Abstract
Renal cell carcinoma (RCC) is a major neoplasm with high incidence in western countries. Tumors are heterogeneous and are composed of differentiated cancer cells, stromal cells, and cancer stem cells (CSCs). CSCs possess two main properties: self-renewal and proliferation. Additionally, they can generate new tumors once transplanted into immunodeficient mice. Several approaches have been described to identify them, through the expression of cell markers, functional assays, or a combination of both. As CSCs are involved in the resistance mechanisms to radio- and chemotherapies, several new strategies have been proposed to directly target CSCs in RCC. One approach drives CSCs to differentiate into cancer cells sensitive to conventional treatments, while the other proposes to eradicate them selectively. A series of innovative therapies aiming at eliminating CSCs have been designed to treat other types of cancer and have not been experimented with on RCC yet, but they reveal themselves to be promising. In conclusion, CSCs are an important player in carcinogenesis and represent a valid target for therapy in RCC patients.
1. Introduction
Renal cell carcinoma (RCC) constitutes the most common form of renal neoplasms, comprising more than 90% of cases in adults of both sexes, with an occurrence 2 to 3 times higher in men than in women. The incidence increases after 40 years of age, as for all tumors of epithelial origin, and decreases after 75 years in both sexes [1, 2]. RCC is classified into several different subtypes based on the pathological features. The most common subtype is clear cell RCC (ccRCC), followed by papillary RCC (pRCC), chromophobe, and collecting duct RCC. The 2013 Vancouver classification includes a total of 17 morphotypes of renal parenchymal malignancy and two benign tumors [3–6]. RCC is becoming more commonly diagnosed worldwide and, consequently, mortality is decreasing in the most developed settings. However, it remains prevalent in low- and middle-income countries, where access to and the availability of optimal therapies are likely to be limited [2]. Surgical management of the primary tumor remains the gold standard of RCC treatment. Nevertheless, RCC high metastatic index and resistance to radiation and chemotherapies have led to the development of new therapeutic agents that target the tumor vasculature or that attenuate the activation of intracellular oncogenic pathways [7].
Tumors are heterogeneous structures composed of different types of cancer cells, each cell population presenting variations in metabolism, receptors, and ligands expression and epigenetic chromatin structure alterations [8–13]. Identifying specific cell types within a tumor that either initiate or maintain tumorigenesis provides valuable information and allows a better understanding of tumor biology, as well as the development of novel treatments. The cell of origin of cancer, or tumor-initiating cell (TIC), is a normal cell that sustains mutations leading to tumor formation [14]. The cells that maintain tumor growth and propagation are the cancer stem cells (CSCs) [15]. However, the use of the TIC or CSC terminology is sometimes redundant, as the distinction between the two populations is blurry. CSCs possess two main characteristics: self-renewal and multipotency capacity. Self-renewal allows unlimited cell division and maintenance of the stem cell pool in the tumor. Multipotency permits CSCs to divide and create a progeny that keeps dividing until they yield terminally differentiated, specialized cells [16]. Additionally, CSCs can by themselves originate a tumor mass indefinitely, following transplant into immunodeficient mice (Figure 1). As a matter of fact, the cancer transplantation assay constitutes the gold standard in identifying CSCs as it can provide evidence of both self-renewal and multilineage potency of CSCs [17]. It consists in implanting a putative CSC population into immunodeficient mice, and if the cells give rise to serially transplantable tumors that recapitulate the cellular heterogeneity of the parental tumors, they can conclusively be qualified of CSCs. On the other hand, TICs can be defined by lineage tracing assays, which allow defining the cell of origin of transformation in mouse models [17]. The use of cell-specific promoters allows distinct cell subpopulations to be labeled, allowing tracking of single-cell-derived clones. This assay permits us to assess the fate of individual cells that undergo transformation and form a tumor and to definitively identify them as TICs. Consecutively, labeled TICs can be sorted and used in serial transplantation to evaluate their CSC properties.
Figure 1.
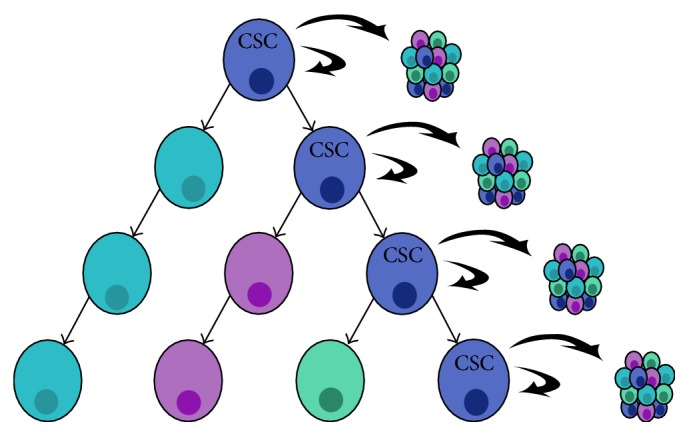
Cancer stem cell model. Tumor cells form a heterogeneous structure and only the cancer stem cells (CSCs) have the ability to self-renew and differentiate into different cell types. CSCs can form new heterogeneous tumors following transplant.
Various hypotheses exist to describe the origin of TICs/CSCs, such as accumulation of several mutations during their lifespan or reprogramming of tumor cells through dedifferentiation by hypoxia and/or epithelial-to-mesenchymal transition (EMT) [18–20]. Several mechanisms confer CSCs resistance to radiation and chemotherapeutic treatments, including their quiescent state, their presence in hypoxic microenvironments, upregulation of damage response mechanisms, and their increased drug efflux potential [16, 21]. Conventional therapy does not target the CSC population in RCC, and despite an initial tumor size reduction the patient relapses. A better identification and characterization of CSCs would allow the development of new drugs to selectively eradicate this population in RCC patients.
2. Cancer Stem Cells in Renal Cell Carcinoma
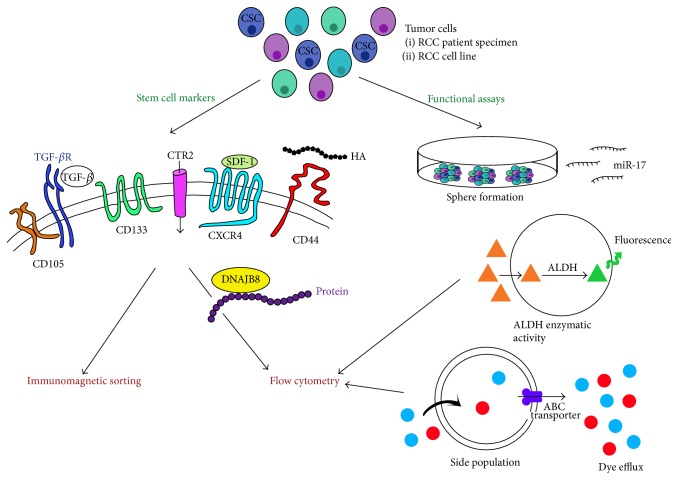
Renal tubular cells have been extensively described as the cellular origin of RCC [19]. Numerous studies have attempted to isolate and characterize a population of CSCs among tubular cells, using either stem cell markers or functional assays (Figure 2) [18, 22, 23].
Figure 2.
Identification of cancer stem cells using stem cell markers and functional assays. Several techniques have been described to identify cancer stem cells (CSCs) and to isolate them by immunomagnetic sorting, flow cytometry, or cell culture. HA: hyaluronic acid; red circle: Hoechst 33342; blue circle: Rhodamine 123; ALDH: aldehyde dehydrogenase; orange triangle: BODIPY-aminoacetaldehyde (substrate); green triangle: BODIPY-aminoacetate.
2.1. Stem Cell Markers
Several molecular markers have been used to identify CSCs and constitute valuable tools for tumor detection and diagnostic, prognostic, and predictive values, as well as determination of therapeutic targets [24]. Of note, no marker has been found so far that would be expressed only in CSCs [25].
2.1.1. CD105
CD105, also called endoglin, is a surface transmembrane molecule that is part of the transforming growth factor-β (TGF-β) receptor complex. It regulates cell proliferation, differentiation, and migration and has an important role in angiogenesis [26]. In 2008, Bussolati et al. proposed CD105 as a marker for tumor-initiating stem cells [27]. Cells from patient specimens of RCC were labeled with CD105 and separated by magnetic sorting. They represented less than 10% of the total tumor cells and expressed the mesenchymal stem cell markers CD44, CD90, CD146, CD73, and CD29 and the mesodermal marker vimentin (VIM), as well as the embryonic stem cell markers NANOG, OCT4, Musashi, and Nestin and embryonic renal marker Pax2, but lacked differentiative epithelial markers such as Cytokeratin. These cells were clonogenic and could generate spheres in specific cell culture medium, which is considered an important characteristic of CSCs. They could differentiate in vitro and in vivo into epithelial cells. CD105+ cells displayed tumor-initiating activity, and as few as 100 cells could generate serially transplantable carcinomas in immunodeficient mice containing few CD105+ tumorigenic cells and a large number of differentiated CD105− cells. They did not express CD133, a known marker of adult human tubular progenitor cells [28, 29]. The same authors also found that extracellular vesicles (EV) released by renal CD105+ CSCs, but not EV derived from a more differentiated CD105− tumor cell population, were able to modify tumor microenvironment and to promote development of a lung premetastatic niche [30]. Recently, they further characterized EV and observed that they favor tumor progression and metastases and are able to modulate the behavior of monocyte-derived dendritic cells and impair T-cell immune response by a mechanism involving HLA-G to the same extent as CSCs [31].
Through automated digital assessment of intratumoral microvascular density, Dubinski et al. showed that CD105 is an unfavorable prognostic marker in ccRCC [32]. Indeed, patients with higher CD105 expression level had significantly shorter progression-free survival as well as a higher tumor stage. CD105 is an important indicator of clinical outcome and could become a valuable therapeutic target following further investigation.
2.1.2. CD133
CD133 is a five-transmembrane domain glycoprotein, and its true function still remains elusive [33]. Human CD133 was first isolated from hematopoietic stem cells by a specific monoclonal antibody that recognized a specific epitope (clone AC133). CD133 currently serves as a useful marker for the isolation of many different types of stem and progenitor cells in adult human tissues, even for clinical purposes [33]. However, only antibodies recognizing the epitopes localized in the second extracellular loop, such as the AC133 and 293C3 clones used to identify human renal progenitors, are suitable for recognition of stem cells and progenitor cells, while other antibodies do not specifically recognize stem cells but also differentiated epithelial cell types [33]. As previously mentioned, CD133 is also a marker of renal progenitor cells in adult human kidney, and resident RCC CD133+ cells have been shown to promote tumor vascularization and angiogenesis [34]. Bruno et al. sorted CD133+ and CD133− cells from RCC patients and showed that CD133+ cells alone did not induce tumor formation in immunodeficient mice, but cotransplanted with cells from the K1 RCC cell line they led to enhanced development and growth of tumors. This effect was not imputed to the tumorigenic nature of CD133+ cells as identical effects were observed using CD133+ cells from normal kidney, and it was speculated that the CD133+ cells present in the tumor could have migrated from healthy kidney tissue. Interestingly, the neovasculature was from human origin, as shown by the presence of human HLA class I and CD31, indicating CD133+ cells as their origin, results corroborated by fluorescence in situ hybridization for expression of human chromosome X [34]. Galleggiante et al. identified and characterized a population of ccRCC-derived CD133+/CD24+ CSCs [35]. Gene expression profile identified copper transport protein 2 (CTR2) as a membrane marker for this neoplastic population. CTR2 in RCC patients had an important role in cisplatin-based resistance. CD133+/CD24+/CTR2+ cells did not express mesenchymal markers and were more undifferentiated than tubular adult renal progenitor cells. These cells presented in vitro self-maintenance and differentiating capabilities and could induce an angiogenic response in vivo. Lindgren et al. isolated a population of ALDHhigh cells from adult human renal cortical tissue, which also expressed CD133 and VIM [19, 29]. They showed that this CD133+/VIM+ population of tubular-committed progenitors demonstrate a significant transcriptional similarity with pRCC and coexpress VIM, keratin 7 (KRT7), and KRT19, a pattern characteristic of pRCC. Additionally, VIM+/KRT7+/KRT19+ cells were observed in cortical adenomata, benign renal neoplasms. These results suggest that pRCC develops from these progenitor cells and that cortical adenomas may represent a benign intermediate step during this oncogenic process.
Clinical significance of CD133 expression in human RCC is inconsistent and varies greatly between studies [24]. da Costa et al. used an anti-CD133 polyclonal antibody, which stained diffusely differentiated epithelial structures in embryonic as well as adult kidneys, and found that patients in the CD133 low-expression group had a higher probability of death from RCC and disease progression [36]. Conversely, D'Alterio et al. used an anti-CD133/1 monoclonal antibody that selectively recognized renal progenitors and did not see any correlation with the clinical pathological features or patient prognosis [37]. However, another two studies which used the anti-CD133 monoclonal antibody showed contrasting results. Zhang et al. observed that CD133 expression correlated with tumor grade, stage, histological type, and tumor location [38]. Prognosis of patients with CD133high, CD44high, positive vasculogenic mimicry, and low microvessel density was worse than that of patients with CD133low, CD44low, negative vasculogenic mimicry, and high microvessel density. Kim et al. reported that high levels of CD133 expression were observed in ccRCC with more differentiated morphology and were associated with a macro-/microcystic pattern, nonsarcomatoid changes, and nonmetastatic disease and therefore would consider it as a favorable prognostic marker [39]. A follow-up study by the same group showed that CD133 may serve as a favorable prognostic marker in pRCC, as it correlated with small tumor size, low Fuhrman nuclear grade, and prolonged disease-specific survival [40]. Further studies on the significance of CD133 expression in RCC are thus necessary.
2.1.3. CXCR4
CXCR4 is an alpha-chemokine receptor specific for stromal-derived factor-1 (SDF-1, also called CXCL12), a molecule endowed with potent chemotactic activity. CXCR4 has been found to be a prognostic marker in various types of cancer and plays a role in the cell proliferation and migration of cancer cells [41]. The CXCL12/CXCR4 axis forms critical communication bridges between tumor cells and stromal cells to create a permissive microenvironment for tumor growth and metastasis. Schrader et al. analyzed CXCL12α/CXCR4 expression and function in human RCC cell lines (A-498, Caki-1, Caki-2, and HA-7), patient RCC samples, and corresponding normal kidney tissue [42]. They observed that none of the four RCC cell lines expressed CXCL12, while A-498 cells expressed CXCR4. More importantly, RCC samples showed a decreased expression of CXCL12 and increased expression of CXCR4, compared to their respective adjacent normal kidney tissue, revealing a role in tumor progression. Gassenmaier et al. compared two RCC cell lines, RCC-26 and RCC-53, in their capacity to form spheres in vitro and to establish tumors in vivo [43]. They observed differences in levels of chemokine expression, as CXCR4 was present only in the more tumorigenic cell line RCC-53. CXCR4+ cells presented CSC characteristics, such as increased resistance to tyrosine kinase inhibitors, higher sphere-forming ability, and tumor growth-inducing potential in vivo, and expressed high levels of stem cell-associated makers NANOG, OCT3/4, and SOX2. Downregulation of CXCR4 expression by small interfering RNA or pharmacological inhibition by AMD3100 hindered sphere formation and reduced the viability of CXCR4+ cells [43]. Recently, Micucci et al. showed that the enhanced self-renewal activity of the CXCR4+ spheres was preceded by the upregulation of hypoxia-inducible factor 2α (HIF2α) in the RCC cell lines Caki-1, Caki-2, 786-O, and 769-P [44]. Knockdown of HIF2α abrogated CXCR4 expression and sphere formation, while inhibition of HIF2α abolished tumor growth in vivo, revealing the crucial role of HIF2α activation in CSC expansion.
Numerous studies show that an elevated CXCR4 expression in tumor samples from RCC patients is correlated with poor outcome [37, 45–47]. Additionally, CXCR4 is significantly related to the biological features of the tumor stage, including stage, Fuhrman grade, and clinical presentation [48].
2.1.4. CD44
The CD44 antigen is a cell surface glycoprotein involved in cell-cell interactions and cell adhesion and migration. A major receptor for hyaluronic acid, CD44, is an extensively described CSC marker in several human carcinomas [49]. In the human embryonic cell line 293T, Debeb et al. described CD44+/CD24− cells with several CSC features [50]. Although the nature of CSCs is still the subject of debate, CD44+ human carcinomas are highly malignant and resistant to therapy, and the presence of CD44 has been shown to confer increased metastatic potential, properties that are frequently associated with CSCs [38, 49]. CD44 is closely associated with proliferation, metastasis, cancer recurrence, and prognosis in RCC [51]. Lim et al. suggested that CD44 expression in RCC provides useful prognostic information both in primary and in metastatic RCC and may help in determining the appropriate therapy [51]. A meta-analysis of the literature performed by Li et al. revealed that elevated CD44 expression is a poor prognostic marker for five-year overall survival and correlates with high Fuhrman grade and recurrence [52]. A possible mechanism of upregulation of CD44 expression in ccRCC has been proposed in 2016 by Ma et al. [53]. They observed that about a third of tumor samples expressed CD44, with no correlation with clinical outcome, but associated with a high density of tumor-associated macrophages. In vitro experiments using RCC cell lines and human macrophages demonstrated that CD44 expression increased following direct coculture with macrophages. Silencing or suppression by NF-κB inhibitors of TNF-α on macrophages abolished the increased CD44 expression in RCC. This study suggests that TNF-α derived from tumor-associated macrophages is linked to CD44 upregulation via NF-κB signaling in ccRCC [53]. Even though the nature of CD44 as CSC marker is unclear, its role in RCC tumorigenicity remains noteworthy and further investigation might reveal interesting clinical applications.
2.2. Functional Assays
Identifying selective CSCs could be a difficult task, and several functional assays have been set up to circumvent this obstacle.
2.2.1. Sphere-Forming Capacity
Zhong et al. proposed to select CSCs from the SK-RC-42 RCC cell line on their capacity to form spheres in a serum-free medium, in the presence of epithelial growth factor and basic fibroblast growth factor [54]. This cell population expressed stem cell markers (OCT4, NANOG, BMI, and β-catenin) and was more resistant to chemotherapeutic agents and radiation than monolayer adherent cells. Sphere-forming cells displayed a distinct immunophenotype, as they did not express MHC-II, CD80, FAS, and natural killer (NK) activating receptors, indicating that they could contribute to T-cell and NK cell immune suppression. Surprisingly, both sphere-forming and monolayer adherent cells expressed comparable levels of CD133, CD44, and CD24 markers, while CD105 levels were higher in monolayer adherent cells [54]. Using the same approach, Lichner et al. showed that RCC spheres exhibit CSC properties, including self-renewal, high tumorigenicity, and differentiation capacity, as well as the expression of OCT4, NANOG, KLF4, and LIN28 [55]. Interestingly, inhibition of miR-17 enhanced RCC sphere formation, while overexpression of miR-17 hampered sphere formation, through modulation of the TGF-β-EMT axis [55, 56]. miR-17 belongs to an oncogenic microRNA (miRNA) cluster which is essential for development and homeostasis [57]. However, the dual oncogenic/tumor suppressor function of miR-17 and the other miRNAs of the family has been suggested in various tumor types, including RCC, and will require close scrutiny [58, 59]. miRNAs have an important role in posttranscriptional gene regulation and have been shown to be required for the maintenance of normal pluripotent embryonic stem cells in mice [60]. They have been described in several other types of solid tumors, such as breast cancer, and further investigation of their role in RCC tumor initiation, therapy resistance, progression, relapse, and metastasis will certainly help understand tumor biology [61].
2.2.2. Side Population Sorting
Addla et al. isolated CSCs by sorting the side population (SP) of cells that take up Hoechst 33342 dye, by flow cytometry analysis [62, 63]. This technique was previously described to identify hematopoietic stem cells (HSCs). HSCs can be distinguished by their ability to form a SP, as they are able to quickly carry out dyes efflux owing to the presence of specific membrane transporters on their surface (such as ABC transporters), a property typically associated with stemness [64]. About 6% of RCC cells could be defined as SP, and these cells expressed CD105 but not CD133 and were able to proliferate and differentiate, as well as grow into spheroids. However, the cells were found to be both in G0 and in G1 phase of the cell cycle, suggesting heterogeneity within the SP. Relationship between SP and CSCs has been questioned in several types of neoplasms, such as in glioblastoma, where SP does not contribute to self-renewal and tumorigenicity attributed to CSCs [65, 66]. Huang et al. applied this method to identify SP in five human RCC cell lines. Flow cytometry analysis revealed that the 769P cell line contained the largest SP, which presented CSC characteristics such as the ability to proliferate, self-renew, and differentiate, as well as strong resistance to chemotherapy and radiotherapy that could be linked to the ABCB1 transporter. In vivo serial tumor transplantation in immunodeficient mice showed that 769P SP cells formed tumors [67]. Hughes et al. suggested combining Hoechst SP detection with synchrotron radiation-Fourier transform infrared spectroscopy in order to measure discrete differences in the biochemistry of small numbers of single cells [68]. They showed that SP cells were very small, consisting of a nucleus and limited cytoplasm, relative to the remaining renal cells. A similar SP identification approach consists in evaluating the cell's ability to carry out toxins efflux using Rhodamine 123 (Rho123). Lu et al. separated Rho123high from Rho123low in the 786-O RCC cell line and showed that the Rho123high population formed a small subset of cells with higher proliferative activity, long-term differentiation potential, resistance to radiation, increased colony-forming capacity, and high tumorigenicity compared to Rho123low cells [69]. Rho123high could therefore be considered to be CSCs, even though they lacked CD105 expression.
Recently, SP cells from the RenCa RCC cell line were genetically modified to knock out (KO) DnaJ (Hsp40) homolog, subfamily B, member 8 (Dnajb8) and investigate its role in the tumorigenicity of RCC [70]. The authors confirmed a previously described role for this heat shock protein in the maintenance of RCC CSCs, as Dnajb8 KO cells showed reduced ratios of SP cells and reduced sphere-forming capacity [70, 71]. In vivo single-cell transplantation assay revealed a role for DNAJB8 in tumor initiation, while in vitro experiments did not indicate a function in stress responses [70]. SP cells could be used as a potent tool to assess gene functions in a wide range of experiments. Additionally, DNAJB8 emerged as a new potential CSC marker. However, no clinical significance data is available so far for either SP cells or DNAJB8.
2.2.3. Aldehyde Dehydrogenase 1 Enzymatic Activity
Recent evidence suggests that enhanced aldehyde dehydrogenase (ALDH) activity is a hallmark of CSCs, assessable by the ALDEFLUOR assay [72]. Debeb et al. cultured and passaged HEK 293T cells as spheres in serum-free stem cell-promoting medium [50]. They observed a larger number of ALDH+ cells in spheres compared to cells growing in monolayer. As mentioned earlier, these cells were also CD44+/CD24− and exhibited a CSC-like phenotype, that is, resistance to radiation and expression of higher levels of stem cell survival signaling including β-catenin, Notch1, and Survivin. Moreover, spheres have increased expression of mesenchymal genes including VIM, N-cadherin, Zeb1, Snail, and Slug as well as prometastatic genes RhoC, Tenascin C, and MTA1. Additionally, levels of miRNAs associated with self-renewal and metastasis formation were significantly decreased in the spheres. 293T cells cultured as spheres represent an important research tool for studying the molecular and biological mechanisms of CSCs and for testing and developing new targets for cancer therapy [50]. In ACHN and Caki-2 RCC cell lines, ALDHhigh cells displayed several CSC properties in vitro, that is, clonogenic and self-renewal ability and increased expression of OCT3/4A, NANOG, and Pax2. ALDHhigh cells had higher tumorigenicity in vivo [73]. Ueda et al. studied the ALDH enzymatic activity of SP from ACHN and KRC/Y RCC cell lines [74]. In the metastatic ACHN cell lines, they observed that ALDH+ cells formed about 15% of the total number of cells and had higher sphere-forming capacity, self-renewal ability, and tumorigenicity than ALDH− cells. Furthermore, SP cells were enriched in ALDH+ cells compared to non-SP cells, suggesting a certain correlation between ALDH enzymatic activity and SP. However, in the primary KRC/Y cell line, only 6.5% of the cells were ALDH+ and this proportion was similar in SP and non-SP. The population of CD133+/VIM+ cells described by Lindgren et al. and mentioned earlier were isolated from ALDHhigh adult human renal cortical tissue, underlining the importance of combining several isolation techniques [29].
While Ozbek et al. reported that ALDH1 expression was correlated with tumor grade but not with tumor stage in patients with RCC, Abourbih et al. observed that ALDH1 expression did not vary significantly based on tumor stage or grade and did not correlate with progression-free survival [75, 76]. Further investigations are necessary to determine the relationship between RCC clinical prognosis and ALDH1.
3. Cancer Stem Cell-Targeted Therapies
As previously stated, tumors from RCC patients show resistance to chemotherapy and radiation and are weakly responsive to immunotherapeutic agents such as interferon α and interleukin-12. While antiangiogenic drugs like tyrosine kinase inhibitors (TKI) have significantly improved outcome in patients with metastatic disease, the majority still presents resistance over time as the tumor develops evasion mechanisms in response to vascular endothelial growth factor (VEGF) inhibition [77]. Varna et al. have demonstrated the involvement of CD133+/CXCR4− CSCs in that process [78]. In perinecrotic areas where CSCs were numerous and predominant, the VEGF inhibitor sunitinib was able to generate resistance to its own therapeutic effect via induced hypoxia, which promotes tumor aggressiveness [18]. Indeed, hypoxia leads to the activation of HIF, which causes adaptive changes within cancer cells and aggressive behavior contributing to tumor progression and resistance and conducting to poor prognosis [44].
Therefore, it might be of great interest to develop new therapeutic drugs or immunotoxins that target CSCs, based on molecular mechanisms that regulate stem cell properties (Figure 3) [16].
Figure 3.
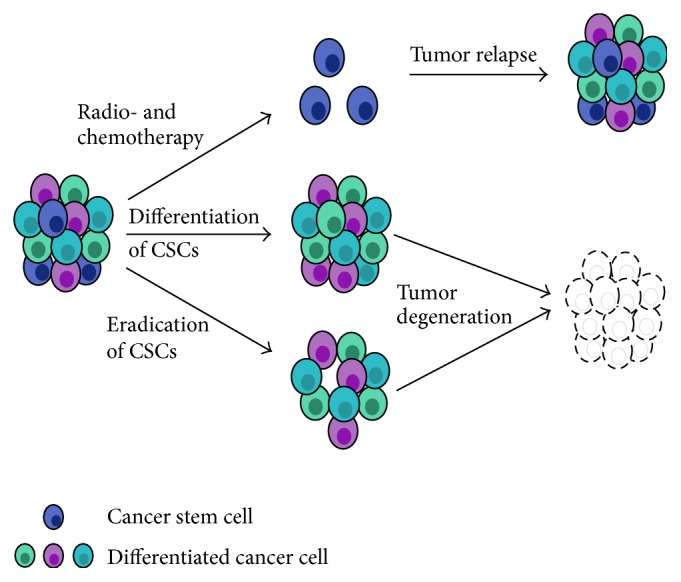
Therapies to target radio- and chemoresistant cancer stem cells. The presence of cancer stem cells (CSCs) in the tumor can lead to a relapse following conventional therapy. Newly developed strategies propose to either differentiate or eradicate CSCs, leading to the degeneration of the tumor.
3.1. Interleukin-15
The identification of reliable inducers of CSC differentiation could facilitate the elaboration of efficient strategies for eradicating CSCs. Azzi et al. demonstrated that interleukin-15 (IL-15), a regulator of kidney homeostasis, could induce the differentiation of CD105+ CSCs from human RCC [23, 79]. Previously published data on IL-15 and IL-15Rα KO mice showed that IL-15 was an autocrine survival factor for renal tubular epithelial cells [80, 81]. CD105+ CSCs treated with IL-15 lost the expression of stem cell markers and tumor-initiating and sphere-forming ability. On the other hand, they gained epithelial markers as well as functional epithelial properties such as polarity and transmembrane resistance [79]. Most importantly, CD105+ CSCs became sensitive to the chemotherapeutic drugs vinblastine and paclitaxel. By contrast, RCC neither secrete the cytokine nor express—both in vivo and in vitro—the IL-15Rγ chain and JAK3 [82]. Another in vitro study showed that IL-15 upregulated E-cadherin expression through ϒc chain signaling pathway on renal epithelial tubular cells and blocked their EMT [83].
A recently completed phase I study recorded on the National Institutes of Health (NIH) registry (NCT01021059 on https://clinicaltrials.gov/) proposed to administer intravenous recombinant human IL-15 in adults with refractory metastatic malignant melanoma and metastatic renal cell cancer. Preliminary results showed the safety and feasibility of the study [84]. IL-15 administration markedly altered the homeostasis of lymphocyte subsets in blood, in particular NK cells and γδ cells and CD8 memory T cells. To reduce toxicity and increase effectiveness of the treatment, dosing strategies have been modified, including continuous intravenous infusions and subcutaneous IL-15 administration.
3.2. mTOR Inhibitors
The mammalian target of rapamycin (mTOR) is a serine/threonine protein kinase, member of the large phosphatidylinositol 3-kinase- (PI3K-) related kinase family. It is known to regulate cell growth and cell proliferation in stem cells [85]. mTOR pathway activation leads to constitutive HIF-1α expression, an important signaling in the pathogenesis of RCC, as well as the expression of cell-cycle regulators c-myc and cyclin D1 [86–89]. mTOR inhibitors have been shown to inhibit both tumor cell proliferation and angiogenesis [90, 91]. Numerous recent studies have demonstrated the links between the PI3K/Akt/mTOR signaling pathway and CSC biology [92]. Interestingly, experiments on neuroblastoma showed that rapamycin targets specifically CSCs [93]. Similar data confirmed that mTOR inhibitors eradicated CSCs in nasopharyngeal carcinoma, colon cancer, and pancreatic cancer [94–96]. However, contrasting studies underline the putative role of CSCs in mTOR inhibitor-mediated resistance to RCC treatment, in particular in ccRCC [86]. A recent study detailed this resistance mechanism in breast cancer [97]. Accumulative evidence shows that mTOR is an important pathway in carcinogenesis, and a better understanding of the effects of mTOR inhibition in RCC will lead to more successful therapies.
On the NIH database, as many as 36 phase I, II, and III clinical trials, ongoing or completed, propose to use mTOR inhibitors to treat RCC. The phase III trial on treatment-resistant patients with metastatic RCC showed safety and benefits of everolimus over placebo (NCT00410124). Multiple combination treatments have been proposed to lower everolimus toxicity while maintaining a sufficient efficiency [91]. The advantage of such an approach is that it can modulate different signaling pathways involved in cancer biology and target both CSCs and cancer cells for a more global effect.
3.3. Bone Morphogenetic Protein 2
The bone morphogenetic protein 2 (Bmp-2) encodes a member of the TGF-β superfamily. It plays an important role in the development of bone and cartilage, as well as in the regulation of various cellular processes including cell differentiation, proliferation, morphogenesis, cellular survival, and apoptosis [98]. BMP-2 has been reported to either stimulate or inhibit tumor growth, depending on cancer type [99]. Wang et al. showed that BMP-2 could inhibit tumorigenicity of CSCs in human osteosarcoma OS99-1 cells, inhibit tumor growth of human RCC, and induce bone formation [100–102]. In a follow-up study, the authors evaluated whether BMP-2 can be used to block the tumor-initiating ability of human renal ALDH+ CSCs in vitro and induce bone formation in vivo [99]. They found that BMP-2 inhibited CSC growth, downregulated the expression of stem cell markers, and upregulated the transcription of osteogenic markers. ACHN and Caki-2 RCC cells implanted in immunodeficient mice developed into large tumors, while animals treated with BMP-2 presented limited growth and important bone formation, indicating BMP-2 as a potential new drug targeting CSCs in RCC. A recent study by Mitsui et al. revealed that impaired regulation of Bmp-2 via epigenetic pathways was associated with RCC pathogenesis and confirms the usefulness of BMP-2 as a molecular marker for designing improved diagnostic and therapeutic strategies for RCC [103]. Several clinical trials involving BMP-2 treatment have been registered, primarily for patients with fractured and degenerative disk disease, but none for cancer patients.
3.4. Other Possible Therapeutic Targets
Numerous other strategies have been proposed to target CSCs but have not been experimented with on RCC so far [25, 104, 105]. They include targeting ATP-driven efflux transporters or cell surface markers, inhibiting CSC-related signaling pathways, inhibiting autophagy signaling in CSCs, increasing bioavailability of CSC-specific agents, delivering CSC-specific chemotherapeutics or nucleic acid drugs, and using CSC-targeted nanocarriers. Among those, a few stand out and deserve attention.
3.4.1. ATP-Driven Efflux Transporter Inhibition
As previously stated, CSCs present high levels of ABC transporters, which might be involved in drug resistance by decreasing the cellular accumulation of therapeutic agents [106]. Several groups have attempted to eradicate CSCs using the low molecular weight inhibitors fumitremorgin C and tryprostatin or monoclonal antibodies such as cyclosporin A, VX710, or tariquidar [104, 107, 108]. Their use in clinical settings has been hindered by low inhibition efficiency and elevated toxicity to healthy cells.
3.4.2. Cell Surface Marker Inhibition
Monoclonal antibodies and inhibitors have been proposed to block CSC surface markers. Jin et al. used an activating antibody directed to CD44 to inject into immunodeficient mice transplanted with acute myeloid leukemia and observed lower leukemic repopulation [104, 109]. Therapies against CD133 have been successfully used to treat lung cancer, glioblastoma, and liver cancer [110–112]. Due to the few experiments performed using this strategy, further studies must be carried out to validate this approach.
3.4.3. Salinomycin Treatment
Salinomycin is a biologically active substance isolated from the culture broth of a strain of Streptomyces albus. Gupta et al. identified salinomycin as a selective inhibitor of human breast CSCs by high-throughput screening [113, 114]. Successive studies demonstrated that salinomycin could kill CSCs in different types of human cancers including gastric cancer, lung adenocarcinoma, osteosarcoma, colorectal cancer, squamous cell carcinoma, and prostate cancer, most likely by interfering with ABC drug transporters, the Wnt/β-catenin signaling pathway, and other CSC pathways. Promising results from preclinical trials in human RCC show that salinomycin is able to effectively eliminate CSCs and to induce partial clinical regression of heavily pretreated and therapy-resistant cancers [113, 115].
3.4.4. Nanomedicine-Based Therapies
Recently, nanomedicine-based therapies have been developed to target CSCs [104]. Nanoparticles can be used as high-capacity carriers for chemotherapeutic or nucleic acid drugs and can accumulate at tumor sites through two different mechanisms: a passive one, that is, enhanced permeation retention due to the higher porosity of the vasculature and impaired lymphatic drainage, or an active one, due to the presence on the nanoparticles of molecules of high affinity for the receptors exclusively overexpressed on the surface of CSCs [116, 117]. As an example, it has been proposed to incorporate salinomycin into nanocarriers in order to lower its toxicity. Yao et al. developed a drug delivery system which targets specifically gastric CSCs [118]. Indeed, chitosan-coated single wall carbon nanotubes loaded with salinomycin functionalized with hyaluronic acid could selectively eliminate CD44+ gastric CSCs in vitro. As a combination therapy, Zhang et al. used octreotide-modified paclitaxel-loaded PEG-b-PCL polymeric micelles and salinomycin-loaded PEG-b-PCL polymeric micelles to eradicate breast cancer cells [119]. While paclitaxel eliminated the bulk of cancer cells, salinomycin targeted CSCs, resulting in a stronger antitumoral activity than either drug separately in vitro and in vivo. Numerous studies in a wide range of cancers showed the efficiency of nanoparticle-based therapies to target CSCs [104].
Nanoparticles are becoming a prominent player in the fight against tumors, as more than 150 clinical trials registered with the NIH propose this innovative technology combined with more conventional therapies to treat cancer.
Despite important progress being made in developing CSC-targeted therapies, very few discoveries have been translated to a clinical setting, underlining the complexity of the mechanisms involved in carcinogenesis. However, the major mobilization of the scientific community suggests that this approach is worth exploring notwithstanding the numerous setbacks.
4. Conclusions
A vast body of evidence supports the existence within renal tumors of CSCs with self-renewal and multidifferentiation properties. Various approaches have been described to successfully isolate and characterize CSCs, leading to the identification of a variety of CSCs and suggesting that several subpopulations of CSCs may coexist within a heterogeneous tumor. Combination of stem cell markers with functional assays permits a better identification of a smaller population enriched in CSCs, but important limitations have been observed [22, 120]. For example, functional assays present important variations in their staining protocols, from dye concentration to incubation time, leading to important discrepancies in the results [66]. Additionally, culture conditions as well as cell detachment methods in in vitro experiments must be optimized and standardized in order not to affect CSC phenotype [121–123]. Despite the aforementioned caveats, identification of CSCs led to a better understanding of carcinogenesis.
It is worth noticing that the techniques developed to isolate and study CSCs are all performed in vitro, eventually followed by implantation of the cells into immunodeficient mice to test their tumor forming capacity. However, most studies involving RCC patient samples failed to verify that CSCs could be xenografted into immunodeficient mice and could produce serially transplantable tumors that recapitulate the original tumoral tissue. Interestingly, Hasmim et al. proposed an alternative strategy for identifying renal CSCs by adapting, to in vitro culture, primary cell suspensions from serial patient-derived xenografts obtained by implanting RCC samples in immunodeficient mice [124]. They isolated 3 different CSC subsets of cells, which presented different characteristics and confirmed the heterogeneity of the CSC population. Indeed, all 3 populations were ALDH+ and formed serial spheroids but expressed different combinations of stem cell markers (CD133, CD105, CD146, CD29, OCT4, NANOG, and Nestin) and the non-CSC tumor marker E-cadherin. Admirably, all 3 subsets developed serial tumors in SCID mice and therefore successfully encompassed all characteristics of CSCs. This approach is gaining attention within the scientific community, as the use of patient-derived xenografts will allow in the future the development of personalized medicine, that is, unique treatment regimens for individual patients [125].
While their clinical significance remains elusive at the moment, CSCs undoubtedly play an important role in tumor resistance to radiotherapy and chemotherapy. To circumvent this drawback in conventional therapy, several research groups are taking advantage of CSCs unique properties to design new drug compounds that selectively target CSCs. A few therapies, such as IL-15 and mTOR administration, are already being tested in clinical trials to treat RCC, providing new prospects for patients.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.Coleman M. P., Esteve J., Damiecki P., Arslan A., Renard H. Trends in Cancer Incidence and Mortality. IARC Scientific; 1993. [DOI] [PubMed] [Google Scholar]
- 2.Znaor A., Lortet-Tieulent J., Laversanne M., Jemal A., Bray F. International variations and trends in renal cell carcinoma incidence and mortality. European Urology. 2015;67:519–530. doi: 10.1016/j.eururo.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Yap N. Y., Rajandram R., Ng K. L., Pailoor J., Fadzli A., Gobe G. C. Genetic and chromosomal aberrations and their clinical significance in renal neoplasms. BioMed Research International. 2015;2015:22. doi: 10.1155/2015/476508.476508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs G., Akhtar M., Beckwith B. J., et al. The Heidelberg classification of renal cell tumours. The Journal of Pathology. 1997;183(2):131–133. doi: 10.1002/(sici)1096-9896(199710)183:2<131::aid-path931>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Delahunt B., Srigley J. R. The evolving classification of renal cell neoplasia. Seminars in Diagnostic Pathology. 2015;32(2):90–102. doi: 10.1053/j.semdp.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Srigley J. R., Delahunt B., Eble J. N., et al. The international society of urological pathology (ISUP) vancouver classification of renal neoplasia. The American Journal of Surgical Pathology. 2013;37(10):1469–1489. doi: 10.1097/pas.0b013e318299f2d1. [DOI] [PubMed] [Google Scholar]
- 7.Jonasch E., Gao J., Rathmell W. K. Renal cell carcinoma. The British Medical Journal. 2014;349 doi: 10.1136/bmj.g4797.g4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son S. H. Y., Kim D.-H., Hong C. M. O., et al. Prognostic implication of intratumoral metabolic heterogeneity in invasive ductal carcinoma of the breast. BMC cancer. 2014;14, article 585 doi: 10.1186/1471-2407-14-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel A. P., Tirosh I., Trombetta J. J., et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H., Laird P. W. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baylin S. B., Jones P. A. A decade of exploring the cancer epigenome—biological and translational implications. Nature Reviews Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd E. A., Grigsby P. W. Intratumoral metabolic heterogeneity of cervical cancer. Clinical Cancer Research. 2008;14(16):5236–5241. doi: 10.1158/1078-0432.CCR-07-5252. [DOI] [PubMed] [Google Scholar]
- 13.Collins L. C., Botero M. L., Schnitt S. J. Bimodal frequency distribution of estrogen receptor immunohistochemical staining results in breast cancer: an analysis of 825 cases. American Journal of Clinical Pathology. 2005;123(1):16–20. doi: 10.1309/hcf0-35n9-wk40-etj0. [DOI] [PubMed] [Google Scholar]
- 14.Visvader J. E. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 15.Kreso A., Dick J. E. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Shukrun R., Pode Shakked N., Dekel B. Targeted therapy aimed at cancer stem cells: wilms' tumor as an example. Pediatric Nephrology. 2014;29(5):815–823. doi: 10.1007/s00467-013-2501-0. [DOI] [PubMed] [Google Scholar]
- 17.Rycaj K., Tang D. G. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Research. 2015;75(19):4003–4011. doi: 10.1158/0008-5472.can-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myszczyszyn A., Czarnecka A. M., Matak D., et al. The role of hypoxia and cancer stem cells in renal cell carcinoma pathogenesis. Stem Cell Reviews and Reports. 2015;11:919–943. doi: 10.1007/s12015-015-9611-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Axelson H., Johansson M. E. Renal stem cells and their implications for kidney cancer. Seminars in Cancer Biology. 2013;23(1):56–61. doi: 10.1016/j.semcancer.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Bu Y., Cao D. The origin of cancer stem cells. Frontiers in Bioscience. 2012;4:819–830. doi: 10.2741/s302. [DOI] [PubMed] [Google Scholar]
- 21.Alison M. R., Lim S. M. L., Nicholson L. J. Cancer stem cells: problems for therapy? The Journal of Pathology. 2011;223(2):147–161. doi: 10.1002/path.2793. [DOI] [PubMed] [Google Scholar]
- 22.Khan M. I., Czarnecka A. M., Helbrecht I., Bartnik E., Lian F., Szczylik C. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Research and Therapy. 2015;6(1, article 178) doi: 10.1186/s13287-015-0177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bussolati B., Dekel B., Azzarone B., Camussi G. Human renal cancer stem cells. Cancer Letters. 2013;338(1):141–146. doi: 10.1016/j.canlet.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Matak D., Szymanski L., Szczylik C., et al. Biology of renal tumour cancer stem cells applied in medicine. Contemporary Oncology. 2015;19(1):A44–A51. doi: 10.5114/wo.2014.47128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida G. J., Saya H. Therapeutic strategies targeting cancer stem cells. Cancer Science. 2016;107(1):5–11. doi: 10.1111/cas.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallas N. A., Samuel S., Xia L., et al. Endoglin (CD105): a marker of tumor vasculature and potential target for therapy. Clinical Cancer Research. 2008;14(7):1931–1937. doi: 10.1158/1078-0432.ccr-07-4478. [DOI] [PubMed] [Google Scholar]
- 27.Bussolati B., Bruno S., Grange C., Ferrando U., Camussi G. Identification of a tumor-initiating stem cell population in human renal carcinomas. The FASEB Journal. 2008;22(10):3696–3705. doi: 10.1096/fj.08-102590. [DOI] [PubMed] [Google Scholar]
- 28.Angelotti M. L., Ronconi E., Ballerini L., et al. Characterization of renal progenitors committed toward tubular lineage and their regenerative potential in renal tubular injury. Stem Cells. 2012;30(8):1714–1725. doi: 10.1002/stem.1130. [DOI] [PubMed] [Google Scholar]
- 29.Lindgren D., Boström A., Nilsson K., et al. Isolation and characterization of progenitor-like cells from human renal proximal tubules. The American Journal of Pathology. 2011;178(2):828–837. doi: 10.1016/j.ajpath.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grange C., Tapparo M., Collino F., et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Research. 2011;71(15):5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 31.Grange C., Tapparo M., Tritta S., et al. Role of HLA-G and extracellular vesicles in renal cancer stem cell-induced inhibition of dendritic cell differentiation. BMC Cancer. 2015;15, article 1009 doi: 10.1186/s12885-015-2025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubinski W., Gabril M., Iakovlev V. V., et al. Assessment of the prognostic significance of endoglin (CD105) in clear cell renal cell carcinoma using automated image analysis. Human Pathology. 2012;43(7):1037–1043. doi: 10.1016/j.humpath.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Yin A. H., Miraglia S., Zanjani E. D., et al. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90(12):5002–5012. [PubMed] [Google Scholar]
- 34.Bruno S., Bussolati B., Grange C., et al. CD133+ renal progenitor cells contribute to tumor angiogenesis. The American Journal of Pathology. 2006;169(6):2223–2235. doi: 10.2353/ajpath.2006.060498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galleggiante V., Rutigliano M., Sallustio F., et al. CTR2 identifies a population of cancer cells with stem cell-like features in patients with clear cell renal cell carcinoma. The Journal of Urology. 2014;192(6):1831–1841. doi: 10.1016/j.juro.2014.06.070. [DOI] [PubMed] [Google Scholar]
- 36.da Costa W. H., Rocha R. M., da Cunha I. W., da Fonseca F. P., Guimaraes G. C., Zequi S. D. C. CD133 immunohistochemical expression predicts progression and cancer-related death in renal cell carcinoma. World Journal of Urology. 2012;30(4):553–558. doi: 10.1007/s00345-011-0769-x. [DOI] [PubMed] [Google Scholar]
- 37.D'Alterio C., Cindolo L., Portella L., et al. Differential role of CD133 and CXCR4 in renal cell carcinoma. Cell Cycle. 2010;9(22):4492–4500. doi: 10.4161/cc.9.22.13680. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y., Sun B., Zhao X., et al. Clinical significances and prognostic value of cancer stem-like cells markers and vasculogenic mimicry in renal cell carcinoma. Journal of Surgical Oncology. 2013;108(6):414–419. doi: 10.1002/jso.23402. [DOI] [PubMed] [Google Scholar]
- 39.Kim K., Ihm H., Ro J. Y., Cho Y. M. High-level expression of stem cell marker CD133 in clear cell renal cell carcinoma with favorable prognosis. Oncology Letters. 2011;2(6):1095–1100. doi: 10.3892/ol.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim K., Ro J. Y., Kim S., Cho Y. M. Expression of stem-cell markers OCT-4 and CD133: important prognostic factors in papillary renal cell carcinoma. Human Pathology. 2012;43(12):2109–2116. doi: 10.1016/j.humpath.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Guo F., Wang Y., Liu J., Mok S. C., Xue F., Zhang W. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35:816–826. doi: 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- 42.Schrader A. J., Lechner O., Templin M., et al. CXCR4/CXCL12 expression and signalling in kidney cancer. British Journal of Cancer. 2002;86(8):1250–1256. doi: 10.1038/sj.bjc.6600221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gassenmaier M., Chen D., Buchner A., et al. CXC chemokine receptor 4 is essential for maintenance of renal cell carcinoma-initiating cells and predicts metastasis. Stem Cells. 2013;31(8):1467–1476. doi: 10.1002/stem.1407. [DOI] [PubMed] [Google Scholar]
- 44.Micucci C., Matacchione G., Valli D., Orciari S., Catalano A. HIF2α is involved in the expansion of CXCR4-positive cancer stem-like cells in renal cell carcinoma. British Journal of Cancer. 2015;113(8):1178–1185. doi: 10.1038/bjc.2015.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G., Badin G., Zhao A., et al. Prognostic value of CXCR4 expression in patients with clear cell renal cell carcinoma. Histology and Histopathology. 2013;28(9):1217–1222. doi: 10.14670/HH-28.1217. [DOI] [PubMed] [Google Scholar]
- 46.Wang L., Chen W., Gao L., et al. High expression of CXCR4, CXCR7 and SDF-1 predicts poor survival in renal cell carcinoma. World Journal of Surgical Oncology. 2012;10, article 212 doi: 10.1186/1477-7819-10-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zagzag D., Krishnamachary B., Yee H., et al. Stromal cell-derived factor-1α and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Research. 2005;65(14):6178–6188. doi: 10.1158/0008-5472.can-04-4406. [DOI] [PubMed] [Google Scholar]
- 48.Ottaiano A. Finding markers for cancer stem cells in renal cell carcinoma: looking beyond CD133. Cell Cycle. 2010;9(22):p. 4431. doi: 10.4161/cc.9.22.13823. [DOI] [PubMed] [Google Scholar]
- 49.Toole B. P. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clinical Cancer Research. 2009;15(24):7462–7468. doi: 10.1158/1078-0432.ccr-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debeb B. G., Zhang X., Krishnamurthy S., et al. Characterizing cancer cells with cancer stem cell-like features in 293T human embryonic kidney cells. Molecular Cancer. 2010;9, article 180 doi: 10.1186/1476-4598-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim S. D., Young A. N., Paner G. P., Amin M. B. Prognostic role of CD44 cell adhesion molecule expression in primary and metastatic renal cell carcinoma: a clinicopathologic study of 125 cases. Virchows Archiv. 2008;452(1):49–55. doi: 10.1007/s00428-007-0530-4. [DOI] [PubMed] [Google Scholar]
- 52.Li X., Ma X., Chen L., et al. Prognostic value of CD44 expression in renal cell carcinoma: a systematic review and meta-analysis. Scientific Reports. 2015;5 doi: 10.1038/srep13157.13157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma C., Komohara Y., Ohnishi K., et al. Infiltration of tumor-associated macrophages is involved in CD44 expression in clear cell renal cell carcinoma. Cancer Science. 2016 doi: 10.1111/cas.12917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong Y., Guan K., Guo S., et al. Spheres derived from the human SK-RC-42 renal cell carcinoma cell line are enriched in cancer stem cells. Cancer Letters. 2010;299(2):150–160. doi: 10.1016/j.canlet.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 55.Lichner Z., Saleh C., Subramaniam V., Seivwright A., Prud'homme G. J., Yousef G. M. miR-17 inhibition enhances the formation of kidney cancer spheres with stem cell/tumor initiating cell properties. Oncotarget. 2015;6:5567–5581. doi: 10.18632/oncotarget.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrocca F., Vecchione A., Croce C. M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor β signaling. Cancer Research. 2008;68(20):8191–8194. doi: 10.1158/0008-5472.can-08-1768. [DOI] [PubMed] [Google Scholar]
- 57.Concepcion C. P., Bonetti C., Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer Journal. 2012;18(3):262–267. doi: 10.1097/ppo.0b013e318258b60a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chow T.-F. F., Mankaruos M., Scorilas A., et al. The miR-17-92 cluster is over expressed in and has an oncogenic effect on renal cell carcinoma. The Journal of Urology. 2010;183(2):743–751. doi: 10.1016/j.juro.2009.09.086. [DOI] [PubMed] [Google Scholar]
- 59.Girgis A. H., Iakovlev V. V., Beheshti B., et al. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer Research. 2012;72(20):5273–5284. doi: 10.1158/0008-5472.CAN-12-0656. [DOI] [PubMed] [Google Scholar]
- 60.Melton C., Judson R. L., Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu C., Tang D. G. MicroRNA regulation of cancer stem cells. Cancer Research. 2011;71(18):5950–5954. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Addla S. K., Brown M. D., Hart C. A., Ramani V. A. C., Clarke N. W. Characterization of the Hoechst 33342 side population from normal and malignant human renal epithelial cells. American Journal of Physiology—Renal Physiology. 2008;295(3):F680–F687. doi: 10.1152/ajprenal.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oates J. E., Grey B. R., Addla S. K., et al. Hoechst 33342 side population identification is a conserved and unified mechanism in urological cancers. Stem Cells and Development. 2009;18(10):1515–1522. doi: 10.1089/scd.2008.0302. [DOI] [PubMed] [Google Scholar]
- 64.Goodell M. A., Brose K., Paradis G., Conner A. S., Mulligan R. C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. Journal of Experimental Medicine. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broadley K. W. R., Hunn M. K., Farrand K. J., et al. Side population is not necessary or sufficient for a cancer stem cell phenotype in glioblastoma multiforme. Stem Cells. 2011;29(3):452–461. doi: 10.1002/stem.582. [DOI] [PubMed] [Google Scholar]
- 66.Wu C., Alman B. A. Side population cells in human cancers. Cancer Letters. 2008;268(1):1–9. doi: 10.1016/j.canlet.2008.03.048. [DOI] [PubMed] [Google Scholar]
- 67.Huang B., Huang Y. J., Yao Z. J., et al. Cancer stem cell-like side population cells in clear cell renal cell carcinoma cell line 769P. PLoS ONE. 2013;8(7) doi: 10.1371/journal.pone.0068293.e68293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hughes C., Liew M., Sachdeva A., et al. SR-FTIR spectroscopy of renal epithelial carcinoma side population cells displaying stem cell-like characteristics. Analyst. 2010;135(12):3133–3141. doi: 10.1039/c0an00574f. [DOI] [PubMed] [Google Scholar]
- 69.Lu J., Cui Y., Zhu J., He J., Zhou G., Yue Z. Biological characteristics of Rh123high stem-like cells in a side population of 786-O renal carcinoma cells. Oncology Letters. 2013;5(6):1903–1908. doi: 10.3892/ol.2013.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamashita M., Hirohashi Y., Torigoe T., et al. Dnajb8, a member of the heat shock protein 40 family has a role in the tumor initiation and resistance to docetaxel but is dispensable for stress response. PLoS ONE. 2016;11(1) doi: 10.1371/journal.pone.0146501.e0146501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishizawa S., Hirohashi Y., Torigoe T., et al. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem-like cells. Cancer Research. 2012;72(11):2844–2854. doi: 10.1158/0008-5472.CAN-11-3062. [DOI] [PubMed] [Google Scholar]
- 72.Marcato P., Dean C. A., Giacomantonio C. A., Lee P. W. K. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10(9):1378–1384. doi: 10.4161/cc.10.9.15486. [DOI] [PubMed] [Google Scholar]
- 73.Wang L., Park P., Zhang H., La Marca F., Lin C. Y. Characterization of renal tumor-initiating cells in human renal carcinoma cell lines. Cancer Research. 2011;71(supplement 8):p. 293. [Google Scholar]
- 74.Ueda K., Ogasawara S., Akiba J., et al. Aldehyde dehydrogenase 1 identifies cells with cancer stem cell-like properties in a human renal cell carcinoma cell line. PLoS ONE. 2013;8(10) doi: 10.1371/journal.pone.0075463.e75463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abourbih S., Sircar K., Tanguay S., et al. Aldehyde dehydrogenase 1 expression in primary and metastatic renal cell carcinoma: an immunohistochemistry study. World Journal of Surgical Oncology. 2013;11, article 298 doi: 10.1186/1477-7819-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ozbek E., Calik G., Otunctemur A., et al. Stem cell markers aldehyde dehydrogenase type 1 and nestin expressions in renal cell cancer. Archivio Italiano di Urologia e Andrologia. 2012;84(1):7–11. [PubMed] [Google Scholar]
- 77.Buczek M., Escudier B., Bartnik E., Szczylik C., Czarnecka A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: from the patient's bed to molecular mechanisms. Biochimica et Biophysica Acta—Reviews on Cancer. 2014;1845(1):31–41. doi: 10.1016/j.bbcan.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Varna M., Gapihan G., Feugeas J.-P., et al. Stem cells increase in numbers in perinecrotic areas in human renal cancer. Clinical Cancer Research. 2015;21(4):916–924. doi: 10.1158/1078-0432.CCR-14-0666. [DOI] [PubMed] [Google Scholar]
- 79.Azzi S., Bruno S., Giron-Michel J., et al. Differentiation therapy: targeting human renal cancer stem cells with interleukin 15. Journal of the National Cancer Institute. 2010;103(24):1884–1898. doi: 10.1093/jnci/djr451. [DOI] [PubMed] [Google Scholar]
- 80.Eini H., Tejman-Yarden N., Lewis E. C., Chaimovitz C., Zlotnik M., Douvdevani A. Association between renal injury and reduced interleukin-15 and interleukin-15 receptor levels in acute kidney injury. Journal of Interferon and Cytokine Research. 2010;30(1):1–8. doi: 10.1089/jir.2009.0005. [DOI] [PubMed] [Google Scholar]
- 81.Shinozaki M., Hirahashi J., Lebedeva T., et al. IL-15, a survival factor for kidney epithelial cells, counteracts apoptosis and inflammation during nephritis. The Journal of Clinical Investigation. 2002;109(7):951–960. doi: 10.1172/jci200214574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giron-Michel J., Azzi S., Ferrini S., et al. Interleukin-15 is a major regulator of the cell-microenvironment interactions in human renal homeostasis. Cytokine and Growth Factor Reviews. 2013;24(1):13–22. doi: 10.1016/j.cytogfr.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 83.Giron-Michel J., Azzi S., Khawam K., et al. Interleukin-15 plays a central role in human kidney physiology and cancer through the γc signaling pathway. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0031624.e31624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conlon K. C., Lugli E., Welles H. C., et al. Redistribution, hyperproliferation, activation of natural killer cells and CD8 T cells, and cytokine production during first-in-human clinical trial of recombinant human interleukin-15 in patients with cancer. Journal of Clinical Oncology. 2015;33(1):74–82. doi: 10.1200/JCO.2014.57.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ochocki J. D., Simon M. C. Nutrient-sensing pathways and metabolic regulation in stem cells. Journal of Cell Biology. 2013;203(1):23–33. doi: 10.1083/jcb.201303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kornakiewicz A., Solarek W., Bielecka Z. F., Lian F., Szczylik C., Czarnecka A. M. Mammalian target of rapamycin inhibitors resistance mechanisms in clear cell renal cell carcinoma. Current Signal Transduction Therapy. 2013;8(3):210–218. doi: 10.2174/1574362409666140206222746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Posadas E. M., Limvorasak S., Sharma S., Figlin R. A. Targeting angiogenesis in renal cell carcinoma. Expert Opinion on Pharmacotherapy. 2013;14(16):2221–2236. doi: 10.1517/14656566.2013.832202. [DOI] [PubMed] [Google Scholar]
- 88.Choo A. Y., Yoon S.-O., Kim S. G., Roux P. P., Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(45):17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu Q. J., Shen H. L., Lin J., et al. Synergistic roles of p53 and HIF1α in human renal cell carcinoma-cell apoptosis responding to the inhibition of mTOR and MDM2 signaling pathways. Drug Design, Development and Therapy. 2016;10:745–755. doi: 10.2147/DDDT.S88779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salvadori M. Antineoplastic effects of mammalian target of rapamycine inhibitors. World Journal of Transplantation. 2012;2:74–83. doi: 10.5500/wjt.v2.i5.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Czarnecka A. M., Kornakiewicz A., Lian F., Szczylik C. Future perspectives for mTOR inhibitors in renal cell cancer treatment. Future Oncology. 2015;11(5):801–817. doi: 10.2217/fon.14.303. [DOI] [PubMed] [Google Scholar]
- 92.Xia P., Xu X.-Y. PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. American Journal of Cancer Research. 2015;5(5):1602–1609. [PMC free article] [PubMed] [Google Scholar]
- 93.Smith K. M., Datti A., Fujitani M., et al. Selective targeting of neuroblastoma tumour-initiating cells by compounds identified in stem cell-based small molecule screens. EMBO Molecular Medicine. 2010;2(9):371–384. doi: 10.1002/emmm.201000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sharma N., Nanta R., Sharma J., et al. PI3K/AKT/mTOR and sonic hedgehog pathways cooperate together to inhibit human pancreatic cancer stem cell characteristics and tumor growth. Oncotarget. 2015;6(31):32039–32060. doi: 10.18632/oncotarget.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen J., Shao R., Li F., et al. PI3K/Akt/mTOR pathway dual inhibitor BEZ235 suppresses the stemness of colon cancer stem cells. Clinical and Experimental Pharmacology and Physiology. 2015;42(12):1317–1326. doi: 10.1111/1440-1681.12493. [DOI] [PubMed] [Google Scholar]
- 96.Yang C., Zhang Y., Zhang Y., et al. Downregulation of cancer stem cell properties via mTOR signaling pathway inhibition by rapamycin in nasopharyngeal carcinoma. International Journal of Oncology. 2015;47(3):909–917. doi: 10.3892/ijo.2015.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhola N. E., Jansen V. M., Koch J. P., et al. Treatment of triple-negative breast cancer with TORC1/2 inhibitors sustains a drug-resistant and notch-dependent cancer stem cell population. Cancer Research. 2016;76(2):440–452. doi: 10.1158/0008-5472.can-15-1640-t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kang M. H., Kim J. S., Seo J. E., Oh S. C., Yoo Y. A. BMP2 accelerates the motility and invasiveness of gastric cancer cells via activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. Experimental Cell Research. 2010;316(1):24–37. doi: 10.1016/j.yexcr.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 99.Wang L., Park P., La Marca F., Than K. D., Lin C.-Y. BMP-2 inhibits tumor-initiating ability in human renal cancer stem cells and induces bone formation. Journal of Cancer Research and Clinical Oncology. 2015;141(6):1013–1024. doi: 10.1007/s00432-014-1883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang L., Park P., Marca F. L., Than K., Rahman S., Lin C.-Y. Bone formation induced by BMP-2 in human osteosarcoma cells. International Journal of Oncology. 2013;43(4):1095–1102. doi: 10.3892/ijo.2013.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang L., Park P., Zhang H., et al. BMP-2 inhibits tumor growth of human renal cell carcinoma and induces bone formation. International Journal of Cancer. 2012;131(8):1941–1950. doi: 10.1002/ijc.27444. [DOI] [PubMed] [Google Scholar]
- 102.Wang L., Park P., Zhang H., et al. BMP-2 inhibits the tumorigenicity of cancer stem cells in human osteosarcoma OS99-1 cell line. Cancer Biology and Therapy. 2011;11(5):457–463. doi: 10.4161/cbt.11.5.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mitsui Y., Hirata H., Arichi N., et al. Inactivation of bone morphogenetic protein 2 may predict clinical outcome and poor overall survival for renal cell carcinoma through epigenetic pathways. Oncotarget. 2015;6(11):9577–9591. doi: 10.18632/oncotarget.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shen S., Xia J.-X., Wang J. Nanomedicine-mediated cancer stem cell therapy. Biomaterials. 2016;74:1–18. doi: 10.1016/j.biomaterials.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 105.Liu H., Lv L., Yang K. Chemotherapy targeting cancer stem cells. American Journal of Cancer Research. 2015;5(3):880–893. [PMC free article] [PubMed] [Google Scholar]
- 106.Gottesman M. M., Fojo T., Bates S. E. Multidrug resistance in cancer: role of ATP-dependent transporters. Nature Reviews Cancer. 2002;2(1):48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 107.Woehlecke H., Osada H., Herrmann A., Lage H. Reversal of breast cancer resistance protein-mediated drug resistance by tryprostatin A. International Journal of Cancer. 2003;107(5):721–728. doi: 10.1002/ijc.11444. [DOI] [PubMed] [Google Scholar]
- 108.Rabindran S. K., Ross D. D., Doyle L. A., Yang W., Greenberger L. M. Fumitremorgin C reverses multidrug resistance in cells transfected with the breast cancer resistance protein. Cancer Research. 2000;60(1):47–50. [PubMed] [Google Scholar]
- 109.Jin L., Hope K. J., Zhai Q., Smadja-Joffe F., Dick J. E. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nature Medicine. 2006;12(10):1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 110.Brescia P., Ortensi B., Fornasari L., Levi D., Broggi G., Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31(5):857–869. doi: 10.1002/stem.1317. [DOI] [PubMed] [Google Scholar]
- 111.Bertolini G., Roz L., Perego P., et al. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(38):16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rountree C. B., Ding W., He L., Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. STEM CELLS. 2009;27(2):290–299. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Naujokat C., Steinhart R. Salinomycin as a drug for targeting human cancer stem cells. Journal of Biomedicine and Biotechnology. 2012;2012:17. doi: 10.1155/2012/950658.950658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta P. B., Onder T. T., Jiang G., et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boehmerle W., Muenzfeld H., Springer A., Huehnchen P., Endres M. Specific targeting of neurotoxic side effects and pharmacological profile of the novel cancer stem cell drug salinomycin in mice. Journal of Molecular Medicine. 2014;92(8):889–900. doi: 10.1007/s00109-014-1155-0. [DOI] [PubMed] [Google Scholar]
- 116.Xia P. Surface markers of cancer stem cells in solid tumors. Current Stem Cell Research and Therapy. 2014;9(2):102–111. doi: 10.2174/1574888X09666131217003709. [DOI] [PubMed] [Google Scholar]
- 117.Gao Z., Zhang L., Sun Y. Nanotechnology applied to overcome tumor drug resistance. Journal of Controlled Release. 2012;162(1):45–55. doi: 10.1016/j.jconrel.2012.05.051. [DOI] [PubMed] [Google Scholar]
- 118.Yao H.-J., Zhang Y.-G., Sun L., Liu Y. The effect of hyaluronic acid functionalized carbon nanotubes loaded with salinomycin on gastric cancer stem cells. Biomaterials. 2014;35(33):9208–9223. doi: 10.1016/j.biomaterials.2014.07.033. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y., Zhang H., Wang X., Wang J., Zhang X., Zhang Q. The eradication of breast cancer and cancer stem cells using octreotide modified paclitaxel active targeting micelles and salinomycin passive targeting micelles. Biomaterials. 2012;33(2):679–691. doi: 10.1016/j.biomaterials.2011.09.072. [DOI] [PubMed] [Google Scholar]
- 120.Panchision D. M., Chen H.-L., Pistollato F., Papini D., Ni H.-T., Hawley T. S. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells. 2007;25(6):1560–1570. doi: 10.1634/stemcells.2006-0260. [DOI] [PubMed] [Google Scholar]
- 121.Pallini R., Casalbore P., Mercanti D., Maggiano N., Larocca L. M. Phenotypic change of human cultured meningioma cells. Journal of Neuro-Oncology. 2000;49(1):9–17. doi: 10.1023/A:1006436903976. [DOI] [PubMed] [Google Scholar]
- 122.Wang Y., Krivtsov A. V., Sinha A. U., et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stingl J., Eirew P., Ricketson I., et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 124.Hasmim M., Bruno S., Azzi S., et al. Isolation and characterization of renal cancer stem cells from patient-derived xenografts. Oncotarget. 2015 doi: 10.18632/oncotarget.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Williams S. A., Anderson W. C., Santaguida M. T., Dylla S. J. Patient-derived xenografts, the cancer stem cell paradigm, and cancer pathobiology in the 21st century. Laboratory Investigation. 2013;93(9):970–982. doi: 10.1038/labinvest.2013.92. [DOI] [PubMed] [Google Scholar]