversion analyses. We initially examined one of these, a mutant here designated MMA. The mutation in MMA originally arose as an intergenic suppressor of a highly deleterious two-amino-acid deletion of the carboxy terminus of the M protein endodomain (Kuo and Masters, 2002). This suppressor (Q437MMA) consisted of a 10-nt deletion in domain N3 that resulted in a frameshift replacing the carboxy-terminal 18 amino acids of N protein with only three residues, MMA. In the MMA mutant, this N3 mutation had been reconstructed in the background of a wild-type M protein, producing a virus with a phenotype indistinguishable from that of the wild type. Infections with two independent isolates of the MMA mutant produced quantities of virions identical to wild type and containing the same relative amounts of N and M protein ( Fig. 3A). Remarkably, the MMA mutant possessed the same packaging-defective trait as the silPS mutant (Fig. 3B).
Fig. 3.
Packaging defect of a subset of MHV domain N3 mutants. The sources and descriptions of the various N protein domain N3 or spacer B mutants are given in the text. MMA(1) and MMA(2) are two independent isolates of the MMA mutant (Alb336 and Alb337; Kuo and Masters, 2002). (A and C) Western blots of purified virions probed with anti-N polyclonal and anti-M monoclonal antibodies. (B and E) Northern blots of RNA isolated from immunopurified virions. (D) Northern blot of total RNA isolated from mock-infected or infected 17Cl1 cells harvested at 15 h postinfection. MHV RNA was detected with a probe specific for the 5′ half of the N gene. (F) Alignment of the carboxy termini of wild-type and mutant MHV N proteins; the GenBank accession number for wild-type MHV-A59 is AY700211. Charged amino acids are noted above the alignment. Mutated residues are indicated in color: blue for viruses with wild-type packaging; red for viruses with defective packaging. The segment of N3 essential for packaging competence is indicated by the box. Arrows denote two aspartate residues critical for interaction between the N and M proteins (Hurst et al., 2005).
This finding led us to screen additional N3 mutants. Three of these were constructed clustered charged-to-alanine mutants: CCA2 (R421A, R425A, R426E), CCA3 (E430A, D431A, R432A), and CCA5 (D446A, E449A, D450A, D451A) (Hurst et al., 2005). The fourth, CCA4rev, had been isolated as a suppressor (rev26) of the lethal pair of charged-to-alanine mutations D440A-D441A. CCA4rev contained the additional mutations I438S and D451NAATLSFIGWSSS, the latter of which was due to a 40-nt insertion in domain N3 that resulted in a frameshift replacing the carboxy-terminal four amino acids of the N molecule by 13 heterologous residues (Hurst et al., 2005). As with the MMA mutant, all of these mutants were fully competent in virion assembly, having phenotypes similar or identical to the wild type and forming virions with a normal complement of N and M proteins (Fig. 3C). Although the MMA and CCA4rev mutants lack the D440 and D441 residues that play a major role in interaction with the M endodomain, we have previously postulated that they were selected through their ability to form compensating hydrophobic interactions with other regions of the M endodomain.
In infected cells, all of the domain N3 mutants synthesized the same sgRNAs, and in similar relative amounts, as those produced by wild-type MHV (Fig. 3D). Mutants CCA2, CCA3, and CCA5 transcribed proportionately less sgRNA4 because they had been constructed by an earlier version of targeted RNA recombination, using a vector lacking a mutation that upregulates transcription of this particular sgRNA (Kuo et al., 2000). Among the additional N3 mutants, only CCA5 was observed to be defective in selective packaging of gRNA (Fig. 3E). Mutants CCA2, CCA3 and CCA4rev all exhibited the same stringent packaging of gRNA displayed by the wild type. To reinforce this outcome, we verified that all mutants, including MMA, contained the wild-type sequences for the N protein CTD and for the PS in nsp15, using the same purified RNA that had been analyzed by Northern blotting. These results thus showed that domain N3 plays a role equally important to that of the CTD in the selective packaging of MHV gRNA. Moreover, comparison of packaging-competent and packaging-defective mutants of domain N3 allowed us to map the packaging role of N3 to a short segment near the carboxy terminus of N, amino acids 442-450 (Fig. 3F). Mutants MMA and CCA5 either lack or are mutated in this segment; mutants CCA2, CCA3, and CCA4rev retain wild-type sequence in this segment, despite harboring other nearby mutations. Notably, this segment is adjacent to, but distinct from the D440-D441 residues critical for the virion assembly interaction of N3 (Hurst et al., 2005, Verma et al., 2006). One additional mutant, N-BPR, which replaced spacer B with a completely heterologous peptide linker (Hurst et al., 2009), was also packaging-defective. This may indicate a requirement for spacer B to participate in packaging, but as discussed below, we believe this role is ancillary.
3. Discussion
The coronavirus N protein NTD and CTD each possess nonspecific RNA-binding capabilities (Chang et al., 2009), in accord with the function of N in the helical encapsidation of the entire 30-kb viral genome. Our previous work (Kuo et al., 2014) as well as data shown here (Fig. 1) suggest that, in addition, the MHV CTD has evolved to specifically recognize a unique gRNA element, the MHV PS. The key finding of the present study is that domain N3, the region of N protein that binds to the M protein endodomain, is also involved in the gRNA packaging process (Fig. 3).
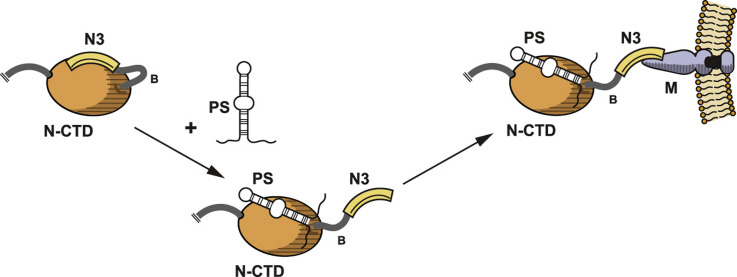
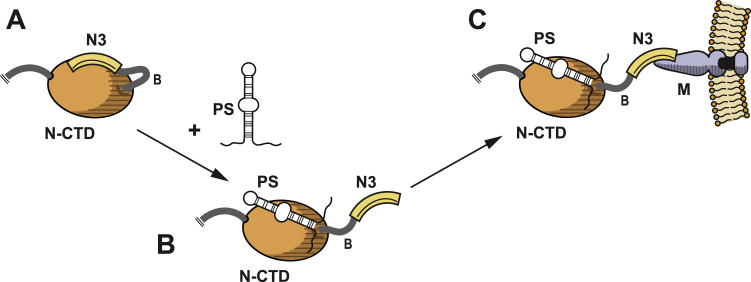
Taken together, these two results lead to a model for the obligatory coupling of gRNA encapsidation to virion assembly ( Fig. 4). We hypothesize that the largely acidic domain N3 is initially bound to the CTD, occluding at least part of the basic RNA-binding surface of the CTD (Chen et al., 2007a). This interaction contributes to the specificity of RNA binding by the CTD, since, among the multiplicity of available RNA species, only the PS has sufficiently high affinity to efficiently dislodge domain N3. Binding of the PS to the CTD releases domain N3, making it accessible for binding to the M endodomain. This nucleation step in gRNA recognition would then be propagated by conformational interactions that allow subsequent N molecules to nonspecifically bind to adjacent sites along the gRNA strand, thereby encapsidating the genome. The binding of additional N molecules, in turn, would free progressively more N3 domains to contact M endodomains, driving virion budding. In this model, sequestering of domain N3 by the CTD thus serves two purposes: first, it imparts to the CTD a higher selectivity for the PS; second, it ensures that N3 is not allowed to bind to the M endodomain until the CTD has bound to gRNA.
Fig. 4.
Model for the dual role of domain N3 in genome packaging and virion assembly. (A) Domain N3 is originally bound to the CTD, either directly occluding the RNA-binding site or else indirectly altering the conformation of the RNA-binding site. (B) Binding of the PS to the CTD displaces domain N3. (C) The released domain N3 is now accessible for binding to the M protein endodomain.
Our data could be taken to support an alternative hypothesis – that domain N3 combines with the CTD to form an extended RNA-binding site for the PS. However, this scenario seems unlikely, since such binding would block, rather than enhance, the association of N3 with the M endodomain. Additionally, the region in N3 to which the packaging role localizes is set among a high density of negatively-charged residues (Fig. 3F). We also note that the binding of N3 to the CTD was anticipated by two prior studies that observed N3 to participate in N-N interactions (Hurst et al., 2009, Lo et al., 2013), although neither postulated that the CTD was the interacting partner of N3.
It is well established for MHV and BCoV that both gRNA and all sgRNAs are co-immunoprecipitated from infected cell extracts by anti-N antibodies (Cologna et al., 2000, Narayanan et al., 2000). If our model is correct, then this would suggest that it is the NTD that is responsible for binding to sgRNAs, by either nonspecific or sequence-specific binding. Evidence has been presented for high-affinity binding of the MHV NTD to the transcription-regulating sequence found at the leader-body junction of all sgRNAs (Grossoehme et al., 2009). However, such sequence-specific binding does not appear to be a general property of coronavirus NTDs (Keane et al., 2012). Consistent with our model, it has been demonstrated that anti-M protein antibody co-immunoprecipitates only the fraction of N protein that is bound to gRNA or to an engineered heterologous RNA containing the MHV PS (Narayanan et al., 2000, Narayanan and Makino, 2001). Although we cannot rule out a direct role for M protein in packaging (Narayanan et al., 2003), our data suggests that the participation of M is secondary, through selecting N protein that has bound to the PS.
The model in Fig. 4 necessarily simplifies a number of details that remain to be clarified. One of these is the stoichiometry of N protein to RNA. In the helical nucleocapsids of nonsegmented negative-strand RNA viruses, one N protein molecule binds from six to nine nucleotides of single-stranded RNA (Green et al., 2006, Alayyoubi et al., 2015, Gutsche et al., 2015). A model for the SARS-CoV nucleocapsid has estimated a ratio of one N molecule per 7 nucleotides of RNA (Chang et al., 2014). It thus seems likely that the 95-nt MHV PS accommodates multiple CTDs, the accumulation of which nucleates cooperative encapsidation of the entire viral genome. A single AA-bulge repeat unit (Fig. 1A) is a good candidate for the minimal relevant substructure within the PS that is recognized by one CTD. A second issue to be resolved is the role of CTD dimerization in PS binding and the propagation of the nucleation event to trigger nonspecific binding of N to the remainder of the genome. CTD dimerization also raises the possibility that N3-CTD association could occur in trans in the dimer, rather than in cis. A third uncertainty is the role of spacer B. The N-BPR mutant lost the ability to selectively package gRNA (Fig. 3E). However, the nearly total absence of charge in spacer B of the N-BPR mutant may prevent the association of its domain N3 with the CTD, causing N3 to be constitutively displayed for binding to the M endodomain. A more directed mutational analysis of spacer B will be required to learn if it has some function beyond serving as a flexible linker. Finally, it should be pointed out that MHV domain N3 contains two residues that are phosphorylation targets, S424 and T428 (White et al., 2007). This could conceivable provide a level of regulation to coordinate the roles of gRNA as a translation and transcription template and as a constituent of progeny virions.
4. Materials and methods
4.1. Cells and viruses
Wild-type MHV-A59 and mutants were propagated in mouse 17 clone 1 (17Cl1) cells; plaque titrations and plaque purifications were performed with mouse L2 cells. Murine cell lines were grown in Dulbecco’s modified minimal essential medium (DMEM) supplemented with 10% fetal bovine serum. The host-range chimeric coronavirus fMHV.v2 (Goebel et al., 2004), which was used for reverse genetics, was grown in feline FCWF cells maintained in low-glucose DMEM containing 10% fetal bovine serum.
4.2. MHV mutant construction
All mutants in this study were created by targeted RNA recombination (Kuo et al., 2000, Goebel et al., 2004). Mutants MN8 and MN10 (Kuo et al., 2016), silPS (Kuo and Masters, 2013), MMA (Kuo and Masters, 2002), CCA2, CCA3, and CCA5 (Hurst et al., 2005), and N-BPR (Hurst et al., 2009) have been described previously. Donor RNAs for generation of the T-CTD-R, T-CTD-R-PS1, and T-CTD-R-PS2 mutants were transcribed from plasmids derived from the vector pPM9, which contains the 3’ end of the MHV-A59 genome, starting from the middle of nsp14, and including a 2.1-kbp deletion that removes TRS2, gene 2a, and most of the HE gene (Kuo and Masters, 2013). The TGEV CTD substitution was initially constructed by PCR from overlapping oligonucleotides and inserted between the unique NgoMIV and BstXI sites of MHV N gene plasmid pCK70XB (Kuo et al., 2014). The chimeric N gene was then shuttled into pPM9 to produce pPM9-T2b, which was used for construction of the original TGEV-CTD mutant. The vector for the T-CTD-R mutant (pT2bR) was subsequently made by transfer of a cDNA fragment encoding the reverting mutation G78R between the unique EagI (M gene) and NheI (N gene) sites in pPM9-T2b. Plasmids pT2bR-PS1 and pT2bR-PS2, transcription vectors for the T-CTD-R-PS1 and T-CTD-R-PS2 mutants, were constructed by insertion of cDNA for nt 1-650 or nt 156-327 of the TGEV genome between the unique SalI and AscI sites downstream of the nsp16 stop codon in pT2bR.
Following targeted RNA recombination and plaque purification, verification of constructed mutants was carried out by reverse transcription of isolated RNA with a random hexanucleotide primer and avian myeloblastosis virus reverse transcriptase (Life Sciences). PCR amplification of cDNA was performed with the Expand High Fidelity PCR System (Roche), and PCR products were purified with QIAquick spin columns (Qiagen) prior to Sanger DNA sequencing. The isogenic wild-type strain used for comparison with TGEV CTD mutants was Alb649 (Kuo and Masters, 2013); the isogenic wild-type strain for all other mutants was Alb741 (Kuo et al., 2014).
4.3. Virus purification
Monolayers of 17Cl1 cells grown in Eagle’s medium containing 10% fetal bovine serum were inoculated at a multiplicity of 1 PFU/cell. Infections were allowed to proceed for 12–16 h, to a point where syncytia formation was maximal but little or no cell lysis or detachment had occurred. Released virus in harvested extracellular medium was precipitated with polyethylene glycol and resuspended in magnesium- and calcium-free phosphate-buffered saline, pH 7.4 (PBS). Virions were sedimented onto cushions of 60% sucrose in PBS by centrifugation at 151,000×g for 2.5 h in a Beckman SW41 rotor at 4 °C. Samples were removed from cushions, diluted with PBS to contain 10% sucrose and layered onto 10–20–40–60% sucrose step gradients. Virion bands were collected from the 20–40% sucrose interface after centrifugation at 151,000×g for 2.5 h in a Beckman SW41 rotor at 4 °C. For the SARS-CoV M chimera set (Fig. 2) and the domain N3 mutant set (Fig. 3), virions were further purified by pulldown with anti-M monoclonal antibody J.1.3 and nProtein A Sepharose beads (GE Healthcare) exactly as described previously (Kuo and Masters, 2013).
4.4. Analysis of viral RNA and protein
RNA was extracted from purified virions or from infected cells with Ultraspec reagent (Biotecx) according to the manufacturer׳s instructions and then isolated with Direct-zol RNA MiniPrep spin columns (Zymo Research). Northern blotting analysis of purified virion RNA was conducted as described in detail previously (Kuo and Masters, 2010). RNA was probed with a PCR product corresponding to nt −17 through +747 of the MHV N gene; the probe was labeled with an AlkPhos Direct kit and was visualized using CDP-Star detection reagent (GE Healthcare).
Proteins from purified virions were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), with prestained protein standards included in adjacent lanes, and analyzed by Western blotting exactly as described previously (Hurst et al., 2009). Proteins were detected with anti-MHV M protein monoclonal antibody J.1.3 (Fig. 1, Fig. 2, Fig. 3) and either anti-MHV N protein monoclonal antibody J.3.3 (Fig. 1) or else anti-MHV N protein rabbit polyclonal antibody (Fig. 2, Fig. 3). Bound antibodies were visualized by enhanced chemiluminescence detection (Pierce). For normalization of purified virions prior to RNA extraction, chemiluminescence was quantitated with a BioRad ChemiDoc XRS+ instrument. Both monoclonal antibodies were generously provided by John Fleming (University of Wisconsin, Madison). Antibody J.1.3 recognizes an epitope in the M protein ectodomain (de Haan et al., 1998); antibody J.3.3 recognizes an epitope in N protein domain N3 (Hurst et al., 2005).
Acknowledgments
We thank the Wadsworth Center׳s Applied Genomics Technology Core Facility for DNA sequencing and the Media and Tissue Culture Core Facility for media preparation.
This work was supported by National Institutes of Health (National Institute of Allergy and Infectious Diseases) Grant R01 AI064603. The funding source had no role in the study design, collection and analysis of data, decision to publish, or preparation of the manuscript.
References
- Alayyoubi M., Leser G.P., Kors C.A., Lamb R.A. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc. Natl. Acad. Sci. USA. 2015;112:E1792–E1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Chen C.M., Chiang M.H., Hsu Y.L., Huang T.H. Transient oligomerization of the SARS-CoV N protein – implication for virus ribonucleoprotein packaging. PLoS One. 2013;8:e65045. doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hou M.H., Chang C.F., Hsiao C.D., Huang T.H. The SARS coronavirus nucleocapsid protein – forms and functions. Antivir. Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.K., Hsu Y.L., Chang Y.H., Chao F.A., Wu M.C., Huang Y.S., Hu C.K., Huang T.H. Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging. J. Virol. 2009;83:2255–2264. doi: 10.1128/JVI.02001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Chang C.K., Chang Y.W., Sue S.C., Bai H.I., Riang L., Hsiao C.D., Huang T.H. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., Olsthoorn R.C. Group-specific structural features of the 5′-proximal sequences of coronavirus genomic RNAs. Virology. 2010;401:29–41. doi: 10.1016/j.virol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.C., van den Born E., van den Worm S.H., Pleij C.W., Snijder E.J., Olsthoorn R.C. New structure model for the packaging signal in the genome of group IIa coronaviruses. J. Virol. 2007;81:6771–6774. doi: 10.1128/JVI.02231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R., Hogue B.G. Identification of a bovine coronavirus packaging signal. J. Virol. 2000;74:580–583. doi: 10.1128/jvi.74.1.580-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cologna R., Spagnolo J.F., Hogue B.G. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology. 2000;277:235–249. doi: 10.1006/viro.2000.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escors D., Izeta A., Capiscol C., Enjuanes L. Transmissible gastroenteritis coronavirus packaging signal is located at the 5′ end of the virus genome. J. Virol. 2003;77:7890–7902. doi: 10.1128/JVI.77.14.7890-7902.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosmire J.A., Hwang K., Makino S. Identification and characterization of a coronavirus packaging signal. J. Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T.J., Zhang X., Wertz G.W., Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., 3rd, Leibowitz J.L., Giedroc D.P. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche I., Desfosses A., Effantin G., Ling W.L., Haupt M., Ruigrok R.W., Sachse C., Schoehn G. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Kuo L., Masters P.S., Vennema H., Rottier P.J. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Koetzner C.A., Masters P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009;83:7221–7234. doi: 10.1128/JVI.00440-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Ye R., Goebel S.J., Jayaraman P., Masters P.S. An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J. Virol. 2010;84:10276–10288. doi: 10.1128/JVI.01287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K.R., Kuo L., Koetzner C.A., Ye R., Hsue B., Masters P.S. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J. Virol. 2007;81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane S.C., Liu P., Leibowitz J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 2002;76:4987–4999. doi: 10.1128/JVI.76.10.4987-4999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Evolved variants of the membrane protein can partially replace the envelope protein in murine coronavirus assembly. J. Virol. 2010;84:12872–12885. doi: 10.1128/JVI.01850-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Masters P.S. Functional analysis of the murine coronavirus genomic RNA packaging signal. J. Virol. 2013;87:5182–5192. doi: 10.1128/JVI.00100-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Koetzner C.A., Hurst K.R., Masters P.S. Recognition of the murine coronavirus genomic RNA packaging signal depends on the second RNA-binding domain of the nucleocapsid protein. J. Virol. 2014;88:4451–4465. doi: 10.1128/JVI.03866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst-Hess K.R., Koetzner C.A., Masters P.S. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J. Virol. 2016:90. doi: 10.1128/JVI.03212-15. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.S., Lin S.Y., Wang S.M., Wang C.T., Chiu Y.L., Huang T.H., Hou M.H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Wu D., Shen C., Chen K., Shen X., Jiang H. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem. Cell Biol. 2006;38:589–599. doi: 10.1016/j.biocel.2005.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M.M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J. Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L., Mateos-Gomez P.A., Capiscol C., del Palacio L., Enjuanes L., Sola I. Transmissible gastroenteritis coronavirus genome packaging signal is located at the 5′ end of the genome and promotes viral RNA incorporation into virions in a replication-independent process. J. Virol. 2013;87:11579–11590. doi: 10.1128/JVI.01836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Makino S. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 2001;75:9059–9067. doi: 10.1128/JVI.75.19.9059-9067.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Maeda A., Maeda J., Makino S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K., Chen C.J., Maeda J., Makino S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 2003;77:2922–2927. doi: 10.1128/JVI.77.5.2922-2927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige P.E., Jr. Follow the yellow brick road: a paradigm shift in virus assembly. J. Mol. Biol. 2016;428:416–418. doi: 10.1016/j.jmb.2015.12.009. [DOI] [PubMed] [Google Scholar]
- van der Most R.G., Bredenbeek P.J., Spaan W.J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J. Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Bednar V., Blount A., Hogue B.G. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 2006;80:4344–4355. doi: 10.1128/JVI.80.9.4344-4355.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S., Lopez L.A., Bednar V., Hogue B.G. Importance of the penultimate positive charge in mouse hepatitis coronavirus A59 membrane protein. J. Virol. 2007;81:5339–5348. doi: 10.1128/JVI.02427-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.C., Yi Z., Hogue B.G. Identification of mouse hepatitis coronavirus A59 nucleocapsid protein phosphorylation sites. Virus Res. 2007;126:139–148. doi: 10.1016/j.virusres.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]