Abstract
In human cells, error-free repair of DNA double-strand breaks requires the DNA pairing and strand exchange activities of RAD51 recombinase. Activation of RAD51 recombination activities requires the assembly of RAD51 presynaptic filaments on the single-stranded DNA that forms at resected DSB ends. Mutations in proteins that control presynaptic filament assembly, such as BRCA2, and in RAD51 itself, are associated with human breast cancer. Here we describe the properties of two mutations in RAD51 protein that derive from human lung and kidney tumors, respectively. Sequence variants Q268P and Q272L both map to the DNA binding loop 2 (L2) region of RAD51, a motif that is involved in DNA binding and in the allosteric activation of ATP hydrolysis and DNA strand exchange activities. Both mutations alter the thermal stability, DNA binding, and ATPase properties of RAD51, however both variants retain intrinsic DNA strand exchange activity towards oligonucleotide substrates under optimized conditions. In contrast, both Q268P and Q272L variants exhibit drastically reduced DNA strand exchange activity in reaction mixtures containing long homologous ssDNA and dsDNA substrates and human RPA protein. Mixtures of wild-type and variant proteins also exhibit reduced DNA strand exchange activity, suggesting that heterozygous mutations could negatively affect DNA recombination and repair processes in vivo. Together, the findings of this study suggest that hypomorphic missense mutations in RAD51 protein could be drivers of genomic instability in cancer cells, and thereby contribute to the etiology of metastatic disease.
Keywords: RAD51, recombination, homology-directed repair, cancer, variant, presynaptic filament
Graphical abstract
1. Introduction
Human RAD51 protein is a DNA pairing enzyme orthologous to the E. coli RecA protein. RAD51 catalyzes DNA pairing and strand exchange reactions that are central to the processes of homologous recombination and homology-directed DNA repair (1). Defects in RAD51-dependent HR/HDR pathways lead to genome instability and are directly associated with human cancer (2).
The biochemical properties of RAD51 include sequence-nonspecific binding to ssDNA and dsDNA, ssDNA-stimulated ATP hydrolysis, and ATP-dependent pairing and exchange of homologous ssDNA/dsDNA segments (3). RAD51 enzymatic activities are activated upon its assembly into a right-handed, helical filament on ssDNA the so-called presynaptic filament. This nucleoprotein filament undergoes dynamic changes in association, dissociation, and conformation in response to ATP binding and hydrolysis (3–5). The ATP-bound form is considered to be the active form of the filament. It has a high helical pitch of 90–130Å and contains ssDNA that is significantly stretched (4,5). The ADP-bound form is considered to be the inactive form of the filament. It has a low helical pitch of 65–85Å. RAD51 interacts with DNA via residues in two disordered loop regions, L1 and L2, which are located in the conserved catalytic core domain (3). L2 is considered to be the primary binding site for ssDNA. The N-terminal end of L2 is capped by a conserved glutamine residue, Gln-268 in human RAD51. Based on comparisons with the E. coli RecA-DNA structures (6), Gln-268 is a key residue in the network of interactions that connects the ssDNA and ATP binding sites of RAD51. Therefore Gln-268 and residues in L2 are critical for the allosteric activation of enzyme activities in response to ssDNA binding. In addition to regulation by the ATPase cycle, the formation and dynamics of the RAD51 presynaptic filament are also regulated by mediator proteins including BRCA2 and the RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3), by helicase/motor proteins including RAD54, BLM and others, and likely by signaling proteins and post-translational modifications (2,3,7,8).
Many human tumors exhibit changes in RAD51 recombination activity. Activity variation may be linked to changes in RAD51 expression levels (9–18), to deleterious mutations in RAD51 itself (19–21), or to defects involving any of several tumor suppressor proteins (BRCA2, PALB2, p53 and others) that interact with RAD51 and regulate its activity (2,3,22–25). Changes in RAD51 expression levels are observed in many types of cancer and may correlate with the progress of the disease and/or with tumor resistance or sensitivity to chemotherapeutic drugs (9–11). RAD51 overexpression has been reported in malignant prostate cancer, small cell lung carcinoma, and invasive ductal breast cancer, where in the latter case the level of RAD51 expression correlates directly with the histological grading of the tumor (10). Conversely, down-regulation of RAD51 is observed in approximately 30% of sporadic breast tumors and cell lines (13–16), and is reported to increase the radiosensitivity of prostate cancer and malignant glioma cells (17,18).
Sequence variants of RAD51 protein have received less study due to their low penetrance. However, recent studies identified three breast cancer-associated RAD51 missense mutations, the somatic variants D149N and G151D, and the germline variant R150Q (19–21). These mutations, from different individuals, occurred in adjacent residues of a conserved Schellman loop motif that occupies a prominent position on the outer surface of the RAD51 presynaptic filament (20). The motif is distant from binding sites for DNA, ATP, BRCA2 and PALB2, but it is near a reported p53 binding site (23–25). All three RAD51 mutants are proficient in DNA pairing and strand exchange, but they form presynaptic filaments with altered physical and biochemical properties (20). The R150Q and G151D variants have low catalytic efficiencies for ssDNA-stimulated ATP hydrolysis. All three variants interact with wild-type RAD51 and can form mixed presynaptic filaments with WT on ssDNA. Mixed G151D/WT presynaptic filaments have biochemical properties that are intermediate between those of filaments containing pure WT or pure G151D protein. These studies demonstrated that cancer-associated RAD51 sequence variants may have functional phenotypes, and that a heterozygous RAD51 variant could still exert an effect on recombination through co-integration with WT protein in presynaptic filaments (20).
In the current study we describe the biochemical properties of two new sequence variants of RAD51 protein, Q268P and Q272L, which were identified in human lung and kidney tumors, respectively. Both mutations affect the DNA binding loop L2 region, including the allosteric switch residue Gln-268. We hypothesized that both of these mutations would affect RAD51 DNA binding and enzymatic functions, a prediction that is borne out by the data. Both variants are hypomorphs with greatly reduced DNA strand exchange activity compared to wild-type. Mixtures of either variant with wild-type also have reduced DNA strand exchange activity, which has important implications for the potentially damaging effects of Q268P and Q272L variants in vivo. Our findings extend our knowledge of the range of RAD51 dysfunctions that may occur in cancer cells, and which could potentially serve as drivers of genomic instability and tumor progression.
2. Materials and Methods
2.1. Reagents
Chemicals, biochemicals, and enzymes were purchased from Sigma-Aldrich unless specifically stated. Restriction enzymes and T4 polynucleotide kinase were purchased from New England Biolabs. All reagents were analytical grade and solutions were made with Barnstead NANO-pure water. TE buffer contained 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA.
2.2. Nucleic acids
Oligonucleotides were purchased from Operon. Circular M13mp18 ssDNA (7.3 kb) was prepared as described (26). Circular φX174 ssDNA (5.4 kb) was purchased from New England Biolabs. Supercoiled φX174 dsDNA (5.4 kbp) was purchased from Promega and was linearized by digestion with PstI restriction endonuclease. Linearized dsDNA molecules were purified using a DNA Clean-up Kit from Thermo Scientific. The concentrations of ssDNA and dsDNA stock solutions were determined by UV absorbance in a Nanodrop Spectrophotometer (Thermo Scientific), according to manufacturer’s instructions, and are expressed in units of micromoles of nucleotide residues per liter. All DNA molecules were stored at −20°C in TE buffer.
2.3. Site-directed mutagenesis of human RAD51 protein
Plasmid pET-15b expressing a His6-tagged version of the human RAD51 protein was a generous gift from Dr. Hitoshi Kurumizaka at Waseda University, Japan. The Q268P and Q272L mutations were introduced separately using the QuikChange (Stratagene) and Q5 (New England Bioloabs) site-directed mutagenesis protocols, respectively. The primers used are shown in Table 1 (oligos 1 and 2 for Q268P; oligos 3 and 4 for Q272L). PCR reactions were carried out according to the QuikChange protocol and the resulting plasmids were sequenced at the University of Vermont Cancer Center DNA Analysis Facility to verify successful mutagenesis.
Table 1.
Lengths and sequences of oligonucleotide primers and substrates.
| Oligo No. | Length (b) | Sequence (5′ → 3′) | Use |
|---|---|---|---|
| 1 | 33 | GTGGTAATCACTAATCCGGTGGTAGCTCAAGTG | Q268P forward primer |
| 2 | 33 | CACTTGAGCTACCACCGGATTAGTGATTACCAC | Q268P reverse primer |
| 3 | 22 | GTGGTAGCTCTAGTGGATGGAG | Q272L forward primer |
| 4 | 21 | CTGATTAGTGATTACCACTGC | Q272L reverse primer |
| 5 | 63 | ACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCAAAACAAAAGGA | Strand exchange partial duplex |
| 6 | 32 | TCCTTTTGTTTTGAGGTCATTTTTGCGGATGG | Strand exchange partial duplex |
| 7 | 31 | ACAGCACCAGATTCAGCAATTAAGCTCTAAG | Strand exchange homol. duplex |
| 8 | 31 | CTTAGAGCTTAATTGCTGAATCTGGTGCTGT | Strand exchange homol. duplex |
| 9 | 25 | AAAAAAAAAAAAAAAAAAAAAAA*A (* denotes position of AF546 label) | DNA binding w/AF546 label |
| 10 | 25 | TTTTTTTTTTTTTTTTTTTTTTTTT | Compl. of #9 for dsDNA binding |
2.4. Purification of human RPA and RAD51 Proteins
Human RPA protein expression plasmid p11d-tRPA was a kind gift from Dr. Marc S. Wold at the University of Iowa. RPA was expressed in E. coli strain BL21 (DE3) and the protein was purified as described (27,28). Human RAD51 wild-type, Q268P, and Q272L proteins were expressed and purified from plasmid pET-15b in Rosetta2 (DE3) E. coli cells (Novagen, Inc., Madison, WI) as described (20). Purified RAD51 proteins were dialyzed into storage buffer consisting of 20 mM HEPES (pH 7.5), 150 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, and 10% glycerol, frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined by UV absorbance using a Nanodrop Spectrophotometer (Thermo Scientific) and an extinction coefficient of 0.34 A280 = 1 mg/mL. All protein stock solutions were >95% pure as determined by SDS-PAGE, nucleic acid-free as determined by A280/A260 ratio, and nuclease-free as determined by agarose gel electrophoresis of treated DNA samples.
2.5. ATPase assays
Steady-state rates of ATP hydrolysis were measured using thin-layer chromatography (TLC) assays. Reactions at 37°C contained 30 mM MES (pH 6.2), 6 mM MgCl2, 30 mM KCl, 1 mM DTT, and 0.1 mg/mL BSA in a final volume of 50 μL. In addition the reaction mixtures contained 2.0 μM wild-type or variant RAD51, 12 μM M13mp18 ssDNA, and 1 mM ATP (containing 10 μCi/mL of α-[32P]-ATP). Reactions were initiated by the addition of ATP + α-[32P]-ATP. Aliquots (8 μL) were removed at various time points and quenched with 8 mM EDTA and 1% SDS (final concentrations). Quenched samples were spotted out at 1 μL volume onto PEI-cellulose TLC plates (20 X 20 cm) at 1 cm intervals. After all the samples were spotted and dried the TLC plates were developed with 0.75 M KH2PO4 and allowed to air dry. The TLC plates were exposed for 1.5 hours to a K-screen (Kodak) and scanned by a Bio-Rad Personal Molecular Imager-FX (University of Vermont Cancer Center DNA Analysis Facility). Quantification of the phosphorimage was performed by Quantity One v4.5.1 (Bio-Rad) software and subsequently fit using Graphpad Prism v5.0 (Graphpad Software Inc).
2.6. DNA strand exchange assays with φX174 substrates
DNA strand exchange reactions using φX174 ssDNA and dsDNA substrates were carried out as described (20), with minor modifications. All reaction steps were carried out at 37ºC. The final volume of each reaction was 50 μL. All concentrations given are final concentrations. RAD51 wild-type, Q268P, or Q272L protein (7.5 μM) was preincubated with 30 μM φX174 ssDNA for 5 min in buffer containing 40 mM Tris-HCl (pH 7.8), 1 mM MgCl2, 100 mM KCl, 1 mM DTT, 2 mM ATP, 8 mM creatine phosphate, and 28 μg/mL creatine phosphokinase. Human RPA protein (2 μM) was added followed by a 5 min incubation, then 100 mM ammonium sulfate was added followed by a 1 min incubation. Reactions were initiated by the simultaneous addition of 30 μM (15 μM basepairs) linear φX174 dsDNA and 4 mM spermidine. At the indicated times, 6.5 μL aliquots were removed and brought to a final concentration of 0.8% SDS and 0.8 mg/mL Proteinase K, then incubated at 37ºC for 15 min. Following addition of 10% glycerol and a trace of bromophenol blue, the samples were loaded onto a 0.8% agarose gel in TAE buffer, and electrophoresed for 16 hrs at 25 V. Gels were stained with 1 μg/mL Sybr Gold and digitally photographed to record a negative image. Results were quantified by measuring the intensity of the joint molecule and nicked circle product bands relative to total DNA by using a Bio-Rad Personal Molecular Imager-FX with Quantity One (Bio-Rad) software (University of Vermont Cancer Center DNA Analysis Facility). Some reactions were modified to contain mixtures of wild-type and variant RAD51 proteins at ratios and total concentrations given in the text.
2.7. DNA strand exchange assays with oligonucleotide substrates
Oligo 8 was 5′-[32P] labeled using T4 polynucleotide kinase (NEB) according to manufacturer’s instructions, and excess label was removed using the Probe Quant G-50 microcolumn kit (GE Healthcare). The partial duplex DNA substrate was constructed by annealing oligo 5 to oligo 6 (11.7 μM molecules, each), and the homologous duplex DNA substrate was constructed by annealing [32P]-oligo 8 to oligo 7 (11.7 μM molecules, each), both in 30 mM HEPES, pH 7.0. In a PCR thermal cycler, annealing mixtures were heated at 95ºC for 5 min, followed by 60 min at 50ºC, and then cooled to 20ºC in 5ºC increments over 30 min. Strand exchange reactions were carried out at 30ºC under the following conditions (stated concentrations represent final concentrations in a final reaction volume of 50 μL): Preincubation mixture contained 30 mM HEPES, pH 7.0, 20 mM KCl, 0.35 μM oligo 5/6 partial duplex DNA, 10.5 μM RAD51 (WT, Q268P, or Q272L), 6.2 mM ATP, and 2 mM MgCl2. Control reactions contained an equivalent volume of RAD51 storage buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercapotethanol, 10% glycerol) in place of protein. After a 5 min preincubation, the concentration of MgCl2 was increased to 18 mM, and the reaction was allowed to preincubate for another 15 min. The reaction was started by adding 0.35 μM [32P]-oligo 8/7 homologous duplex DNA. Aliquots of 10 μL each were removed at 0, 30, 60, and 90 min and quenched by adding to 10 μL of stopping solution to yield final concentrations of 60 mM Tris-HCl, pH 7.5, 2% SDS, 50 mM EDTA, 2.5 mg/mL proteinase K, and 1x Promega Blue/Orange Loading Dye, then kept at room temperature prior to electrophoresis. Samples were run on a 4–20% TBE PAGEr Gold Precast Polyacrylamide gel (Lonza) and exposed to a K-screen for visualization. Images were taken using Bio-Rad Personal Molecular Imager-FX (Vermont Cancer Center DNA Analysis Facility), and data was quantified using Quantity One v4.5.1 (Bio-Rad).
2.8. Electrophoretic mobility shift assays
The properties of RAD51 and variant complexes with circular φX174 ssDNA and linear φX174 dsDNA were examined by electrophoretic mobility shift. Protein-dsDNA complexes were assembled in buffer containing 24 mM HEPES (pH 7.5), 1 mM MgCl2, 30 mM NaCl, 1 mM dithiothreitol, 0.4 mM 2-mercaptoethanol, 0.02 mM EDTA, 0.1 mg/mL BSA, 2% glycerol, and 1 mM ATP. For protein-ssDNA complex assembly, the NaCl concentration was increased to 45 mM. RAD51 wild-type, Q268P, or Q272L proteins (0–8 μM depending on experiment) were incubated with 20 μM φX174 ssDNA or 20 μM (10 μM basepairs) φX174 dsDNA for 20 min at 37ºC. Samples were loaded onto a 0.8% agarose gel in Tris-acetate-EDTA buffer, then electrophoresed for 16 hrs at 25 V. The gel was stained with 1 μg/mL Sybr Gold, trans-illuminated with UV light, and digitally photographed.
2.9. AlexaFluor 546 DNA binding assay
RAD51-DNA interactions were measured using DNA substrates labeled with the fluorophore AlexaFluor 546 where a fluorescence quenching effect occurs upon recombinase binding (29). Titration experiments were conducted by adding increasing amounts of RAD51 WT, Q268P, or Q272L protein to either 2 μM (nucleotides) of AlexaFlour 546-labeled ssDNA (oligo 9) or 2 μM basepairs of AlexaFluor 546-labeled dsDNA (oligo 9/10). Titrations were carried out at room temperature and in the presence/absence of 2 mM ATP in binding buffer containing 30 mM Tris, pH 7.5, 10 mM MgCl2, and 0.1 mM DTT. After each addition of protein, the mixture was allowed to incubate for 30 seconds before taking the fluorescence reading. Amplitudes of fluorescence quenching were quantified by determining the fraction of the fluorescence signal that was quenched by the addition of RAD51 at each concentration (dilution factors were taken into account). Amplitude values were plotted against concentration of RAD51. The binding curves were fitted to the Hill equation using Graphpad. For all binding experiments, the fluorophore was excited at 554 nm and fluorescence was monitored between 560–600 nm. All slit widths were set at 5 nm.
2.10. Thermofluor assay for thermal stability of proteins
The thermofluor stability assay was conducted as previously described with minor changes (30,31). Samples were assembled directly into a 96-well plate (USA Scientific) where each well contained assay buffer (30 mM Tris, pH 7.5; 10 mM magnesium chloride; 0.1 mM dithiothreitol), 5 μM RAD51, 25x Sypro Orange (Invitrogen), and either 0 or 2 mM ATP. Once assembled, the plate was sealed with an optical adhesive film (USA Scientific). Temperature was controlled using an AB 7500 Fast Sequence Detection System (UVM Advanced Genome Technologies Core) where the temperature ranged from 25°C to 95°C with a 1°C increment and a 1 min dwell time per cycle. Normalized fluorescence intensities were plotted versus temperature and melting temperatures (Tm) were calculated using the build-in Boltzmann equation in GraphPad (31). The data presented here are representative of three independently conducted trials with similar observations.
2.11. Bioinformatic and structural analyses
Human RAD51 variants were catalogued and annotated by the University of Vermont Molecular Bioinformatics Shared Resource. Variants were identified using Ensembl Variation, which pulls data from 36 data sources, such as dbSNP, COSMIC, and ESP. Variants are predicted to be of low, moderate, or high impact using SNPEff (32). As missense variants, Q268P and Q272L are predicted to be of moderate impact. Zygosity and validation status were taken from the COSMIC database. Sequences of Homo sapiens RAD51 (Hs RAD51; CAG38796.1), Saccharomyces cervisiae Rad51 (Sc Rad51; CAA45563.1), and Escherichia coli RecA (Ec RecA; P0A7G6.2, 3CMW_G) proteins in Figure 1 were aligned using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). All structural figures of human RAD51 and yeast Rad51 proteins were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.). Structures of monomeric human RAD51 core domain and core domain-BRC4 complex were derived from PDB ID no. 1N0W (33). Structures of filamentous S. cerevisiae Rad51 protein were derived from PDB ID no. 3LDA (34).
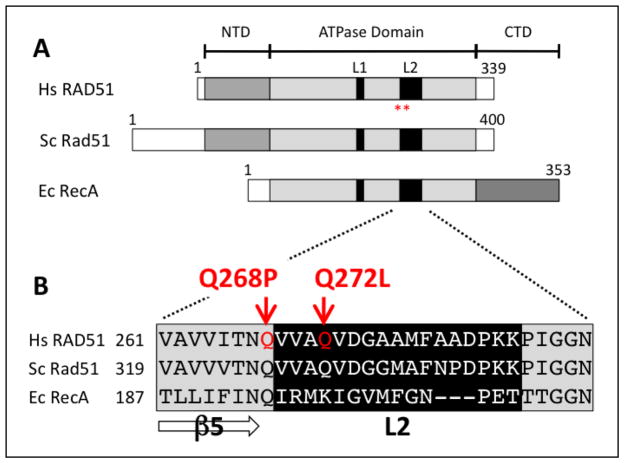
Figure 1. Locations of Q268P and Q272L mutations in the primary structure of human RAD51 protein.
(A) The Q268P and Q272L mutations (positions denoted by red asterisks) occur in the conserved RecA homology domain of RAD51, shown here in alignment with the yeast Rad51 and E. coli RecA proteins. (B) Protein sequence alignment in the vicinity of DNA binding loop L2.
3. Results
3.1. Cancer affiliation of RAD51 variant proteins Q268P and Q272L
A survey of 183 lung adenocarcinoma tumors identified one sample with an 803A>C substitution mutation in the coding sequence of the RAD51 gene, resulting in a Q268P missense mutation in the RAD51 protein (sample ID LUAD-D01751; COSMIC ID COSM338187) (35,36). The zygosity of the mutation was unknown and the somatic status of the mutation was unconfirmed. The source was a tumor sample taken from a 50 year-old European female with a history of heavy smoking, who was diagnosed with stage I-B lung cancer.
A survey of 499 kidney renal clear cell carcinoma tumors identified one sample with an 815A>T substitution mutation in the coding sequence of the RAD51 gene, resulting in a Q272L missense mutation in the RAD51 protein (sample ID TGCA-CJ-4920-01; COSMIC ID COSM470584) (36,37). The mutation was confirmed somatic but its zygosity was unknown. The source was a primary tumor sample taken from a 64 year-old female who died of kidney cancer 5 months after diagnosis.
3.2. Structural context of Q268P and Q272L mutations
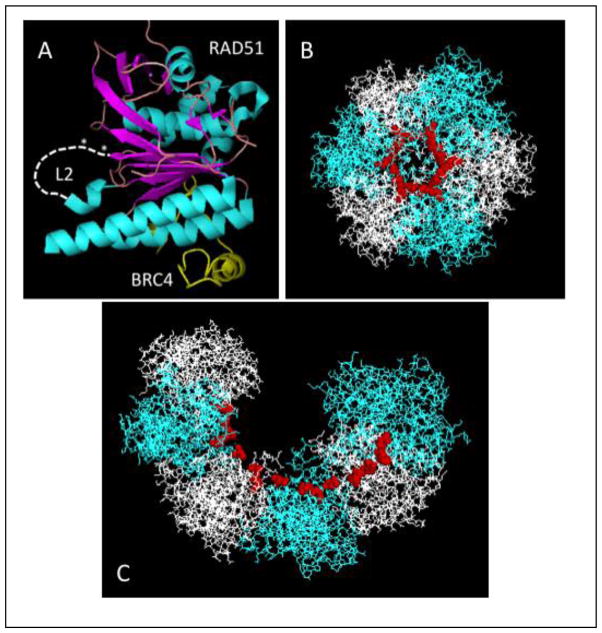
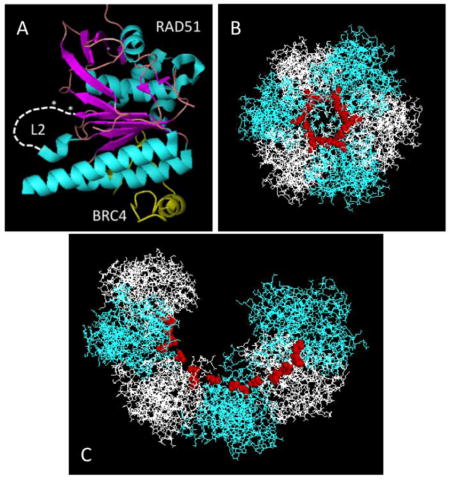
The Q268P and Q272L mutations both affect the DNA binding region of RAD51 protein. Specifically, both mutations occur in the vicinity of DNA binding loop 2 (L2) (Fig. 1A-B). Based on comparisons of the human RAD51 catalytic domain structure (Fig. 2A) to the yeast Rad51 and E. coli RecA filament structures (6,34,38), the L2 loop forms part of the inner surface of the open helix of the RAD51 filament. This is illustrated in Figure 2B–C, where conserved residues representing the N- and C-terminal attachment sites for L2 are highlighted as red spheres mapped onto the yeast Rad51 filament structure. L2 forms at least part of the primary DNA binding site that interacts with ssDNA within the presynaptic filament (3,6).
Figure 2. Locations of the L2 loop and mutations in the RAD51 protomer and filament.
(A) The approximate locations of DNA binding loop L2 (white dashed line) and of the Q268P and Q272L mutations (white asterisks) are indicated in the context of the human RAD51-BRC4 structure (PDB ID no. 1N0W). (B) – (C) One helical turn (6 protomers) of the structure of the yeast Rad51 filament (PDB ID no. 3LDA) is shown with alternating protomers in blue and white and with attachment points for the L2 loop (including Gln-326, the equivalent of Gln-268 in human RAD51) shown as red spheres. The view in (B) is looking down the helical axis, while that in (C) is shown at an oblique angle.
The Q272L mutation occurs within L2, and affects a glutamine residue that is highly conserved among eukaryotic Rad51 proteins (Fig. 1B). L2 residues including Gln-272 are disordered both in the crystal structure of monomeric human RAD51 catalytic domain (33) and in structures of yeast Rad51 filaments (34,38), none of which contain bound DNA or nucleotide. However the position of L2 is highly conserved in the primary, tertiary, and quaternary structures of all RecA family recombinases, including RAD51 (Figs. 1–2). In the E. coli RecA-ssDNA-ADP-AlF4 structure, L2 is ordered and residues within it make direct contacts with ssDNA (6). It is therefore reasonable to hypothesize that Gln-272 in human RAD51 is involved in ssDNA binding within the presynaptic filament, and that the Q272L mutation might therefore affect filament assembly, activity, or turnover.
The Q268P mutation occurs at the N-terminal end of L2 and involves a glutamine residue that is highly conserved throughout the RecA/Rad51 recombinase family (Fig. 1B). Gln-268 links L2 to beta strand 5 (β5), which forms part of the mostly parallel, twisted β-sheet scaffold of the RecA fold in the catalytic domain of RAD51 (Fig. 2A). A variety of biochemical data indicate that the glutamine at this position acts as an allosteric switch to communicate signals between the DNA and ATP binding sites of the enzyme. In the E. coli RecA-ssDNA-ADP-AlF4 structure, the equivalent residue (Gln-194) forms part of a hydrogen-bonded network that connects the γ-phosphate of ATP to L2 and ssDNA (6). Mutation of E. coli RecA Gln-194 to any other amino acid abolishes recombination and LexA repressor cleavage activities in vivo (39). It is therefore reasonable to hypothesize that Gln-268 in human RAD51 is involved in linking DNA binding to the ATPase and DNA strand exchange activities of the enzyme, and that the Q268P mutation might therefore affect these processes.
3.3. Thermal stability defect of RAD51 Q268P and Q272L variant proteins
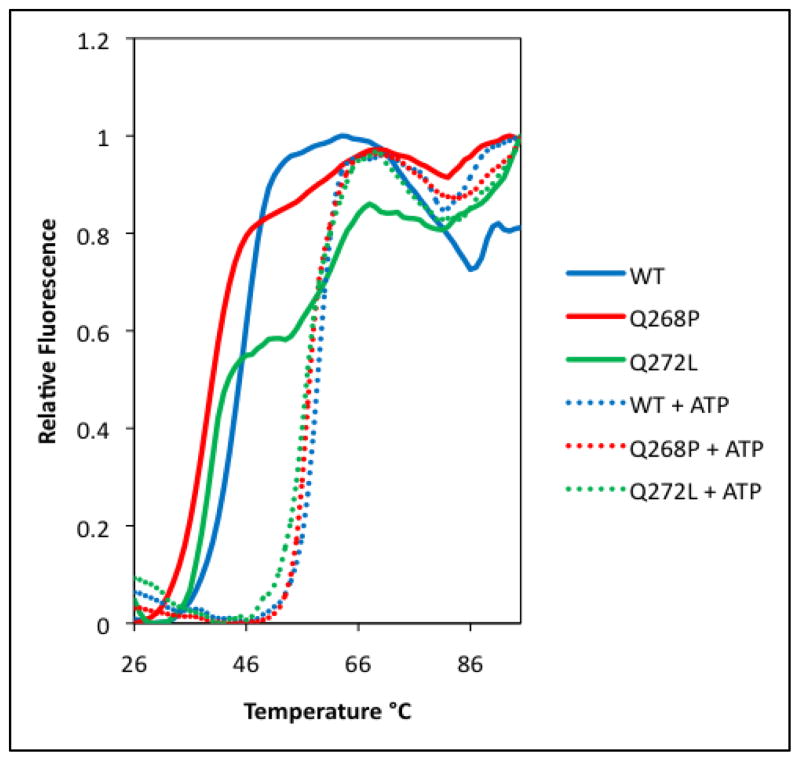
The thermofluor assay (30,31) was used to compare the thermal stabilities of human RAD51 Q268P and Q272L with that of WT. The fluorescence intensity of Sypro Orange dye is related to the hydrophobicity of its environment. Temperature-induced unfolding of RAD51 allows the dye to interact with the interior hydrophobic regions of the protein, yielding a melting curve from the increased fluorescence. The melting curves in Figure 3 show that in the absence of ATP, wild-type RAD51 has one distinguishable transition at 44.4±0.2°C. When ATP is present, the Tm is increased to 59.4±0.9°C. This suggests that the recombinase exists in two stable and distinguishable conformational states, with and without ATP, where the latter is more stable than the previous. This finding is consistent with previous observations that ATP binding alters the susceptibility of RAD51 to partial proteolysis (20). In the presence of ATP, the variants Q268P and Q272L exhibit melting temperatures similar to that of wild type, 57.5±0.2°C and 56.8±0.1°C respectively (Fig. 3). However their thermal profiles in the absence of ATP are quite different from that of wild-type; both appear to contain two melting transitions instead of one (Fig. 3). The first transition occurs at 38.7±0.2°C (Q268P) or 39.2±0.1°C (Q272L), representing a destabilization of >5°C compared to the ATP-free form of WT. The second transition of Q272L occurs at 58.2±3.1°C, which resembles the ATP-bound form of WT, even though in this case there is no ATP present. The second transition of Q268P occurs at 53.2±1.6°C, which is destabilized by ~6ºC compared to the ATP-bound form of WT. The presence of two transition states in a melting curve is indicative of a high degree of flexibility between two conformational states (31). This suggests that the protein dynamics of these two RAD51 variants are different from those of WT. With WT, the transition between the two conformations is ATP-dependent. In contrast, Q268P and Q272L appear to sample both conformational states without ATP. The addition of ATP captures the variants in the more stable conformation.
Figure 3. Thermal stabilities of RAD51 wild-type (WT), Q268P and Q272L variant proteins in the presence and absence of ATP.
Thermofluor assays to determine melting curves and Tm values (± standard deviation) were as described in Materials and Methods. Relative fluorescence is normalized to the highest signal in each curve. Solid lines represent melting curves in the absence of ATP for RAD51 WT (blue), Q268P (red), and Q272L (green), respectively. Dotted lines represent melting curves in the presence of 2 mM ATP for RAD51 WT (blue), Q268P (red), and Q272L (green), respectively.
3.4. DNA binding defect of Q268P and Q272L variant proteins
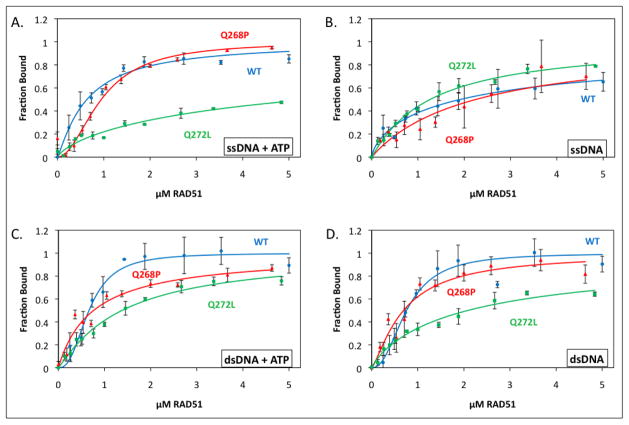
The DNA binding properties of Q268P and Q272L were investigated and compared to WT using two complementary assays: First, the interactions of RAD51 proteins with long, mixed-sequence ϕX174 ssDNA and dsDNA molecules were examined qualitatively by electrophoretic mobility shift assay (Fig. 4). Second, the affinities of RAD51 proteins for homopolymeric ssDNA and dsDNA olignucleotides were determined by monitoring the quenching of AlexaFluor 546 fluorescence that occurs upon recombinase binding to the fluor-conjugated oligos (29). The amplitude of fluorescence change is a measure of the fractional saturation of DNA with protein, which allows calculation of apparent Kd values from titration data (Fig. 5; Table 2).
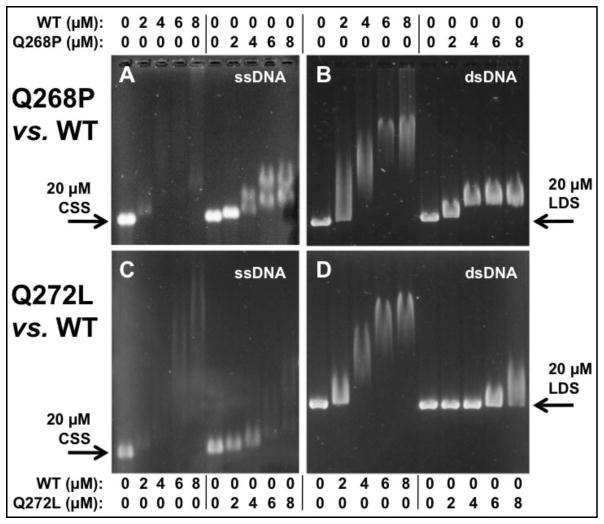
Figure 4. Electrophoretic mobility shift assays for the binding of RAD51 variant and wild-type proteins to ϕX174 ssDNA and dsDNA in the presence of 1 mM ATP.
(A–B) Concentrations of RAD51 WT or Q268P as indicated were incubated with 20 μM ϕX174 circular ssDNA (CSS) or linear dsDNA (LDS) then electrophoresed as described in Materials and Methods. (C–D) Binding reactions identical to Panels A–B except that RAD51 Q272L replaced Q268P.
Figure 5. Quantitative ssDNA and dsDNA binding data for RAD51 variants and wild-type in the presence and absence of ATP.
Binding of RAD51 protein to AlexaFluor 546-labeled ssDNA and dsDNA homopolymers was monitored by fluorescence quenching as described in Materials and Methods. Blue circles denote wild-type RAD51, red triangles denote Q268P, and green squares denote Q272L. (A) Protein binding to ssDNA in the presence of 1 mM ATP. (B) Protein binding to ssDNA in the absence of ATP. (C) Protein binding to dsDNA in the presence of 1 mM ATP. (D) Protein binding to dsDNA in the absence of ATP. Details of AF546-labeled ssDNA and dsDNA lattices plus other reaction conditions are described in Materials and Methods.
Table 2.
Apparent dissociation constants for DNA bindinga.
| Kd (nM) | RAD51-WT | RAD51-Q268P | RAD51-Q272L |
|---|---|---|---|
| ssDNA | 1900 ± 1400 | 2500 ± 2300 | 1300 ± 310 |
| ssDNA + ATP | 640 ± 85 | 950 ± 92 | N.D.b |
| dsDNA | 760 ± 92 | 690 ± 160 | 2100 ± 1300 |
| dsDNA + ATP | 660 ± 54 | 760 ± 280 | 1400 ± 360 |
Apparent Kd values were estimated from curve fits of data in Figure 4, as described in Materials and Methods.
Not Determined-- Measuring Kd required unattainably high protein concentrations.
EMSA data for the interactions of Q268P, Q272L, and WT with ϕX174 ssDNA in the presence of ATP are shown in Figure 4A,C. Both variants show large qualitative differences in their interactions with ssDNA compared to WT. As previously reported (20), RAD51 WT forms high and low mobility complexes with ssDNA, resulting in a bimodal smear pattern on non-denaturing electrophoretic gels as the ssDNA is saturated with protein (Fig. 4A). Q268P also forms high and low mobility complexes on ssDNA, but these complexes appear as relatively tight bands rather than smears, which migrate faster than the corresponding WT-ssDNA complexes (Fig. 4A). This result could indicate that Q268P has lower affinity for ssDNA than WT, but it could also reflect changes in filament morphology. Q272L complexes with ϕX174 ssDNA also exhibit faster electrophoretic mobility than the corresponding WT-ssDNA complexes (Fig. 4C), indicating lower affinity for ssDNA in the presence of ATP and/or differences in filament morphology.
Q268P and Q272L also exhibit large differences from WT in their interactions with ϕX174 dsDNA in the presence of ATP (Fig. 4B,D). The Q268P complex appears to saturate at the expected 1:3 ratio of protein to basepairs, however the saturated complexes migrate much faster than WT-dsDNA complexes (Fig. 4B), suggesting differences in filament morphology or stiffness (see Discussion). Q272L appears to interact weakly with ϕX174 dsDNA in the presence of ATP, since essentially no mobility shift is observed at protein concentrations less than 6 μM (Fig. 4D).
Quantitative analysis of protein-ssDNA interactions in the presence of ATP reveals that Q268P has slightly lower affinity for ssDNA than WT (45% increase in apparent Kd), and that Q272L has much lower affinity for ssDNA than WT (Fig. 5A, Table 2). The precise change in the apparent Kd for Q272L versus WT could not be determined because unattainably high concentrations of the former were required to saturate the ssDNA under these conditions. These results appear to be consistent with the EMSA data in Figure 4A,C, and suggest that reduced affinity for ssDNA in the presence of ATP may account for at least part of the reduced electrophoretic mobility shift of Q268P- and Q272L-ssDNA complexes compared to WT complexes. The affinities of both WT and Q268P for ssDNA were markedly lower in the absence of ATP (Fig. 5B, Table 2), although the apparent Kd values also have large errors because of the difficulty in saturating the ssDNA. The opposite was observed with Q272L, which had a lower apparent Kd for ssDNA in the absence of ATP than in its presence (Fig. 5B, Table 2). As a result, the ssDNA binding curves for all three protein variants are similar in Figure 5B. Therefore the most significant impacts of the Q268P and Q272L mutations on ssDNA binding activity occur in the presence of ATP.
Quantitative analysis demonstrates that Q272L has weaker affinity for dsDNA than WT in both the presence and absence of ATP (Fig. 5C,D; Table 2), which is consistent with the EMSA results in Figure 4D. In contrast, Q268P shows no significant differences in affinity for dsDNA compared to WT in the quantitative fluorescence assay (Fig. 4C–D, Table 2). This result along with the EMSA data (Fig. 4) suggests that an important effect of the Q268P mutation is to alter the morphology of RAD51 filaments on either dsDNA or ssDNA, an effect that is observed even in the presence of ATP.
Details of the quantitative DNA binding data in Figure 5 indicate some additional differences between the tumor variant proteins and wild-type RAD51. The binding of Q268P to ssDNA in the presence of ATP appears to be sigmoidal (Fig. 5A), suggesting either an increase in cooperativity for this variant compared to WT, or that Q268P exhibits more than one binding mode for the fluorescent oligo. On the other hand, Q268P and Q272L both exhibit loss of sigmoidicity compared to WT when binding to dsDNA ± ATP (Fig. 5C–D).
3.5. ATPase defect of Q268P and Q272L variant proteins
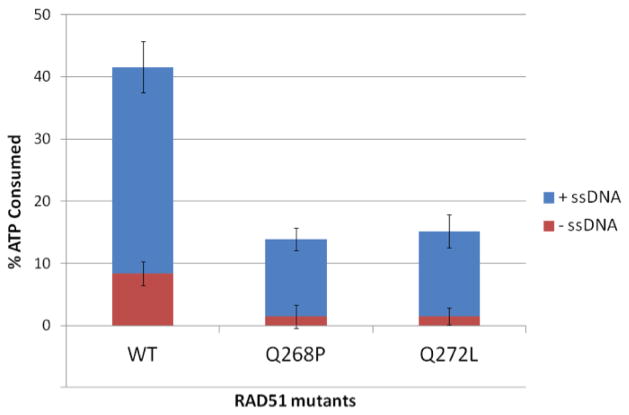
In addition to altered DNA binding properties, both Q268P and Q272L variants exhibit enzymatic deficiencies compared to wild-type. Wild-type RAD51 protein has an intrinsic ATPase activity; at saturating ATP concentration, the addition of a 2-fold excess of ssDNA (assuming a binding site size of 3 nucleotide residues per protein monomer) stimulates this activity by approximately 4-fold (Fig. 6). Both Q268P and Q272L exhibit very low levels of intrinsic ATPase activity that are reduced by greater than 4-fold compared to WT (Fig. 6). The ATPase activities of both variants are stimulated by ssDNA, but the ssDNA-stimulated activity levels are still ~2.5-fold lower than that observed for WT when the same amount of ssDNA is added (Fig. 6). The deficiencies in ssDNA-stimulated ATPase activities of the variants may be due to their lower intrinsic activities, to changes in their interactions with ssDNA, or both.
Figure 6. Intrinsic and ssDNA-stimulated ATPase activities of RAD51 wild-type, Q268P, and Q272L proteins.
ATPase assays were carried out as described in Materials and Methods. Red blocks represent intrinsic ATPase activities in the absence of ssDNA. Blue blocks represent activities observed upon addition of a 2-fold excess of ϕX174 ssDNA (assuming 3 nucleotide residues per monomer).
3.6. DNA strand exchange defect of Q268P and Q272L variant proteins
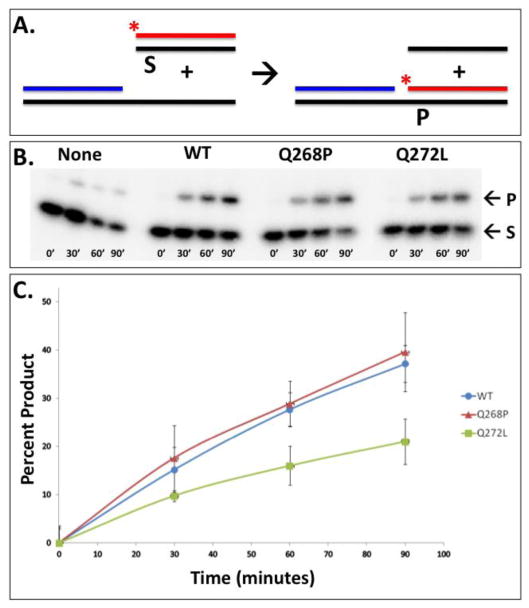
The intrinsic recombination activities of the variants were measured under optimized conditions (30ºC, absence of RPA protein and mediator proteins, oligonucleotide substrates lacking stable ssDNA secondary structure) as shown schematically in Figure 7A. Results demonstrate that Q268P and Q272L both retain intrinsic recombination activity (Fig. 7B,C). Q268P catalyzes this reaction at approximately the same rate as WT (Fig. 7C). Q272L catalyzes this reaction at a 2-fold lower rate than WT (Fig. 7C).
Figure 7. Intrinsic DNA strand exchange activities of RAD51 WT, Q268P, and Q272L proteins with oligonucleotide substrates.
(A) In the presence of RAD51 and ATP, radiolabeled homologous duplex (S) and tailed duplex substrates, are converted into heteroduplex (P) and ssDNA products. Tailed duplex (63/32-mer) was constructed from oligos 5/6, and homologous duplex (31/31-mer) was constructed from oligos 7/8 (Table 1). (B) Phosphorimage of non-denaturing PAGE reactions contained no protein (“None”; left), RAD51 WT (center left), Q268P (center right), or Q272L (right). (C) Quantified results of reactions shown in Panel B; error bars representing results from 3 identical experiments. The background rate from the no-protein control reaction (“None” in Panel B) was subtracted from all data.
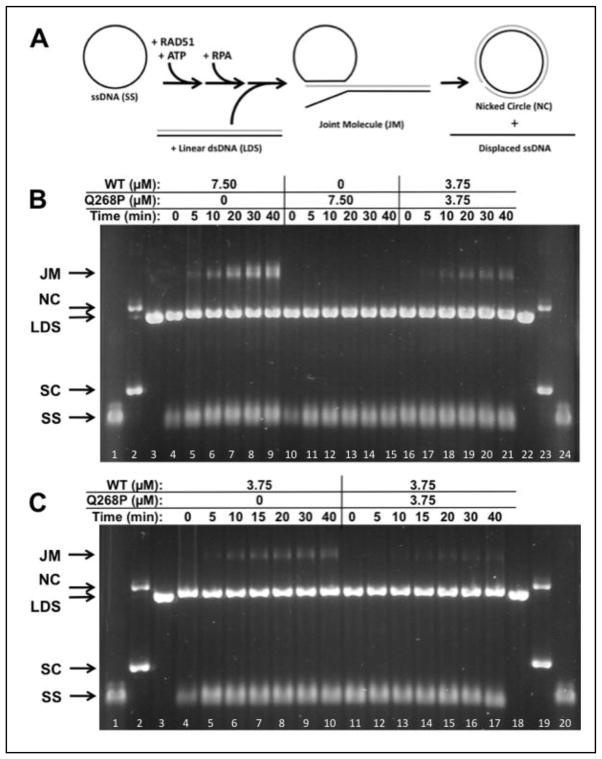
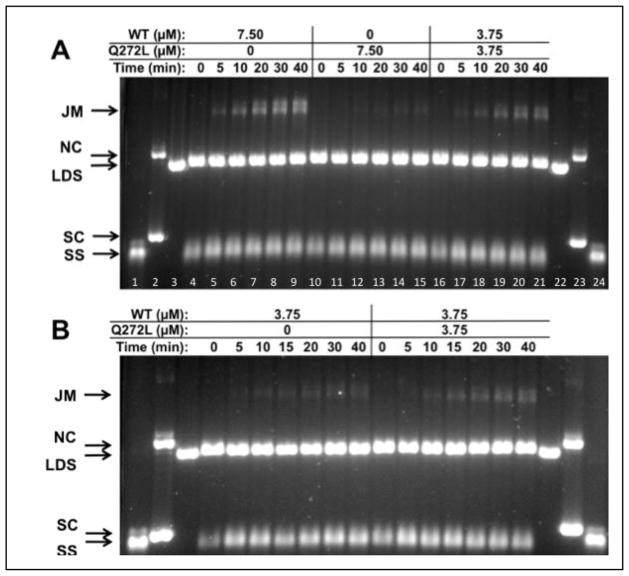
A more stringent test of recombination activity is the strand exchange assay with full-length (5.4 kb/kbp) ϕX174 DNA substrates, a schematic of which is shown in Figure 8A. Reactions with RAD51 WT protein require human RPA protein, which must be added to reaction mixtures after pre-incubation of RAD51 with ssDNA and ATP at 37ºC, and prior to the addition of homologous dsDNA to start the reaction. The reaction with 7.50 μM RAD51 WT protein generates joint molecule (JM) products that increase steadily over a timecourse of 40 min (Fig. 8B, left). The equivalent reaction with 7.50 μM Q268P generates no JM products over the same time period (Fig. 8B, center). Therefore Q268P protein lacks DNA strand exchange activity under stringent conditions of DNA length and RPA presence.
Figure 8. DNA strand exchange activity of RAD51 Q268P versus WT with homologous ϕX174 DNA substrates and RPA protein.
(A) Reaction schematic. See Materials and Methods for details. (B) Timecourses of reactions containing a total of 7.50 μM RAD51 protein (WT and/or Q268P) at the indicated concentrations. ssDNA (SS), dsDNA (LDS), nicked (NC), and supercoiled (SC) ϕX174 DNA markers (lanes 1–3, 22–24) and joint molecule (JM) products are indicated next to the gel. Lanes 4–9: Reaction with WT only; Lanes 10–15: Reaction with Q268P only; Lanes 16–21: Reaction with 1:1 mixture of WT and Q268P. (C) Timecourses of reactions containing 3.75 μM WT RAD51 in the absence/presence of equimolar Q268P as indicated. Lanes 1–3, 18–20: Markers. Lanes 4–10: Reaction with WT only; Lanes 11–17: Reaction with mixture of WT and Q268P. All other experimental conditions were as described in Materials and Methods.
Unlike Q268P, the Q272L variant does promote a small amount of DNA strand exchange when it is present at 7.50 μM concentration (Fig. 9A, middle), but the rate of joint molecule production is severely reduced compared to the rate seen with 7.50 μM RAD51 WT (Fig. 9A, left). Therefore the Q272L mutation severely reduces but does not eliminate DNA strand exchange under stringent conditions of DNA length and RPA presence.
Figure 9. DNA strand exchange activity of RAD51 Q272L versus WT with homologous ϕX174 DNA substrates and RPA protein.
Reactions were carried out as described in Materials and Methods and as shown schematically in Figure 8A. (A) Timecourses of reactions containing a total of 7.50 μM RAD51 protein (WT and/or Q272L) at the indicated concentrations. The positions of SS, LDS, NC, and SC (supercoiled ϕX174 DNA) markers (lanes 1–3, 22–24) are indicated next to the gel, along with the position of JM products. Lanes 4–9: Reaction with WT only; Lanes 10–15: Reaction with Q272L only; Lanes 16–21: Reaction with 1:1 mixture of WT and Q272L. (B) Timecourses of reactions containing 3.75 μM WT RAD51 in the absence/presence of equimolar Q272L as indicated. Lanes 1–3, 18–20: Markers. Lanes 4–10: Reaction with WT only; Lanes 11–17: Reaction with mixture of WT and Q272L.
3.7. Reduced strand exchange activity in reactions with mixtures of wild-type and variant RAD51 proteins
A DNA strand exchange reaction containing equimolar Q268P and WT (3.75μM each, 7.50 μM total enzyme concentration) proceeded at a detectable but greatly reduced rate compared to the reaction with 7.50 μM WT alone (Fig. 8B). 7.50 μM RAD51 is sufficient to saturate all of the input ssDNA in the reaction, while 3.75 μM RAD51 is sub-saturating given a binding site size of 3 nucleotides per monomer. This result indicates that the inactive Q268P variant cannot be “activated” through incorporation into mixed complexes with RAD51 WT on ssDNA. The subsaturating WT concentration (3.75 μM) itself reduces the rate of DNA strand exchange (Fig. 8C, left; compare with Fig. 8B, left), but to a lesser extent than occurs with the equimolar mixture (3.75 μM each) of Q268P and WT (Fig. 8C, right). Therefore the presence of Q268P in the reaction actually inhibits the DNA strand exchange activity of WT.
A DNA strand exchange reaction containing equimolar Q272L and WT (3.75 μM each, 7.50 μM total enzyme concentration) also proceeded at a reduced rate compared to the reaction with 7.50 μM WT alone (Fig. 9A). Thus, like Q268P, the Q272L variant cannot be “activated” by co-occupying ssDNA with WT enzyme. Unlike Q268P, however, Q272 does not appear to inhibit WT RAD51. This is illustrated by the data in Figure 9B, where the rate of strand exchange in a mixture of 3.75 μM Q272L plus 3.75 μM WT (Fig. 9B, right) exceeds that of the reaction with 3.75 μM WT alone (Fig. 9B, left), suggesting that the Q272L and WT rates are additive.
4. Discussion
Tumor-derived variants Q268P and Q272L affect the DNA binding loop 2 (L2) region of RAD51 protein, and both exhibit strongly aberrant biochemical properties compared to the wild-type enzyme. Both variants exhibit defects in thermal stability, DNA binding activity, and ATPase activity, and these defects contribute to severe defects in DNA strand exchange activity under stringent conditions. Mixtures of either variant with wild-type also exhibit reduced DNA strand exchange activity compared to wild-type alone, suggesting that heterozygous expression of either variant could lead to recombination defects in cells.
The thermal stability data (Fig. 3) indicate that in wild-type RAD51 the transition from a thermolabile to a thermostable conformation is tightly coupled to ATP binding. However in Q268P and Q272L both conformations (or similar ones) appear to exist in equilibrium in the absence of ATP, as evidenced by the biphasic melting curves in Figure 3. Furthermore the thermolabile forms of both variants have significantly lower Tm values than WT. Keeping this in mind, it is interesting that the intrinsic ATPase activities of both variants are depressed to levels that are barely detectable above background, and that even the ssDNA-stimulated ATPase activities are reduced 2.5-fold in the variants compared to wild-type (Fig. 6). These results suggest that the conformational change necessary to activate ATP hydrolysis is inefficient in Q268P and Q272L, which could be related to their relatively unstable protein folds compared to WT.
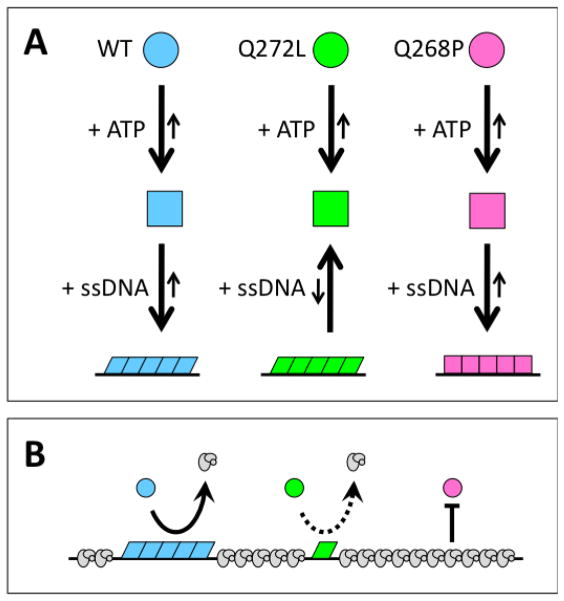
Both Q268P and Q272L variants exhibit intrinsic DNA strand exchange activity under highly optimized conditions, (lower temperature (30ºC), absence of RPA protein, oligonucleotide ssDNA and dsDNA substrates designed to minimize ssDNA secondary structure) (Fig. 7). The intrinsic strand exchange activity of Q268P is indistinguishable from WT, but Q272L has 2-fold lower activity under the same conditions (Fig. 7). However both variants exhibit severe defects in DNA strand exchange reactions performed under more stringent conditions (higher temperature (37ºC), presence of RPA protein, ϕX174 DNA substrates) (Figs. 8–9). Q272L retains a trace of DNA strand exchange activity under these conditions, while Q268P appears to be completely defective. The properties of Q272L and Q268P suggest that the strand exchange defects of both variants may be caused by their inabilities to form stable or competent presynaptic filaments on ssDNA under stringent conditions. This model is described schematically in Figure 10.
Figure 10. Model for the effects of RAD51 Q272L and Q268P mutations on ssDNA binding and RPA displacement.
The ligand-free forms of RAD51 wild-type, Q272L, and Q268P proteins are indicated by blue, green, and pink circular symbols, respectively. (A) Proposed defects in presynaptic filament assembly. ATP binding induces similar conformational changes in all three RAD51 species, as indicated by the square symbols. These ATP-bound forms have different ssDNA binding properties, however. Wild-type RAD51 interacts strongly with ssDNA and forms competent presynaptic filaments in the extended conformation, as indicated by the blue parallelograms. Q272L interacts weakly with ssDNA and forms competent but unstable presynaptic filaments, as indicated by the green parallelograms. Q268P retains relatively strong interactions with ssDNA but forms filaments that do not adopt a competent conformation, as indicated by the pink squares. (B) Proposed defects in RPA displacement. RPA heterotrimers are shown in gray. In the presence of ATP, wild-type RAD51 displaces RPA from ssDNA to form a competent presynaptic filament. Q272L does this very inefficiently because of its low affinity for ssDNA. Q268P fails to displace RPA because it cannot adopt a competent filament conformation. See text for further discussion.
The Q272L variant is a weak DNA binder (Fig. 10A) based on its relatively inefficient saturation of ϕX174 ssDNA and dsDNA molecules in the presence of ATP (Fig. 4C,D), and on its elevated apparent Kd values for both ssDNA and dsDNA oligonucleotides in the presence of ATP (Fig. 5, Table 2). The decrease in DNA binding affinity associated with Q272L is consistent with the position of Gln-272 within L2 and with the known contribution of this loop to the DNA binding surface of RecA/Rad51-family recombinases (3,6). Since the Q272L mutation substitutes a non-polar residue for a polar one, the results of this study demonstrate that hydrogen bonding potential in the side chain of residue 272 is necessary for normal, high-affinity interactions of RAD51 with DNA.
In contrast, the Q268P variant exhibits only a modest increase in apparent Kd for an ssDNA oligo in the presence of ATP compared to WT, and its apparent Kd for a dsDNA oligo in the presence of ATP is unchanged compared to WT (Fig. 5, Table 2). Q268P exhibits different apparent cooperativities or binding modes for both ssDNA and dsDNA compared to WT, however (Fig. 5). In addition, Q268P complexes on ϕX174 ssDNA and dsDNA in the presence of ATP exhibit strikingly faster electrophoretic mobility than the corresponding complexes with WT RAD51 (Fig. 4A,B). The Q268P mutation does not alter the surface charge or isoelectric point of RAD51 protein, therefore the rapid electrophoretic mobility of Q268P-DNA filaments is best explained by a mutation-induced change in filament morphology (Fig. 10A), perhaps to a less stiff/more flexible conformation that would more easily pass through the gel matrix under electromotive force. This conclusion is consistent with the postulated function of residue Gln-268, based on conservation with E. coli RecA (6,39), as an allosteric signaling residue between the ATP and DNA binding sites of RAD51. The Q268P mutation would be predicted to disrupt this network of hydrogen bonds, which may explain the failure of Q268P-ssDNA complexes to adopt the correct conformation in the presence of ATP.
The low affinity of Q272L for ssDNA may prevent this variant from displacing RPA protein (Fig. 10B), a necessary step in presynaptic filament assembly (3), and/or from maintaining stable filaments on longer stretches of ϕX174 ssDNA that are rich in secondary structure. Similarly, Q268P may fail to form competent presynaptic filaments under stringent conditions due to incorrect filament morphology (Fig. 10B). Evidence suggests that the presynaptic displacement of ssDNA-binding proteins from ssDNA requires ATP-linked conformational changes and/or ATP hydrolysis by the recombinase (40–42). Furthermore the ATP binding and hydrolytic cycle is linked to DNA stretching and the formation of an extended conformation of the RAD51-ssDNA filament (3–5). Data from the current study indicate that Q268P is an inefficient ATPase and that it forms abnormal, rapidly migrating complexes on DNA (Fig. 4). This could reflect an inability of Q268P-ssDNA filaments to adopt the extended conformation, which could in turn prevent Q268P from displacing RPA and forming an active presynaptic filament on ssDNA (Fig. 10B).
An alternative explanation for the DNA strand exchange defects of Q268P and Q272L under stringent conditions is that their filaments on ϕX174 ssDNA might be defective in promoting homologous pairing with ϕX174 dsDNA, due to the length and complexity of the DNA sequences, low affinity for dsDNA (in the case of Q272L), or inappropriate filament conformation (in the case of Q268P). A defect in homologous pairing would not rule out defects in the presynaptic phase of recombination as discussed above, and indeed defects in both phases could be linked.
Previous studies established that RAD51 missense variants D149N, R150Q, and G151D can interact with WT protein and form mixed filaments with intermediate properties on ssDNA or dsDNA (20). Results of the current study demonstrate that when presynaptic filaments are assembled from a 1/1 mixture of WT and variant proteins and a stoichiometric amount of ϕX174 ssDNA (w.r.t. potential RAD51 binding sites), a reduced rate of DNA strand exchange occurs compared to otherwise identical reactions containing WT protein alone (Figs. 8B and 9A). The two most logical explanations for these results, which are not necessarily mutually exclusive, are the following: (1) That the Q268P and Q272L variants also form mixed filaments with WT that have reduced DNA strand exchange activity; or (2) That the ssDNA is under-saturated with RAD51 protein due to deficiencies of the Q268P and Q272L variants in ssDNA binding and/or RPA displacement. There is reason to speculate that the first mechanism may apply to Q268P, since its presence appears to inhibit the DNA strand exchange activity of WT enzyme (Fig. 8C), and since its ssDNA binding affinity remains relatively high as discussed above. This raises the possibility of a dominant negative effect of Q268P in vivo, through integration with WT into hybrid presynaptic filaments. In contrast, the residual DNA strand exchange activity of Q272L appears to be additive with that of WT enzyme (Fig. 9B), which could be consistent with either mechanism.
Given the biochemical defects of the tumor-associated Q268P and Q272L variants described in this study, it seems highly probable that these mutations would interfere with homology-directed repair of DNA double-strand breaks and other lesions in vivo. The biochemistry predicts a detrimental effect of either variant on HDR even if it is expressed in a heterozygous state with wild-type RAD51. Therefore hypomorphic RAD51 variants join a list of conditions including BRCA1, BRCA2, and PALB2 mutations and altered expression of RAD51 that may compromise the HDR pathway. Since HDR is a desirable target for chemotherapeutic approaches in the treatment of cancer, further research on somatic and germline variants of RAD51 protein is clearly warranted.
Highlights.
RAD51 tumor variants Q268P and Q272L occur in the DNA binding loop 2 (L2) region.
Variants show abnormal thermal stability, DNA binding, and ATPase activities.
Variants and variant/wild-type mixtures are defective in DNA strand exchange.
Results provide new insights on roles of conserved L2 residues in RAD51 function.
Results suggest Q268P and Q272L could be drivers of genome instability in vivo.
Acknowledgments
This work was supported by National Institutes of Health research grant no. P01CA098993/P3 to SWM, and by an Internal Grant Program award to SWM from the University of Vermont College of Medicine. The sponsors had no involvement in study design, the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
Abbreviations
- HR
homologous recombination
- HDR
homology-directed (DNA) repair
- DSB
DNA double-strand break
- dsDNA
double-stranded DNA
- ssDNA
single-stranded DNA
- SSB
ssDNA binding protein
- WT
wild-type
Footnotes
Conflicts of interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Morrical S. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harbor Perspect Biol. 2015;7:a016444. doi: 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Ehmsen KT, Heyer WD, Morrical SW. Presynaptic filament dynamics in homologous recombination and DNA repair. Crit Rev Biochem Mol Biol. 2011;46:240–270. doi: 10.3109/10409238.2011.576007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanLoock MS, Yu X, Yang S, Lai AL, Low C, Campbell MJ, Egelman EH. ATP-mediated conformational changes in the RecA filament. Structure. 2003;11:187–196. doi: 10.1016/s0969-2126(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 7.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 8.Amunugama R, Fishel R. Homologous recombination in eukaryotes. Prog Mol Biol Transl Sci. 2012;110:155–206. doi: 10.1016/B978-0-12-387665-2.00007-9. [DOI] [PubMed] [Google Scholar]
- 9.Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- 10.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Krüger S, Feller AC, Lopens A, Diedrich K, Schwinger E, Stürzbecher HW. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair (Amst) 2002;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maacke H, Hundertmark C, Miska S, Voss M, Kalthoff H, Stürzbecher HW. Autoantibodies in sera of pancreatic cancer patients identify recombination factor Rad51 as a tumour-associated antigen. J Cancer Res Clin Oncol. 2002;128:219–222. doi: 10.1007/s00432-001-0321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson CA, Ramos L, Villaseñor MR, Anders KH, Press MF, Clarke K, Karlan B, Chen JJ, Scully R, Livingston D, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa K, Honda K, Inamoto T, Shinohara H, Yamauchi A, Suga K, Okuyama T, Shimada T, Kodama H, Noguchi S, et al. Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss in BRCA1-associated cases. Clin Cancer Res. 1999;5:1249–1261. [PubMed] [Google Scholar]
- 15.Yoshikawa K, Ogawa T, Baer R, Hemmi H, Honda K, Yamauchi A, Inamoto T, Ko K, Yazumi S, Motoda H, et al. Abnormal expression of BRCA1 and BRCA1-interactive DNA-repair proteins in breast carcinomas. Int J Cancer. 2000;88:28–36. [PubMed] [Google Scholar]
- 16.Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet. 1995;9:444–450. doi: 10.1038/ng0495-444. [DOI] [PubMed] [Google Scholar]
- 17.Collis SJ, Tighe A, Scott SD, Roberts SA, Hendry JH, Margison GP. Ribozyme minigene-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res. 2001;29:1534–1538. doi: 10.1093/nar/29.7.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohmori T, Yang JL, Price JO, Arteaga CL. Blockade of tumor cell transforming growth factor-betas enhances cell cycle progression and sensitizes human breast carcinoma cells to cytotoxic chemotherapy. Exp Cell Res. 1998;245:350–359. doi: 10.1006/excr.1998.4261. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Yano K, Matsuo F, Saito H, Katagiri T, Kurumizaka H, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, et al. Identification of Rad51 alteration in patients with bilateral breast cancer. J Hum Genet. 2000;45:133–137. doi: 10.1007/s100380050199. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Morrical MD, Donigan KA, Weidhaas JB, Sweasy JB, Averill AM, Tomczak JA, Morrical SW. Tumor-associated mutations in a conserved structural motif alter physical and biochemical properties of human RAD51 recombinase. Nucleic Acids Res. 2015;43:1098–1111. doi: 10.1093/nar/gku1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida T, Takizawa Y, Sakane I, Kurumizaka H. Altered DNA binding by the human Rad51-R150Q mutant found in breast cancer patients. Biol Pharm Bull. 2007;30:1374–1378. doi: 10.1248/bpb.30.1374. [DOI] [PubMed] [Google Scholar]
- 22.Richardson C. RAD51, genomic stability, and tumorigenesis. Cancer Lett. 2005;218:127–139. doi: 10.1016/j.canlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 23.Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic binding site in p53. J Biol Chem. 2005;280:8051–8059. doi: 10.1074/jbc.M411176200. [DOI] [PubMed] [Google Scholar]
- 24.Yoon D, Wang Y, Stapleford K, Wiesmüller L, Chen J. P53 inhibits strand exchange and replication fork regression promoted by human Rad51. J Mol Biol. 2004;336:639–654. doi: 10.1016/j.jmb.2003.12.050. [DOI] [PubMed] [Google Scholar]
- 25.Bucchop S, Gibson MK, Wang XW, Wagner P, Sturzbecher HW, Harris CC. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 1997;25:3868–3874. doi: 10.1093/nar/25.19.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Miniatis T. Molecular cloning: A Laboratory Manual. Cold spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 27.Binz SK, Dickson AM, Haring SJ, Wold MS. Functional assays for replication protein A (RPA) Methods Enzymol. 2006;409:11–38. doi: 10.1016/S0076-6879(05)09002-6. [DOI] [PubMed] [Google Scholar]
- 28.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. Erratum in: J. Biol. Chem. 269, 16519. [PubMed] [Google Scholar]
- 29.Maher RL, Morrical SW. Coordinated binding of single-stranded and double-stranded DNA by UvsX recombinase. PLoS One. 2013;8:e66654. doi: 10.1371/journal.pone.0066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Fang J, Rand KD, Silva MC, Wales TE, Engen JR, Beuning PJ. Conformational dynamics of the Escherichia coli DNA polymerase manager proteins UmuD and UmuD'. J Mol Biol. 2010;398:40–53. doi: 10.1016/j.jmb.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Villanueva N, Rould MA, Morrical SW. Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant. Nucleic Acids Res. 2010;38:4889–4906. doi: 10.1093/nar/gkq209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway AB, Lynch TW, Zhang GS, Fortin CW, Fung LS, Symington PA, Rice PA. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 39.Kelley JA, Knight KL. Allosteric regulation of RecA protein function is mediated by Gln194. J Biol Chem. 1997;272:25778–25782. doi: 10.1074/jbc.272.41.25778. [DOI] [PubMed] [Google Scholar]
- 40.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 41.Fung CW, Fortin GS, Peterson SE, Symington LS. The rad51-K191R ATPase-defective mutant is impaired for presynaptic filament formation. Mol Cell Biol. 2006;26:9544–9554. doi: 10.1128/MCB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, Berger CL, Morrical SW. Kinetics of presynaptic filament assembly in the presence of single-stranded DNA binding protein and recombination mediator protein. Biochemistry. 2013;52:7878–7889. doi: 10.1021/bi401060p. [DOI] [PMC free article] [PubMed] [Google Scholar]