Abstract
Cellular RNA binding proteins incorporated into virions during HIV-1 assembly promote the replication efficiency of progeny virions. Despite its critical role in bolstering virion infectivity, the molecular basis for incorporation of DHX9/RNA helicase A (RHA) to virions remains unclear. Here, cell-based experiments demonstrate truncation of segments of the HIV-1 5′-untranslated region (5′-UTR) that are distinct from the core encapsidation sequence, eliminated virion incorporation of RHA, indicating that RHA recruitment is mediated by specific interactions with the HIV-1 5′-UTR. In agreement with biological data, isothermal titration calorimetry determined the dimer conformation of the 5′-UTR binds one RHA molecule per RNA strand, and the interaction is independent of nucleocapsid protein binding. Nuclear magnetic resonance spectra employing a deuterium-labeling approach enabled resolution of the dimeric 5′-UTR in complex with the RHA N-terminal domain. The structure of the large molecular mass complex was dependent on RHA binding to a double-stranded region of the primer binding site (PBS)-segment of the 5′-UTR. A single A to C substitution was sufficient to disrupt biophysical conformation and attenuate virion infectivity in cell-based assays. Taken together, our studies demonstrate the structural basis for HIV-1 genomic RNA to recruit beneficial cellular cofactor to virions. The support of progeny virion infectivity by RHA is attributable to structure-dependent binding at the PBS-segment of HIV-1 5′-UTR during virus assembly.
Keywords: Dimeric 5′, UTR, NMR, Virus infectivity, RNA binding domain, PBS-segment
Graphical abstract
INTRODUCTION
HIV-1 is a retrovirus absolutely dependent on host RNA binding proteins (RBPs) to sponsor the steps in biogenesis of infectious virions. Investigations of necessary human RBPs have identified DHX9/RNA helicase A (RHA) amongst beneficial host co-factors [1, 2]. RHA activity originates in the virus-producing cell, albeit late in the virus replication cycle, and regulates post-transcriptional fate of the proviral RNA [3–8].
Proteomic studies of HIV-1 virions and tandem affinity purification of HIV-1 Gag binding proteins led to the discovery that RHA is a component of virions [5, 9]. Downregulation of RHA in host cells does not affect dimerization or packaging of virion genomic RNA (gRNA) [5], but reduced infectivity by a factor of 2–3 in cultured cells [5]. Similar global infectivity loss was also observed in infection assays with primary cells [10]. More specifically, the RHA-deficient virions were less efficient in reverse transcription due to deficient primer extension [5, 7]. Exogenously expressed RHA can be recruited to HIV-1 virions and rescue downregulation of endogenous RHA by siRNAs. Functional rescue was not observed when either of the two dsRNA binding domains (dsRBDs) were missing in the exogenous RHA [8], consistent with necessity for the amino-terminal RNA binding domain, as shown previously in study of another retroviral RNA [11].
Subsequently, the in virio stoichiometry of RHA was demonstrated to be proportional to the diploid genomic RNA (1:1) [12], and diminishing RHA in virion producer cells diminished the infectivity of progeny virions on primary cells [10]. The findings implicated RHA interaction with HIV-1 to be a therapeutic target for disabling HIV-1 replication and attenuating HIV-1 pathogenesis in patients. Previous studies have shown that RHA associates with Gag in the presence of gRNA [5], but the molecular details for RHA recruitment into virions remain an important open issue to resolve.
RHA is a multi-domain member of the DExH-box superfamily 2 proteins that facilitates virus infections and plays an emerging role in the innate response [2]. As a ubiquitous RBP, RHA can associate with nascent transcripts undergoing transcription by RNA polymerase II [13], and facilitate cytoplasmic translation of select viral and cellular mRNAs [6]. Prior studies demonstrated the amino-terminal domain is necessary for recognition of cognate RNAs at the 5′-untranslated region (5′-UTR) and binding specificity is attributable to the two dsRBDs (Fig. 1a), whereas other domains of RHA are insufficient for selective interaction with cognate mRNA [11]. Furthermore, the accumulation of RHA in virions was diminished for vector RNAs containing truncations in the 5′-UTR [5].
Fig. 1.
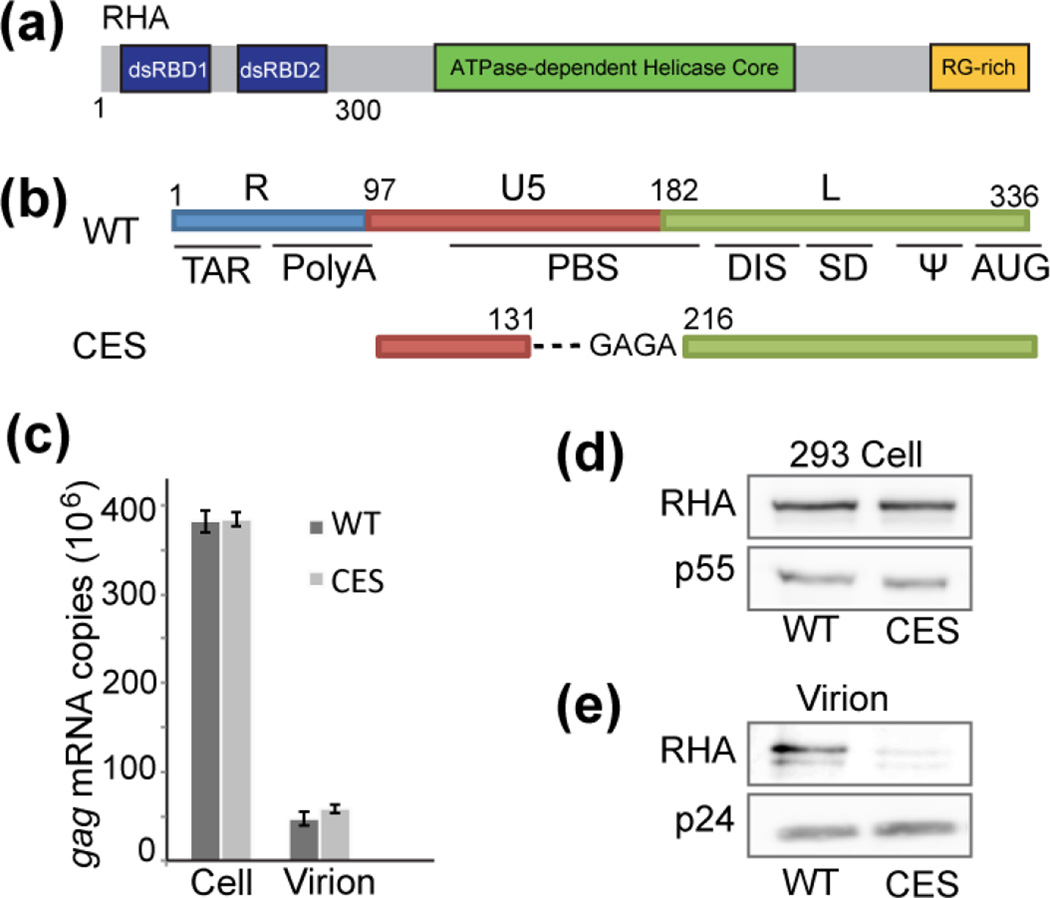
RHA assembles with gRNA containing WT 5′-UTR but not CES. (a) Schematic representation of RHA, which contains two dsRBDs in the N-terminal domain (RHAN300). (b) Schematic representation of the HIV-1 5′-UTR WT and CES in HIV-1 molecular clones that were evaluated for RHA recruitment to assembling virions. HEK293 cells were transfected with either molecular clone for 48 h followed by collection of cell lysates and cell-free supernatant media. Virions were collected by centrifugation over a 20% sucrose pad and quantified by Gag ELISA. (c) The abundance of HIV-1 gRNA in the cell and packaged into equivalent virions was quantified by RT-qPCR with gag-specific primers. (d) Cell-associated RHA and Gag p55 were visualized by Western blot. (e) Equivalent Gag units were subjected to SDS-PAGE and virion-associated RHA and Gag p24 were visualized by Western blot. The results are representative of three independent experiments.
The HIV-1 5′-UTR coordinates many essential events during early and late stages in virus replication, including proviral RNA synthesis, splicing and translation to viral structural proteins. The 5′-UTR contains sequence elements necessary for dimerization of virion precursor RNA and packaging of genomic ribonucleoprotein particles (gRNP) into virions, and provides template to initiate reverse transcription in subsequent infection (see review in [14]). Recent nuclear magnetic resonance (NMR) studies suggest that the 5′-UTR coordinates gRNA dimerization with other viral functions by means of a monomer-dimer RNA switch [15]. In the monomeric 5′-UTR, residues spanning the gag start codon (AUG) form a hairpin while the GC-rich dimer-promoting sequence (DIS) is sequestered by base pairing with the Unique-5 region (U5). Alternatively, the dimer conformation of 5′-UTR places AUG and U5 in the long-range interaction, which exposes DIS for inter-strand dimerization that exposes high-affinity binding sites for the nucleocapsid (NC) domain of the Gag precursor protein, which is necessary to package gRNA during virion assembly [15]. The residues between U5 and AUG have been identified as the core encapsidation signal (CES, Fig. 1b), whereas TAR, Poly(A), and PBS-segment are dispensable to this process [16, 17]. Thus, the previously reported RHA deficiency in defective virions can be explained by two possible hypotheses: either RHA is incorporated to gRNP with RNA elements in the dimeric 5′-UTR, or it assembles with other RNA elements, but the gRNA level in virion was drastically reduced due to the lack of packaging signal, CES.
In this study, we report that CES efficiently directs gRNA packaging, but fails to recruit RHA during virion assembly, indicating that cell-endogenous RHA is recruited by nonessential packaging elements in the 5′-UTR. By combining cell-based approaches with biophysical measurements, we demonstrate the N-terminal domain of RHA binds within the PBS-segment of the dimeric 5′-UTR. Our effort identifying key determinants for virus incorporation of RHA provides insights into the gRNA-mediated recruitment of beneficial host co-factor during virus assembly.
RESULTS
RHA is recruited by RNA elements in the 5′-UTR
Functional recruitment of RHA during the HIV-1 virus assembly is required to support virion infectivity, as RHA-deficient virions have diminished infectivity [5, 10]. The Kleiman group reported that the amino-terminal dsRBD1 and dsRBD2 are necessary for RHA incorporation to virions, whereas activity of the core helicase domain is not necessary [7, 8]. Similar cognate RNA binding activity of RHA dsRBDs has been reported and demonstrated lysine to alanine mutations in dsRBDs abolished RHA binding to retroviral and junD 5′-UTRs; moreover, exogenously expressed dsRBDs inhibited the translation-modulation function of endogenous RHA [11]. The requirement for the N-terminal dsRBDs during virus assembly suggests RHA directly interacts with gRNA, very likely through recognition of structured RNA elements.
In previous work, reduced levels of RHA in viral particles were observed when packaging of gRNA was impaired [5], indicating a gRNA-mediated host factor recruitment mechanism. Cell-based immunoprecipitation assay detected RHA co-precipitating two RNA regions in the HIV-1 gRNA: the 5′-UTR and the Rev Response element (RRE) [8]. While Rev/RRE activity is known to transactivate nuclear export of unspliced virion precursor RNA [4], the 5′-UTR coordinates many essential viral functions and is necessary to package gRNA [14]. Thus we hypothesized that RHA assembles with the 5′-UTR, as this would ensure its incorporation into virions with unspliced virion precursor RNA.
Structured elements in the dimeric 5′-UTR can be grouped as packaging necessary (CES) and packaging dispensable (TAR, Poly(A) and PBS-segment). To compare RHA levels in HIV-1 NL4-3 virions containing an intact 5′-UTR (WT) against those with only CES, HEK293 cells were transfected with molecular clones containing WT (pNL4-3) or CES 5′-UTR (pNL4-3 MSMdenv) (Fig. 1b). Because the CES 5′-UTR is lacking the Tat transactivation responsive sequence (TAR), viral gene transcription is driven by a constitutive promoter (cytomegalovirus) and HIV-1 RNA is synthesized independently of Tat/TAR transactivation [18]. Although gRNA packaging remained unaffected and Gag expression levels were similar to WT (Fig. 1c and d), RHA was almost undetectable in the CES virions when we probed for the presence of RHA by Western blot with similar amounts of virions (Fig. 1e). These data demonstrate that CES and downstream residues, including RRE, are insufficient to incorporate RHA.
To directly assess that RRE is not necessary for the recruitment, we replaced RRE with the simian Mason-Pfizer monkey virus constitutive transport element (CTE) to facilitate nuclear export by host nuclear export proteins instead of Rev/RRE transactivation [19, 20]. As expected, RHA was observed in the CTE-substituted virions (Fig. S1), further demonstrating that RRE is dispensable to RHA incorporation during viral particle assembly. Thus, our data indicate that RHA assembles with RNA elements in the 5′-UTR that are not essential for directing gRNA packaging.
Dimeric 5′-UTR binds one RHAN300 per RNA strand
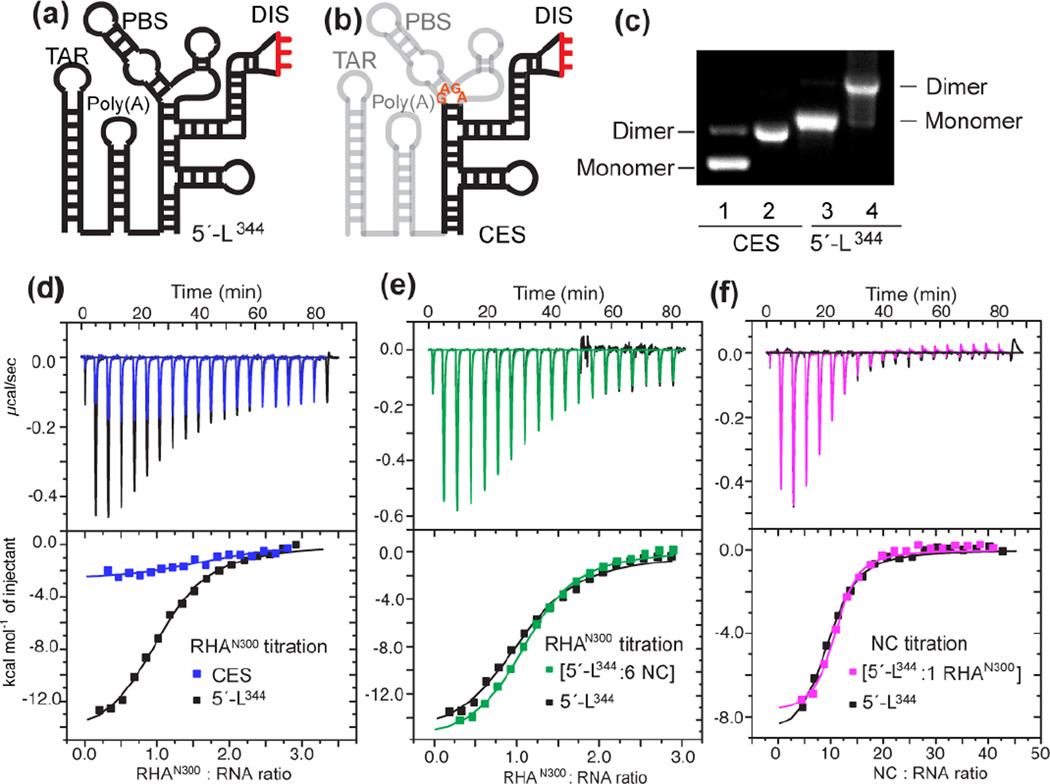
Since both dsRBDs are required for direct interactions with the HIV-1 5′-UTR RNA [8, 11], the first 300 amino acids of RHA (RHAN300) containing both dsRBD1 and dsRBD2 were expressed in E.coli and the recombinant protein was purified for in vitro studies (see Material and Method). An AUG-truncated 5′-UTR (5′-L344, transcript 1–344, Fig. 2a) transcript was synthesized and recapitulated the dimeric 5′-UTR structure during viral genome packaging (Fig. 2c) [15, 16]. To characterize the interactions between RHAN300 and 5′-L344, isothermal titration calorimetry (ITC) was carried out by titrating RHAN300 into pre-dimerized 5′-L344 (Fig. 2d). Nonlinear least squares fitting of the isothermal data by a one-site binding model indicates that 5′-L344 binds one RHAN300 per RNA strand with an affinity of Kd = 0.61 ± 0.05 µM (Table 1). The 1:1 stoichiometry indicates that each 5′-UTR dimer binds two RHA molecules, which agrees with previous cell-based results that HIV-1 virions co-package two RHA molecules on average [12]. When we titrated RHAN300 into CES, (Fig. 2b), we observed a significantly lower affinity (Fig. 2d and Table 1), in good agreement with the RHA immunoblot results (Fig. 1d) that CES is not sufficient for RHA recruitment. The ITC stoichiometry and affinity measurements with RHAN300 are highly consistent with the cell-based data, demonstrating that the N-terminal domain of RHA plays the leading role in recognizing specific RNA elements during HIV-1 assembly. Our in vitro assays with the N-terminal domain of RHA and appropriate 5′-UTR RNA segments recapitulate the cell-based results, validating that the simplified in vitro system is appropriate for testing in vivo conditions.
Fig. 2.
The specific interaction between the dimeric 5′-L344 and RHANTD occurs independently of NC-binding. (a-b) Secondary structures of 5′-L344 and CES are shown. (c) Dimerization of CES and 5′-L344 were validated on 2% native agarose gel. Lane 1 and 3, indicated transcript dissolved in H2O and loaded to gel demonstrated mobility of monomer. Lane 2 and 4, indicated transcript incubated with ITC buffer at 37°C overnight demonstrated reduced mobility of dimer. (d) Overlay of ITC isotherms collected for RHAN300 titrations with 5′-L344 (black) and CES (blue). (e) Superposition of the isotherms collected for RHAN300 titration with premixed [5′-L344:NC] complex (green) and the titration of RHAN300 into 5′-L344 (black). (f) Superposition of the isotherms collected for NC titration with premixed [5′-L344: RHAN300] complex (pink) and the NC titration with 5′-L344 (black).
Table 1.
Thermodynamic parameters for binding of RHAN300 to HIV-1 RNA constructs
| N (stoichiometry) |
Kd (uM) | ΔH (kcal/mol) | -TΔS (kcal/mol) |
ΔG (kcal/mol) |
|
|---|---|---|---|---|---|
| 5′-L344 | 1.1 ± 0.1 | 0.61 ± 0.05 | −15.0 ± 0.90 | 6.42 ± 0.87 | −8.62 ± 0.04 |
| 5′ L 344-A140C | 0.8 ± 0.1 | 1.97 ± 1.06 | −13.8 ± 0.96 | 5.79 ± 1.27 | −7.97 ± 0.34 |
| CES a | N/A b | <MD c | N/A b | N/A b | N/A b |
ITC measurement of the CES: RHAN300 interaction is not reported because the binding is very weak and the isotherms with such a shallow binding curve (Fig. 2d) are not analyzable. Attempted fitting resulted in an estimated C-value less than 0.5, out of the range for a good ITC measurement (5 < C < 500) [37].
N/A = not applicable
<MD = less than minimally detectable
HIV-1 genome packaging is initiated by Gag recognition of the packaging signal located in the dimeric 5′-UTR. This step involves a collection of Gag molecules binding to the dimeric 5′-UTR through the NC domain of Gag [15, 16, 21]. Thus, the possibility exists that RHA interacts with the NC:5′-UTR complex during virus assembly. Since CES contains the major NC binding sites to direct gRNA packaging, and given the fact that CES does not accommodate RHAN300, we speculated that RHA does not compete with NC for binding sites in the dimeric 5′-UTR. To test this hypothesis, 5′-L344 was pre-mixed with NC at 1:6 ratio (6 NC per RNA strand) prior to RHAN300 titration The ITC isothermal profiles were very similar to the RHA: 5′-L344 titration data (Fig. 2e), indicating that the RHA binding site in 5′-L344 does not overlap with the NC binding sites. Moreover, no additional heat was observed in the titration with the NC pre-mixed sample, indicating that no detectable NC:RHA interaction occurred under our experimental conditions. Conversely, pre-forming the 5′-L344:RHAN300 complex prior to NC titration did not change the ITC isothermal profiles (Fig. 2f). Thus, our data indicate that RHA binding to the dimeric 5′-UTR occurs independently of NC binding.
PBS-segment is the major RHA binding site
To precisely map the RHA binding site within the dimeric 5′-UTR, 2D NOESY spectra were collected for [5′-L344]2 (∼230 kDa) and [5′-L344: RHAN300]2 (∼290 kDa). To enhance quality of the NMR spectra derived from these large complexes, the 5′-L344 RNA was prepared with the H8 of adenosine and guanosine specifically deuterated (see Material and Method). This labeling strategy eliminates signals from the C8 proton, and achieves better spectra quality with enhanced sensitivity and resolution by diluting the magnetic environment (Fig. S2).
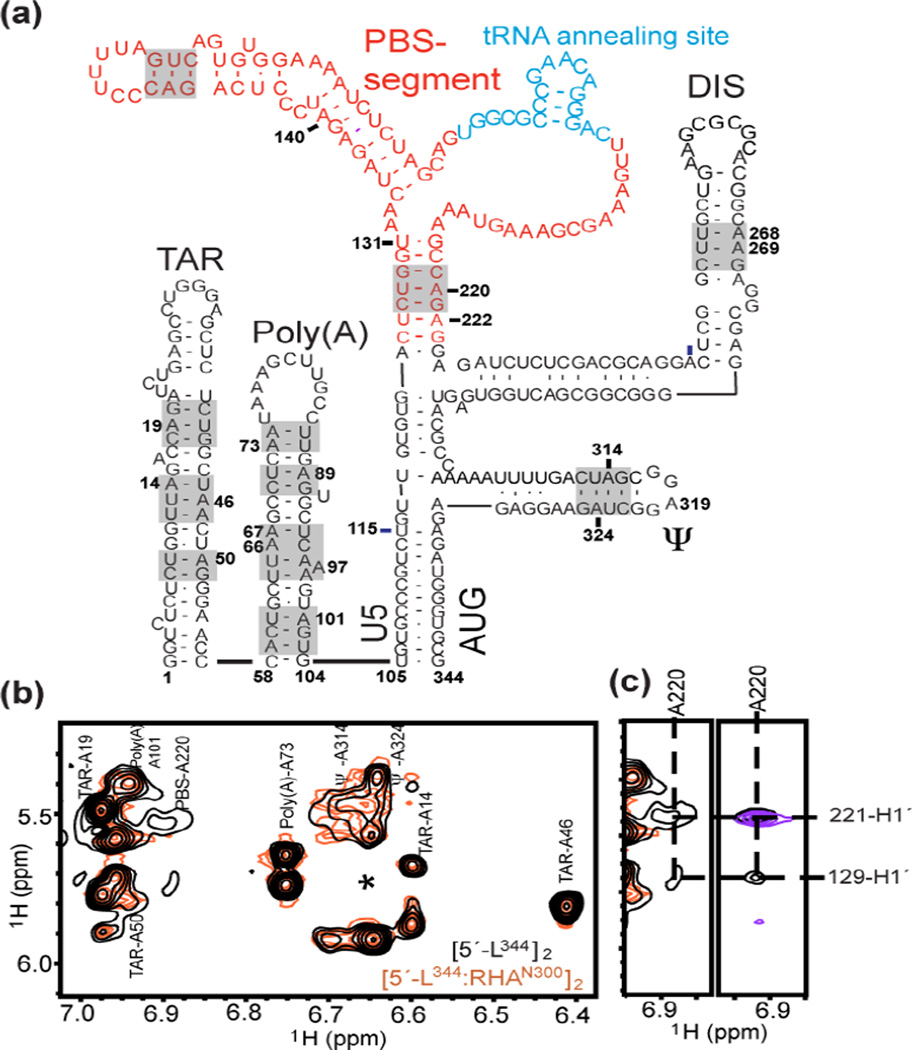
The fingerprint adenosine-H2 signals in 6.4–7.3 ppm were well-dispersed and the spectral quality is comparable with data collected for more intensively deuterated samples reported previously [16]. Thus, we were able to detect characteristic adenosine-H2 signals and some outlier peaks that are informative for structural analysis. In the aromatic region, site-specific chemical shift perturbations were observed upon RHAN300 titration (Fig. S3). Although not fully assignable, these shifted peaks were not present in the NOESY spectrum collected for [5′-L344-ΔPBS]2 (PBS-segment residues 132–216 were substituted by a GAGA tetraloop, Fig. 2a), whereas other cross-peak patterns remained unchanged. These data demonstrate that PBS-segment does not affect the structure of adjacent segments in the dimeric 5′-UTR. The fact that the only signals affected upon RHA titration were the PBS-segment signals indicates this segment was specifically recognized by RHAN300. Expected structural elements within the dimeric 5′-L344 (Fig. 3a, gray boxed regions) were verified by the NMR assignments in the A-H2 fingerprint region, which were made by referencing the previously reported chemical shifts of the segmented HIV-1 RNAs [16, 17]. A220 is the only residue in the PBS-segment that possesses an H2 chemical shift within the fingerprint region (6.4–7.3 ppm). The slow molecular rotational motion of PBS-segment A220 was illustrated by its broad line width as compared to signals belonging to TAR and Poly(A), which are relatively mobile. While most of the NMR signals maintained similar chemical shifts and line widths upon RHAN300 titration, the A220-H2 signals became broad beyond detection (Fig. 3b). Interestingly, in the NOESY spectrum collected for the PBS-segment (residues 125–223), the cross peak of A220-H2 and G129-H1′ disappeared or shifted upon RHAN300 titration (Fig. 3c, right panel), indicating PBS G129 and A220 were on or close to the RHAN300 binding site. In the case of [5′-L344]2, specific binding of RHAN300 to these residues drastically slowed molecular tumbling and resulted in extremely broad signals that became undetectable, further indicating that PBS-segment is the specific RHA binding site within the dimeric 5′-UTR.
Fig. 3.
RHAN300 binds to the PBS-segment of the dimeric 5′-UTR. (a) Sequence and secondary structure of 5′-L344 are shown. The PBS-segment is highlighted in red, and the 18-nt sequence complementary to tRNALys3 is highlighted in cyan. The base pairs in the gray-boxed region were verified by assignment of outlier peaks. (b) Assignments of the adenosine H2 signals are shown. Broad PBS-A220 signals were observed in [5′-L344]2 (black), and their line widths were beyond detection in the [5′-L344:RHAN300]2 complex (orange). The asterisk denotes the A268 H2 signals that only show at very low levels. (c) Left panel, zoom-in view of the PBS-A220 H2 signals. Right panel, the cross peak of A220-H2 and G129-H1′ in the PBS segment RNA (black) spectrum disappeared or shifted upon RHAN300 titration (purple), indicating that RHAN300 bound to the PBS-segment within the dimeric 5′-L344.
We also collected NMR spectra of RHAN300 titration into TAR-Poly(A) (transcript 1–104) and compared the signals to [5′-L344:RHAN300]2. Many signals in the adenosine H2 fingerprint region gave rise to chemical shift perturbations and signal broadening (Fig. S4). However, these changes were not observed in the [5′-L344:RHAN300]2 spectrum, indicating that TAR-Poly(A) residues do not serve as RHA binding sites in the intact dimeric 5′-UTR. Taken together, with the ITC data demonstrating that CES does not contain a high affinity RHA binding site (Fig. 2d), the NMR data indicate that PBS-segment residues encompass the dominant RHA binding site within the intact dimeric 5′-UTR.
Structural mutation A140C in PBS-segment concordantly decreases RHA binding and virion infectivity
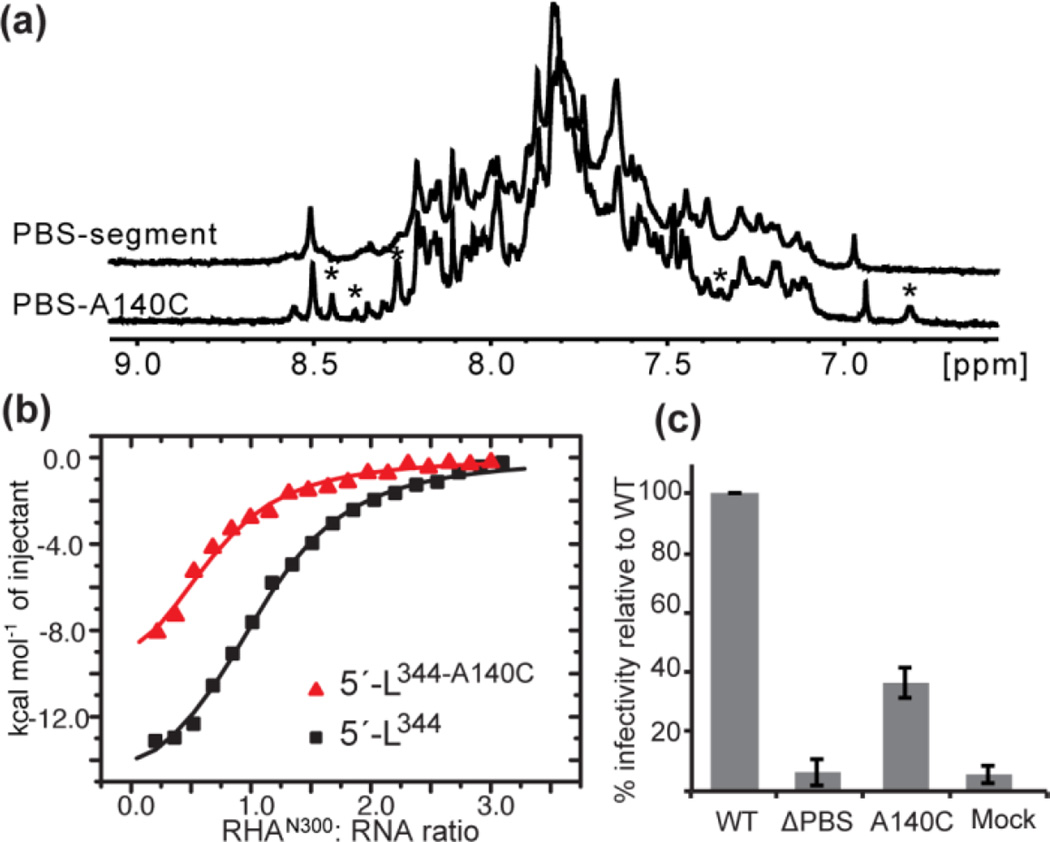
To address the functional recruitment of RHA to PBS-segment, we introduced a series of mutations and performed ITC. The single nucleotide change A140C in the PBS-segment (PBS-A140C) was sufficient to significantly reduce the affinity of RHA. Although NMR data indicate that the direct RHAN300:PBS-segment interactions occur at or close to G129 and A220, these residues may have overlapping functions as they are within the primer activation signal (PAS) stem important for reverse transcription initiation [22–24]. Thus, A140C, which is not part of the tRNALys3 annealing site [25, 26], was suitable to avoid possible interference of other important reverse transcription activities attributable to the PAS [22–24], or the tRNA-like element region that recruits Lysyl-tRNA synthetase (LysRS) [27].
The A140 residue is highly conserved across clades and strains of HIV-1. In <1% cases there is an A-to-G substitution, which alters predicted A140-U172 base complementarity to G-U, consistent with a structural conservation in PBS-segment (HIV Sequence Compendium, 2014). As predicted, the A140C mutation in PBS-segment leads to local structure variation and may introduce structural heterogeneity of PBS-segment, as indicated by a different 1D proton pattern shown in Fig. 4a. Consequently, the RHAN300 binding affinity is reduced (Fig. 4b). The same mutation A140C was introduced in the HIV-1 infectious molecular clone pNL4-3 and the effect on virus infectivity was evaluated in the TZM-bl reporter cell line. The A140C mutant exhibited an approximately 60% decrease in infectivity, which is similar in magnitude to the previously reported reduction in primer extension on gRNA extracted from RHA-deficient virions [7] (Fig. 4c). The reduced infectivity of A140C virions may reflect the failure of proper RHA loading onto PBS-segment during virus assembly, leading to deficient primer extension activity in the progeny virions.
Fig. 4.
A single nucleotide substitution A140C, precludes RHA binding to PBS-segment, and diminishes infectivity of virions. (a) One-dimension 1H spectra for PBS and PBS-A140C. While many of the major sharp signals remained unchanged, a few new small peaks were detected as denoted by asterisks (*), demonstrating local structural alternations in the PBS segment RNA. (b) The A140C mutation moderately reduces RHAN300 binding (red) relative to 5′-L344 (black). (c) Infectivity of WT, ΔPBS, and A140C virions on TZM-bl cells was measured by Luciferase assays. Equivalent amounts of virions based on Gag ELISA were used to infect TZM-bl cells. The A140C mutation led to ∼ 60% reduction in infectivity as depicted in the graph (p≤0.001). ΔPBS and Mock serve as negative controls. Assays were performed in triplicate in three replicated experiments.
We then performed cell-based assays to determine whether or not the decrease in A140C virion infectivity is due to the reduced affinity of RHA to A140C RNA. We hypothesized partial depletion of RHA would diminish infectivity of WT and A140C virions whereas a significant depletion of RHA would lead to WT and A140C virions with similar infectivity levels. On the other hand, if the defect in A140C virions is independent of RHA the difference in virion infectivity between WT and A140C virions will stay the same upon depletion of RHA levels in the producer cells.
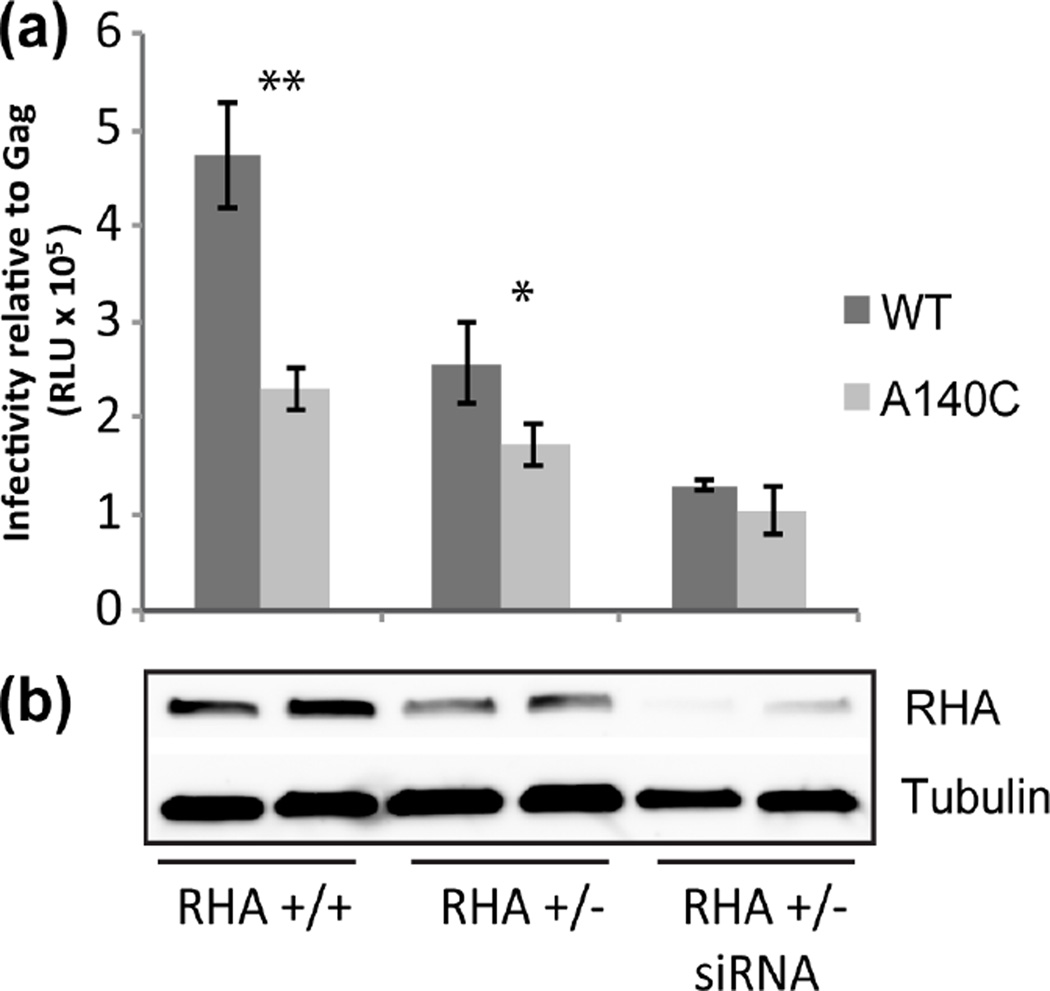
To progressively deplete RHA in virus-producing cells, we took a two-step approach. First, we implemented CRISPR/Cas9 to disrupt one allele of RHA (293 RHA +/−) [28] and diminished endogenous RHA levels were observed when evaluated by immunoblot (Fig 5b). No viable cells were obtained upon disruption of both alleles consistent with prior demonstration RHA is an essential gene [29]. Second, we treated the 293 RHA +/− cells with siRNAs to downregulate residual RHA (Fig 5b). WT or A140C virions were produced in WT HEK293 cells, 293 RHA +/− cells, and 293 RHA+/− cells that were treated with RHA siRNA. Virion infectivity was measured by luciferase assay in the reporter TZM-bl cells (Fig 5a). RHA immunoblot showed decrease in RHA levels in cells with only one RHA allele (+/−) compared with cells containing two RHA alleles (+/+) and further decrease when the 293 RHA +/− cells were treated with RHA siRNAs (Fig. 5b).
Fig. 5.
Infectivity of A140C virus was not sensitive to depletion of RHA in virus producer cells. (a) HEK293, 293 RHA +/− or siRNA treated 293 RHA +/− were transfected to produce WT and A140C virions. (a) Equivalent amounts of virions based on Gag ELISA were used to infect TZM-bl cells and virion infectivity was measured by Luciferase assays. The graph represents average of three independent experiments (±SD). RLU = relative light units; ** p=0.002; * p=0.04. (b) RHA and Tubulin levels in virus producer cells were assayed by immunoblot and a representative blot is shown.
WT virions produced in RHA +/− cells were less infectious compared to WT virions produced in 293 RHA +/+ cells. Further downregulation of RHA by siRNA treatment resulted in a further decrease in virion infectivity. The decrease in virion infectivity directly correlated with the decrease in RHA levels. The A140C virions displayed a similar trend between 293 RHA +/+ cells and 293 RHA +/− cells, albeit the reduction was not as pronounced as WT (Fig. 5a). A140C virions exhibited significantly decreased infectivity levels relative to WT when produced in 293 RHA +/+ (p=0.002) or 293 RHA +/− cells (p=0.04), but no difference in infectivity when produced in RHA deficient cells (RHA +/− treated with RHA siRNA) (p=0.17). The results show that in the absence of RHA, WT and A140C virions have similar infectivity confirming the A140C defect is due to diminished binding affinity of PBS-segment for RHA.
DISCUSSION
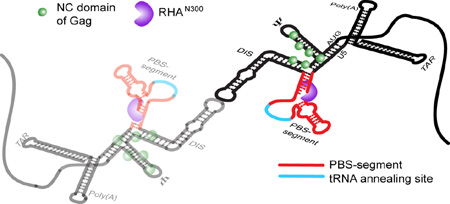
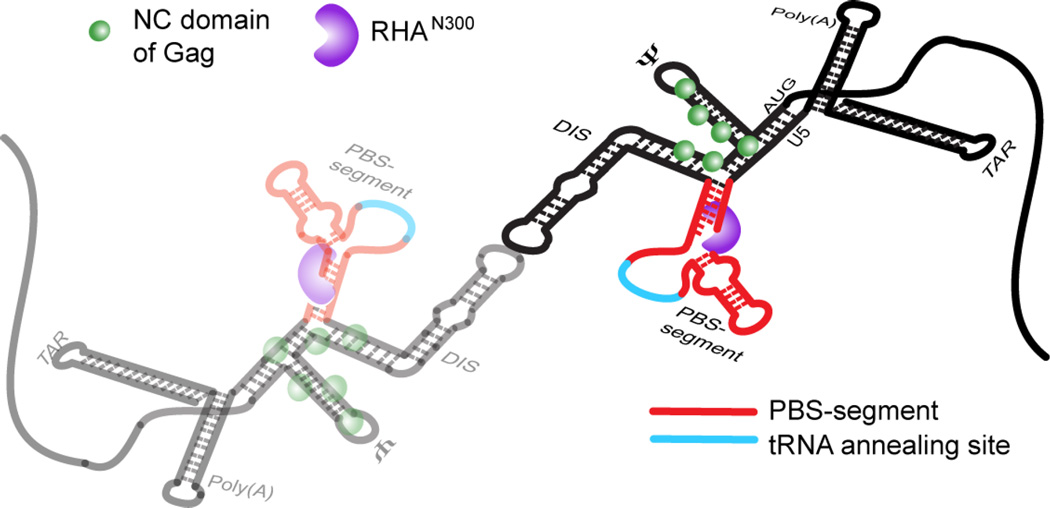
By integrating results from cell-based assays and biophysical analyses, this study identified the structural determinants necessary to incorporate RHA into virions in support of HIV-1 infectivity. Results of cell-based experiments demonstrated that the 5′-UTR is necessary for RHA incorporation to virions, and supported the hypothetical role of RNA elements outside of CES. Candidate RNA elements for RHA binding to the 5′-UTR were screened for RHA binding affinity by ITC, and then we collected NMR data for intact dimeric 5′-UTR complexes [5′-L344]2 (∼230 kDa) and [5′-L344:RHAN300]2 (∼290 kDa). Site-specific chemical shift perturbations were detected, revealing the position of the RNA:protein interaction. Characteristic adenosine cross-peak patterns in TAR, Poly(A), and the PBS-segment were observed, which triggered us to deemphasize RHAN300 binding to TAR and Poly(A) along with CES. Through the N-terminal RNA binding domain, RHA binds the diploid HIV-1 gRNA in one-to-one stoichiometry on the PBS-segment, as modeled in Fig. 6. RHA interaction occurs independently of Gag binding via its NC domain and is spatially consistent with the demonstrated role of RHA in tRNA priming crucial to initiate reverse transcription [5, 7, 8].
Fig. 6.
Model for the 1:1 stoichiometry of RHA bound to dimeric HIV-1 5′-UTR, which is spatially consistent with RHA’s known role in primer placement during initiation of reverse transcription. The RHA N-terminal double-stranded RNA-binding domains, shown in purple, are recruited to double-stranded residues in the PBS-segment, which is juxtaposition to CES-bound Gag p55 NC domain during assembly in virus producer cells. The PBS-segment is highlighted in red with the tRNALys3-annealing site highlighted in cyan; the rest of the 5′-UTR sequence is shown in black. The N-terminal domain of RHA binds the dimeric 5′-UTR independently of NC binding.
Previously, Xing and coworkers measured primer extension efficiency indicative of reverse transcription initiation using deproteinized RNA extracted from newly released virions to provide the source of tRNALys3 annealed to the PBS-segment of the gRNA. They reported a 40%-50% reduction in primer extension when using the RNA template extracted from RHA-downregulated virions, indicating that RHA’s role in the early stage of reverse transcription involves proper tRNALys3 placement within the PBS-segment [7]. In line with their findings, the decreased infectivity of A140C virions underscores the importance of maintaining the proper folding of the PBS-segment to recruit RHA, despite the fact that it undergoes a drastic structural rearrangement upon tRNALys3 annealing.
A140 is a highly conserved residue outside of the proposed tRNALys3 annealing site and PAS region. Phylogenetic evidence, as well as our NMR data, reinforce the critical role of A140 and U172 base pairing to maintain the native PBS-segment folding, and subsequently ensure the functional-important interactions with cellular factors, including RHA. The A140C mutation led to partial misfolding of PBS-segment, but not the PAS stem as demonstrated by our NMR data. The structural alternations resulted in disruption of RHA binding and diminished HIV-1 infectivity. A combination of CRISPR/Cas9 and RHA siRNA downregulation has enabled us to vary cellular RHA levels and investigate differences in the infectivity of progeny virions. While the infectivity of WT virions reduced drastically in response to diminished RHA levels, the infectivity of A140C virions remained relatively insensitive. When there was only basal expression of RHA in producer cells, the infectivity of WT virions and A140C virions plateaued at the similar level, demonstrating equivalence between the A140C mutation and RHA depletion to inefficient recruitment of RHA during assembly. In the alternative case, in which the A140C mutation involved RHA-independent defects, A140C virion infectivity would have remained lower than WT regardless of RHA levels. Our results demonstrated the A140C defect is due to diminished binding affinity of PBS-segment for RHA.
The 5′-UTR utilizes a monomer:dimer RNA switch to coordinate interdependent activities during viral replication [15], which complicates biophysical and biological investigations. For example, an HIV transcript containing an intact AUG hairpin (NL4-3, transcript 1–356) exists in equilibrium of monomer and dimer conformations. Truncation of half AUG hairpin (transcript 1–344) strongly favors the U5:AUG long-range interaction and thus stabilizes the 5′-UTR dimeric conformation. Further truncation to completely remove AUG (transcript 1–327) destabilizes dimeric conformation and stabilizes the 5′-UTR in monomeric conformation [15]. Therefore, strategic design of inclusive RNA coordinates was essential for identifying the RHA binding site by biophysical methods. Specific RHA:5′-UTR interactions were not detectable compared to control RNAs in a previous fluorescence polarization assay performed at relatively weak ion strength buffers; similar affinity Kd values were observed between RHA and various HIV-1 RNA segments of similar size (300–400 nt), including RRE, which was not necessary for recruiting RHA into virions (Fig. S1) [8]. In the present study, the dimeric conformation of the in vitro RNA transcript demonstrated high affinity binding to RHAN300 and, in agreement with Xing et al., TAR-Poly(A) or CES did not exhibit specific interactions with RHA N300. Our results demonstrate the structural basis for HIV-1 RNA binding to host RBPs during virion assembly. Moreover, and in agreement with a recent biophysical study demonstrating RHA’s specific recognition of the spleen necrosis virus 5′-UTR [11], our ITC and NMR data strongly argue against RHA being a non-specific dsRBD helicase.
RHA exhibited strong binding affinity to the HIV-1 5′-UTR during viral replication in a co-immunoprecipitation assay with transfected cell lysate [8]. This outcome should be considered as a sum of the RHA:5′-UTR interactions that include RHA-mediated regulation of HIV-1 protein synthesis and RHA recruitment during assembly of infectious virions. RHA promotes HIV-1 mRNA translation by interacting with the post-transcriptional control element (PCE) located in the 5′-UTR [6, 10]. HIV-1 PCE was identified as the first ∼181 nt of 5′-UTR, which seems to overlap with the PBS-segment we mapped as the RHA binding site during virus assembly. However, the RHA:5′-UTR interaction for virion incorporation and translation activation should be different, as the 5′-UTR utilizes a monomer-dimer switch to regulate translation (Boeras and Boris-Lawrie, unpublished data) and gRNA packaging [15]. Although no high-resolution structural information is currently available for the monomeric 5′-UTR, SHAPE probing of the monomeric and dimeric 5′-UTR separated by polyacrylamide gel electrophoresis has detected nucleotide activity differences in the PBS-segment [30], consistent with the RHA:5′-UTR interactions for virion incorporation and translation activation being different. Thus RHA is likely capable of distinguishing the HIV-1 5′-UTR monomer from the dimer conformation, and with cellular RBPs, to form catalytic messenger ribonucleoprotein complexes for translation and dimeric gRNPs for packaging. RHA might be one of the host factors involved in determining the fate of the HIV-1 primary transcript, and unraveling of this critical role warrants continued investigation of RHA:5′-UTR interactions.
MATERIAL AND METHOD
Cell lines, transfections and infections
HEK293 cells, 293 RHA +/− cells, and TZM-bl cells were maintained in EMEM (Invitrogen) supplemented with 10% Fetal Bovine Serum (Gibco) and 1% Antibiotic-Antimycotic (Gibco). HEK293 and 293 RHA +/− cells were transfected using the XtremeGene (Roche) reagent at a ratio of 1:3 DNA to reagent following the manufacturer’s instructions. TZM-bl cells were plated in 96-well plates and infected with equivalent amounts of virus supernatant based on p24 ELISA, in triplicate. Cells were assayed for luciferase activity two days post infection.
Plasmids
The CES-containing plasmid was created by inserting CES into pMSM-dEnv [18]. pMSM-dEnv lacks most of the viral 5′-UTR, including TAR, Poly(A) and CES. Transcription is driven by a CMV promoter and occurs independently of Tat/TAR activity. CES was generated by PCR using pNL4-3 as template and primers with terminal XbaI and ApaI restriction sites, and then inserted into pMSM-dEnv by restriction with XbaI and ApaI and ligation. To replace PBS-segment with a GAGA tetraloop, PCR was performed with antisense primers complementary to regions flanking the PBS-segment and template pNL4-3 and the PCR product was self-ligated to achieve PBS-segment deletion. The A140C substitution in PBS-segment was introduced into pNL4-3 by site-directed mutagenesis using the QuickChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and the following primers KB2273 5′-TAA AAG GGT CTG AGG GAG CTC TAG TTA CCA GAG TC-3′ and KB2274 5′-GAC TCT GGT AAC TAG AGC TCC CTC AGA CCC TTT TA-3′. All plasmid sequences were verified by sequence analysis at the university core facilities.
RHA downregulation by CRISPR/Cas9 and siRNA
The CRISPR guide RNA sequence (5′-CAG GCA GAA ATT CAT GTG TG-3′) was synthesized by IDT as two complementary primers with 5′ and 3′ overhangs that match the BsmBI overhangs produced in the lentiCRISPR v2 plasmid. Lentiviral particles containing the lentiCRISPR vector payload were produced in 293FT cells (Invitrogen) and were used to transduce fresh 293FT cells. Transduced cells were cultured for 2 days, and then cultured with 1 µg/ml puromycin for 4 days. Single cell isolates were isolated by limiting dilution and expanded to obtain sufficient cells for screening. Genomic DNA was extracted from the single cell clones and the RHA target sequence was amplified with primers (5′-CAT GAA GTT CAA ATC ATA CCG T-3′) and (5′-CAA GAA TTG CTC TGC TTT CTA A-3′). The PCR products were directly sequenced and analyzed to determine precise identity.
The RHA-specific siRNAs are complementary to the 3’ UTR of human RHA and have been used to downregulate RHA in previous studies of HIV-1 in 293 cells [7]. The siRNAs were transfected into 293 RHA +/− cells using Lipofectamine 2000 per manufacturer instructions (Invitrogen). Twenty-four hours later, the cells were co-transfected with HIV pNL4-3 infectious molecular clone or pNL4-3 A140C. Two days later, cell-free supernatant media were collected and used to assay virion infectivity on TZM-bl cells. Also, cell lysates were prepared and subjected to RHA immunoblot (Vaxron) with Tubulin loading control.
To quantify the progeny virion infectivity, TZM-bl cells were lysed in 100 µL Cell lysis buffer (Promega) and 20 µL was mixed with 100 µL luciferase assay reagent (Promega). Luminescence was measured with a GloMax Luminometer (Promega).
Quantification of RHA in HIV-1 virions
Virions were produced in HEK293 cells by transfection with the indicated plasmids for 36–48 hours and cell-free media were loaded over 1 ml of 20% sucrose and subjected to ultracentrifugation in an SW41 rotor at 40000rpm for 2 hours. Pelleted virions were resuspended in RIPA buffer (50mM Tris, 150mM NaCl, 1% NP40, 0.25% deoxycholic acid, 1 mM EDTA) and the amount of Gag protein in the pellet was quantified by HIV-1 Gag p24 ELISA (Zeptometrix). Equivalent virion Gag p24 amounts were subjected to SDS-PAGE and Western blot by anti-RHA (Vaxron) antibody for RHA detection and antiserum 24-2 against HIV-1 Gag p24 (AIDS Reagents Program) for HIV-1 Gag detection.
RNA extraction and quantification
RNA was extracted from cell lysate and virion pellets in Trizol (Invitrogen) by manufacturer instructions, and collected by the RNAeasy clean-up protocol (Qiagen). Equivalent volumes of RNA preparation were reacted with random hexamer primers and Omniscript Reverse Transcriptase (Qiagen) to generate cDNA. Real-time PCR was performed using gag-specific primers KB1614 and KB1615 [31] and CyberGreen (BioRad).
Recombinant protein expression and purification
The DNA fragment encoding the RHAN300 (RHA residues 1–300) [11] was cloned into a modified pMCSG7 vector that incorporates an N-terminal His6-Mocr for enhanced protein expression and solubility [32]. The recombinant RHAN300 protein was expressed in E.coli BL21-CodonPlus (DE3)-RIPL cells (Agilent Technologies) induced with 0.3 mM IPTG at 37°C for 5 h. The harvested bacteria were lysed in buffer A (40 mM NaH2PO4, pH 7.5, 300 mM NaCl). Then the fusion protein was isolated on a Cobalt column (HisPur Cobalt Resin, Thermo Scientific) for affinity chromatography. RHAN300 was eluted with Buffer A supplemented with 120 mM imidazole. Pooled fractions were dialyzed against dialysis buffer (30mM Tris, pH 7.5, 100 mM NaCl, 2 mM EDTA, 5 mM β-Mercaptoethanol) supplemented with TEV protease (1 mg protease/ 50 mg of RHAN300) for His6-Mocr tag cleavage at 4°C for 20 h. After dialysis, the preparation was loaded to an anion exchange column (Hiprep Q HP 16/10, GE Healthcare) for removing the His6-Mocr tag and contamination of nucleic acids. The untagged proteins were purified by size exclusion chromatography using a Superdex 75 column (GE Healthcare) equilibrated with ITC Buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 300 mM KCl, 1 mM MgCl2, 1 mM β-Mercaptoethanol). RHAN300 was concentrated to 6 mg/ml for storage at −80°C.
Preparation of RNA samples
The template for in vitro transcription 5′-L344 (transcript 1–344) was prepared from pUC19 plasmid with insertion of PCR amplicon generated by primers (5′-CGT TGT AAA ACG ACG GCC AGT GAA TTC TAA TAC GAC-3′) and (5′-GGT CGA CGG ATC CGT ATC CGC TAG CTC TCG CAC CCA TCT CTC TCC-3′) and pNL4-3 template DNA. A 3′ BciVI site was introduced for DNA plasmid linearization without introducing non-native 3′- residues during RNA synthesis by T7 polymerase. The mutant 5′-L344-A140C was derived from the 5′-L344 plasmid by site-directed mutagenesis (Agilent, QuickChange II XL Kit) and confirmed by DNA sequencing in the University of Missouri DNA Core Facility.
Template plasmids of 5′-L344 and 5′-L344-A140C were amplified in DH5α cells, and extracted by Plasmid Mega Kit (Qiagen). After BciVI (NEB) digestion, the linearized DNA templates were extracted by phenol-chloroform and precipitated with ethanol. The DNA pellet was lyophilized, dissolved and then washed with sterile MilliQ water by centrifugal filter column (Amicon). The solution of DNA templates were finally concentrated over 1500 ng/µl and stored at 4°C. Transcription templates of TAR-Poly(A) (transcript 1–104) and PBS-segment (transcript 125–233 with two non-native GC base pairs to stabilize the bottom stem) were prepared by PCR from a template plasmid 5′-L344. Transcription template of CES was amplified by PCR using the 5′-LΔTAR-ΔPolyA-ΔPBS plasmid as template [16].
RNA samples were synthesized by in vitro transcription with T7 RNA polymerase as described previously [33]. Optimized transcription conditions were screened by varying the amount of MgCl2, NTPs and DNA templates for each RNA sample. The transcripts were then purified by denaturing polyacrylamide gel electrophoresis and extracted by an Elutrap Electroelution System (Whatman). To prepare site specific H8-deuterated purines, natural abundant ATP and GTP were dissolved in D2O with ∼5 equivalents of trimethylamine and exchanged at 65°C for 1 day (GTP) and 5 days (for ATP) [34]. H8-deuterated 5′-L344 and 5′-L344-ΔPBS were prepared by incorporating H8-deuterated ATP and GTP with regular CTP and UTP in T7 transcriptions, and followed by standard RNA purification procedures.
Isothermal titration calorimetry
All ITC experiments were carried out using a VP-ITC MicroCalorimeter (MicroCal, GE Healthcare) at 30°C. The concentrated RHAN300 protein (65 µM) in ITC Buffer was loaded into the injection syringe. RNA samples (4 µM) were pre-dimerized in ITC buffer at 37°C overnight before loading to the sample cell. Data were collected in the high feedback mode with 20 serial injections of RHAN300. The thermodynamic parameters of the RHAN300-RNA interaction were processed and fitted using one-site binding model (Microcal Origin, version 7). Each value of the binding parameters summarized in Table 1 was an average of three independent titrations.
NMR Spectroscopy
RNA samples were pre-dimerized in ITC buffer at 37°C prior to NMR data acquisition on a Bruker Avance III 800 MHz spectrometer equipped with TCI cryoprobe. Two-dimensional 1H-1H NOESY spectra were collected for [5′-L344]2 and [5′-L344:RHAN300]2 in D2O at 308 K with a mixing time of 80 ms. NMR data were processed with NMRPipe and NMRDraw [35], and analyzed with NMRView [36].
Supplementary Material
Highlights.
Host DHX9/RHA is recruited in HIV-1 virions to support infectivity.
Cell-based assays identified the 5′-UTR necessary to RHA recruitment to virions.
NMR data of the complex (290 kDa) pinpointed PBS as the RHA binding site
A PBS point mutation that precluded RHA binding attenuated virion infectivity
Our work sheds light on the RHA involvement in the early viral replication events.
Acknowledgments
This research was supported by the NIH National Institute of General Medical Sciences P50 GM 103297 to KBL and XH. We are grateful for plasmids: pFL-RHA provided by C.G. Lee (UMDNJ); pMSM-dEnv provided by AT. Panganiban (TNRPC), LentiCRISPR v2 from Feng Zhang (Addgene plasmid #52961), NL43Rev(−)R(−).S by B. Felber (NCI), and the AIDS Reagent Repository for antisera.
Abbreviations used
- HIV-1
human immunodeficiency virus type 1
- RHA
RNA helicase A/DHX9
- 5′-UTR
5′-Untranslated Region
- gRNA
genomic RNA
- NC
nucleocapsid
- RBP
RNA binding protein
- gRNP
genomic ribonucleoprotein particles
- PBS
primer binding site
- CES
core encapsidation signal
- ITC
isothermal titration calorimetry
- NMR
nuclear magnetic resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 2.Ranji A, Boris-Lawrie K. RNA helicases: emerging roles in viral replication and the host innate response. RNA biology. 2010;7:775–787. doi: 10.4161/rna.7.6.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii R, Okamoto M, Aratani S, Oishi T, Ohshima T, Taira K, et al. A Role of RNA Helicase A in cis-Acting Transactivation Response Element-mediated Transcriptional Regulation of Human Immunodeficiency Virus Type 1. The Journal of biological chemistry. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 4.Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, et al. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol Cell Proteomics. 2012;11:M111. doi: 10.1074/mcp.M111.015313. 015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy BB, Hu J, Guo X, Russell RS, Guo F, Kleiman L, et al. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J Biol Chem. 2006;281:12625–12635. doi: 10.1074/jbc.M510596200. [DOI] [PubMed] [Google Scholar]
- 6.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 7.Xing L, Liang C, Kleiman L. Coordinate roles of Gag and RNA helicase A in promoting the annealing of formula to HIV-1 RNA. J Virol. 2011;85:1847–1860. doi: 10.1128/JVI.02010-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xing L, Niu M, Kleiman L. In vitro and in vivo analysis of the interaction between RNA helicase A and HIV-1 RNA. Journal of virology. 2012;86:13272–13280. doi: 10.1128/JVI.01993-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos S, Obukhov Y, Nekhai S, Bukrinsky M, Iordanskiy S. Virus-producing cells determine the host protein profiles of HIV-1 virion cores. Retrovirology. 2012;9:65. doi: 10.1186/1742-4690-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic acids research. 2010;38:1686–1696. doi: 10.1093/nar/gkp1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranji A, Shkriabai N, Kvaratskhelia M, Musier-Forsyth K, Boris-Lawrie K. Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J Biol Chem. 2011;286:5328–5337. doi: 10.1074/jbc.M110.176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma A, Boris-Lawrie K. Determination of host RNA helicases activity in viral replication. Methods Enzymol. 2012;511:405–435. doi: 10.1016/B978-0-12-396546-2.00019-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima T, Uchida C, Anderson SF, Lee CG, Hurwitz J, Parvin JD, et al. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 14.Coffin JM, Hughes SH, Varmus H. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 15.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, et al. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, et al. RNA structure Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zolotukhin AS, Valentin A, Pavlakis GN, Felber BK. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, et al. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci U S A. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol. 2002;76:2329–2339. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huthoff H, Bugala K, Barciszewski J, Berkhout B. On the importance of the primer activation signal for initiation of tRNA(lys3)-primed reverse transcription of the HIV-1 RNA genome. Nucleic Acids Res. 2003;31:5186–5194. doi: 10.1093/nar/gkg714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ooms M, Cupac D, Abbink TE, Huthoff H, Berkhout B. The availability of the primer activation signal (PAS) affects the efficiency of HIV-1 reverse transcription initiation. Nucleic Acids Res. 2007;35:1649–1659. doi: 10.1093/nar/gkm046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS biology. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seif E, Niu M, Kleiman L. In virio SHAPE analysis of tRNA(Lys3) annealing to HIV-1 genomic RNA in wild type and protease-deficient virus. Retrovirology. 2015;12:40. doi: 10.1186/s12977-015-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. Rna. 2013;19:219–229. doi: 10.1261/rna.036681.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. 2016 doi: 10.1038/onc.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenyon JC, Prestwood LJ, Le Grice SF, Lever AM. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013;41:e174. doi: 10.1093/nar/gkt690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS pathogens. 2012;8:e1002612. doi: 10.1371/journal.ppat.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: a novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein expression and purification. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Yu P, LeProust E, Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 36.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods in molecular biology. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 37.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Analytical biochemistry. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.