Abstract
The MDM2 protein plays an important role in the regulation of cell proliferation and apoptosis via ubiquitination and proteasome‐mediated degradation of p53. The genetic polymorphism rs2279744 (c.309T>G) of the MDM2 gene is reportedly associated with susceptibility and/or prognosis in various cancers. In this study, we investigated the risk factors for worse survival in patients with lung adenocarcinoma (AC). We examined the association between c.309T>G and the prognosis of lung cancer by retrospectively reviewing 453 lung cancer patients. We studied both, clinicopathological and genetic characteristics, including the c.309T>G, p53 Arg72Pro, EGFR,KRAS, and p53 mutations. Associations between these factors and survival outcome were analyzed using Cox proportional hazards models. The frequencies of MDM2 polymorphisms were T/T, 20.8%; T/G, 48.6%, and G/G, 30.7%. The overall survival (OS) of AC patients with pathological stage I disease and the MDM2 T/T genotype was significantly shorter than that of those with the T/G or G/G genotypes (P = 0.02). Multivariate analysis revealed that the MDM2 T/T genotype was an independent, significant prognostic factor (hazard ratio [HR] = 2.23; 95% confidence interval [CI]: 1.07–4.65; P = 0.03). The MDM2 T/T genotype was predictive of poorer survival in a Japanese population. Genotyping for this polymorphism might predict the clinical outcomes of stage I AC patients.
Keywords: Lung cancer, MDM2, p53, polymorphism, prognosis, SNP, SNP309
Introduction
Lung cancer is the leading cause of cancer‐related mortality worldwide 1. Although the overall incidence of lung cancer has been declining, particularly in Western countries, an increase in the proportion of lung adenocarcinoma (AC) is evident 2. The 5‐year survival rates for completely resected lung cancer were 86.8% (pathological [p‐] stage IA) and 73.9% (p‐stage IB) in Japan 3. A certain number of p‐stage I lung cancers paradoxically relapse even after surgical resection of the primary lesion and histopathological absence of any lymph node metastasis. This indicates that a small proportion of early‐stage lung cancers have highly metastatic characteristics. Therefore, screening possible high‐risk patients for disease recurrence is necessary to provide tailored medicine.
The p53 gene is a well‐known tumor suppressor that is frequently mutated in non‐small‐cell lung cancer (NSCLC) patients 4. p53 encodes a sequence‐specific DNA‐binding transcription factor targeting various genes that govern specific cellular processes 5. The MDM2 protein plays an important role in regulating cell proliferation and apoptosis by mediating ubiquitination and proteasome‐mediated degradation of p53 after binding directly to the latter protein; MDM2 has an E3 ubiquitin ligase activity 6, 7. A single‐nucleotide polymorphism (SNP) in the MDM2 promoter region, a T‐to‐G change at nucleotide c.309 (rs2279744) in the first intron (c.309T>G), increases the binding affinity toward stimulatory protein 1 (Sp1), causing higher‐level MDM2 expression 8. Also, cells harboring homozygous 309G alleles express higher levels of MDM2 protein, thereby reducing the tumor‐suppressing activity of p53 8. In humans, c.309T>G is associated with earlier onset of tumor formation in both hereditary and sporadic cancers 9. Recently, another antagonizing MDM2 polymorphism, SNP285, has been reported10 among Caucasians. SNP285 has been reported to nullify the effect of SNP309 and to reduced risk of breast, endometrial, and ovarian cancer. Molecular epidemiological studies of the c.309T>G polymorphisms in terms of lung cancer susceptibility11, 12, 13 have yielded contradictory findings. We recently reported that c.309T>G was not associated with lung cancer susceptibility in a Japanese population 14. The effects of c.309T>G on lung cancer survival have reported first in 200715 and remain controversial 12, 15, 16, 17, 18, 19, 20. So far, seven studies have analyzed the association between c.309T>G of the MDM2 gene and lung cancer prognosis. The G allele was reported to be a poor prognosis factor in Caucasians and Asians 15, 16. However, recently, some reports17, 20concluded that the T allele was a poor prognosis factor in Asians. Furthermore, three reports found no association between SNP309 and lung cancer survival in Asian 18, 19, Caucasian and African‐American 12.
In this study, we investigated whether c.309T>G of the MDM2 gene is closely associated with survival outcome of surgically resected NSCLC together with other clinicopathological and genetic characteristics.
Patients and Methods
Study population
To carry out this clinical research, we obtained approval from the Institutional Review Board of the Ethical Committee for Human Genome Analysis at Gunma University, and written informed consent from all the patients who participated. We analyzed 453 consecutive lung cancer patients (stages I–III) surgically treated between January 2003 and December 2012 at the Department of Thoracic and Visceral Organ Surgery, Gunma University Graduate School of Medicine, Gunma, Japan. Patients who had undergone preoperative therapies (chemotherapy and/or radiation therapy) and had a history of lung cancer were excluded. History of cancer and smoking were documented using a chart review before surgery. Never smokers were defined as individuals with a lifetime exposure to fewer than 100 cigarettes. Other patients were defined as smokers these include both former and current smoker. Disease staging was used to divide the patients into two groups: those of stages I and II–III. All the pathological factors, including pleural, vascular, and lymphatic invasion, were documented from the pathologic analysis at Gunma University Hospital. Cases that were positive for vascular invasion or lymphatic invasion were defined as lymphovascular invasion (LVI) positive. All the patients were reclassified according to the 7th edition of the International Union against Cancer (UICC) tumor‐node‐metastasis (TNM) staging system 21. The type of treatment after cancer recurrence was chosen by each individual physician. Overall survival (OS) was determined as the time from tumor resection to death from any cause. Disease‐free survival (DFS) was defined as the time between tumor resection and the first disease progression or death. All research followed the principles of the Declaration of Helsinki.
SNP genotyping
Peripheral venous blood samples were collected, and DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The DNA was used for SNP typing of c.309T>G and p53 Arg72Pro polymorphisms. Genotyping of c.309T>G was carried out using the Duplex SmartAmp method as described previously 22. p53 Arg72Pro was genotyped using polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) based on a previous report 23. Subsequently, the samples representing each genotypic pattern were used as controls in each assay.
Gene mutation analysis
Tissue samples from patients were isolated from surgically resected lung tumors. Lung cancer tissues were immediately frozen after surgical removal and stored at −80°C until DNA extraction using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI). The genomic DNA was used as a template for mutation analysis of EGFR, KRAS, and p53. KRAS and EGFR mutations were analyzed by sequencing as described previously 24, 25. Mutations in exons 5–8 of p53 were detected by direct sequencing 26. Briefly, primers used in the reactions were E5‐6S (5′‐TGCCCTGACTTTCAACTCTG‐3′) and E5‐6AS (5′‐AGTTGCAAACCAGACCTCAGG‐3′) for exons 5 and 6, and E7‐8S (5′‐CTTGCCACAGGTCTCCCCAA‐3′) and E7‐8AS (5′‐TCTCCTCCACCGCTTCTTGT‐3′) for exons 7 and 8. All the p53 mutations were confirmed by sequencing of both DNA strands.
Statistical analyses
Probability values less than 0.05 indicated a statistically significant difference. Differences in the distributions between groups were examined by Pearson χ 2 tests. Kaplan–Meier curve and the log‐rank test were used to estimate differences in survival. Hazard ratios (HRs) from univariate Cox regression analysis were used to determine the association between clinic‐pathological features and OS. Variables with statistically significant differences in univariate analysis were entered into multivariate analysis. Multivariate Cox proportional hazards regression was used to evaluate independent prognostic factors. All the statistical analyses were performed using SPSS Statistics version 20 (IBM Co., NY, USA).
Results
Demographics of patients according to MDM2 genotype
Table 1 depicts the characteristics of the entire study population. The study population was composed of 260 males and 193 females of a median age 68 years (range, 33–87 years). The genotype frequencies of MDM2 polymorphisms were as follows: T/T, 20.8%; T/G, 48.6%; and G/G, 30.7%. The frequency of the MDM2 309G allele was 0.55, consistent with previously described values for Asian lung AC patients 22.
Table 1.
Patients characteristics
| n | % | |
|---|---|---|
| Sex | ||
| Women | 193 | 42.6 |
| Men | 260 | 57.4 |
| Age | ||
| Mean ± SD | 68.1 ± 9.5 | |
| Smoking status | ||
| Never smoker | 179 | 59.5 |
| Smoker | 274 | 60.5 |
| Surgical procedure | ||
| ≥Lobectomy | 388 | 85.7 |
| ≤Segmentectomy | 65 | 14.3 |
| Pathological stage | ||
| I | 322 | 71.1 |
| II | 60 | 13.2 |
| III | 71 | 15.7 |
| T factor | ||
| T1 | 218 | 48.1 |
| T2 | 194 | 42.8 |
| T3 | 39 | 8.6 |
| T4 | 2 | 0.4 |
| N factor | ||
| N0 | 348 | 76.8 |
| N1 | 42 | 9.3 |
| N2 | 63 | 13.9 |
| Histology | ||
| AC | 328 | 72.4 |
| SQ | 107 | 23.6 |
| Others | 18 | 4.0 |
| MDM2 SNP309 | ||
| TT | 94 | 20.8 |
| TG | 220 | 48.6 |
| GG | 139 | 30.7 |
| Adjuvant chemotherapy | 123 | 27.2 |
| Chemotherapy (postrecurrence) | 69 | 15.2 |
| Radiation therapy (postrecurrence) | 51 | 11.3 |
Patients who had undergone preoperative therapies (chemotherapy and/or radiation therapy) and had a history of lung cancer were excluded. AC, adenocarcinoma; SQ, squamous cell carcinoma.
Survival analysis
The median follow‐up time was 56.5 months (range, 1.1–150 months). The 5‐year OS and DFS rates of the total study population were 73.7% (95% confidence interval [CI]: 69.2–78.2) and 66.1% (95% CI: 61.4–20.8), respectively. The percentages of patients treated via chemotherapy and/or radiotherapy after recurrence did not significantly differ (P = 0.156; Pearson χ 2 test). Table 2 shows the results of HR adjusted for age, sex, stage, histology, treatment (chemotherapy after tumor recurrence), and smoking status. Although no association was observed between this polymorphism and the population as a whole, the MDM2 T/T genotype was significantly associated worse DFS and OS among AC patients and p‐stage I AC patients.
Table 2.
Hazard ratios for survival data according to MDM2 genotypes. T/G + G/G (Reference: HR = 1.0) vs. T/T
| DFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Histology | Pathological stage | N | Event | HR | 95% CI | P | Event | HR | 95% CI | P | |
| All | Stage I‐III | a | 453 | 153 | 1.15 | 078–1.69 | 0.48 | 118 | 1.01 | 0.64–1.58 | 0981 |
| AC | b | 328 | 99 | 1.93 | 1.21–3.07 | 0.01 | 73 | 2.05 | 1.19–3.51 | 0.01 | |
| SQ | b | 107 | 54 | 0.71 | 0.32–1.53 | 0.38 | 45 | 0.50 | 0.20–1.28 | 0.15 | |
| AC | Stage I | b | 246 | 47 | 2.11 | 1.13–3.95 | 0.02 | 35 | 3.00 | 1.44–6.24 | 0.003 |
| Stage II, III | b | 82 | 52 | 1.78 | 0.87–3.67 | 0.12 | 38 | 1.33 | 0.54–3.30 | 0.54 | |
| SQ | Stage I | b | 63 | 24 | 0.75 | 0.21–2.67 | 0.66 | 18 | 0.34 | 0.04–2.67 | 0.30 |
| Stage II, III | b | 44 | 30 | 0.65 | 0.22–1.92 | 0.44 | 27 | 0.65 | 0.19–2.17 | 0.48 | |
P < 0.05 are shown in bold. DFS, disease‐free survival; OS, overall survival; AC, adenocarcinoma; SQ, squamous cell carcinoma.
Hazard ratio adjusted for age, sex, stage, histology, treatment (chemotherapy after tumor recurrence), and smoking status.
Hazard ratio adjusted for age, sex, treatment (chemotherapy after tumor recurrence), and smoking status.
Subgroup analysis of stage I AC patients
Among stage I AC patients, a significant association was found between smoking status, pleural invasion, or LVI and c.309T>G (T/T vs. T/G + G/G).(Table 3). Figures 1 and 2. show Kaplan–Meier survival curves of AC p‐stage I patients according to MDM2 genotype for DFS and OS, respectively. The OS of patients with the T/T genotype was shorter than the OSs of patients with the G/G or T/G genotypes (P = 0.021; log‐rank test). The 5‐year OS and DFS rates of the total population were 86.4% (95% CI: 81.7–91.1) and 80.2% (95% CI: 74.5–85.9), respectively. Together with Kaplan–Meier analysis, we compared the T/T genotype with G allele carriers (T/G + G/G) using univariate and multivariate analyses. The results of univariate analysis for OS are summarized in Table 4. The HR for death in the T/T group relative to the T/G + G/G group was 2.20 (95% CI: 1.10–4.36; P = 0.025). Similarly, the clinicopathological factors (gender, age, smoking history, differentiation, LVI, pleural invasion, and EGFR mutation) significantly affected survival. Conversely, the status of the KRAS and p53 mutations and p53codon72 were not significant upon univariate analysis (Table 4). Multivariate analyses for OS revealed that MDM2 T/T genotype was a significant independent risk factor (HR = 2.23; 95% CI: 1.07–4.65; P = 0.033), together with male gender (HR = 5.69; 95% CI: 1.78–18.2; P = 0.003), older age (HR = 2.39; 95% CI: 1.12–5.09; P = 0.002) and LVI (HR = 1.58; 95% CI: 1.01–2.47; P = 0.044) (Table 4).
Table 3.
Patient characteristics of stage I AC patients according to MDM2 genotypes
| MDM2 genotypes | T/T vs. T/G vs. G/G | T/T vs. T/G + G/G | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | T/T | T/G | G/G | P a | P a | ||||
| All cases | 246 | 58 | 23.6 | 115 | 46.7 | 73 | 29.7 | – | – |
| Sex | |||||||||
| Women | 134 | 27 | 20.3 | 65 | 48.9 | 41 | 20.8 | 0.42 | 0.23 |
| Men | 113 | 31 | 27.4 | 50 | 44.2 | 32 | 28.3 | ||
| Age in years | |||||||||
| <70 | 135 | 33 | 24.4 | 63 | 46.7 | 39 | 28.9 | 0.92 | 0.76 |
| ≥70 | 111 | 25 | 23.6 | 52 | 46.8 | 34 | 30.6 | ||
| Smoking status | |||||||||
| Never smoker | 140 | 24 | 17.1 | 73 | 52.1 | 43 | 30.7 | 0.02 | 0.01 |
| Smoker | 106 | 34 | 32.1 | 42 | 39.6 | 30 | 28.3 | ||
| p53 Arg72Pro | |||||||||
| Arg/Arg | 90 | 20 | 22.2 | 42 | 46.7 | 28 | 31.1 | 0.90 | 0.93 |
| Arg/Pro | 115 | 28 | 24.3 | 56 | 48.7 | 31 | 27.0 | ||
| Pro/Pro | 41 | 10 | 24.4 | 17 | 41.5 | 14 | 34.1 | ||
| Performance statusb | |||||||||
| 0 | 208 | 49 | 23.6 | 101 | 48.6 | 58 | 27.9 | 0.30 | 1.00 |
| 1–2 | 21 | 5 | 23.8 | 7 | 33.3 | 9 | 42.9 | ||
| Surgical procedure | |||||||||
| Lobectomy | 201 | 49 | 24.4 | 91 | 45.3 | 61 | 30.3 | 0.61 | 0.70 |
| Segmentectomy | 45 | 9 | 20.0 | 24 | 53.3 | 12 | 26.7 | ||
| Differentiation | |||||||||
| Well | 137 | 30 | 21.9 | 69 | 50.4 | 38 | 27.7 | 0.44 | 0.55 |
| Moderate or poorly | 109 | 28 | 25.7 | 46 | 42.2 | 35 | 32.1 | ||
| T factor | |||||||||
| T1 | 158 | 35 | 21.5 | 71 | 44.9 | 53 | 33.5 | 0.19 | 0.35 |
| T2 | 88 | 24 | 27.3 | 44 | 50.0 | 20 | 22.7 | ||
| Pleural invasion | |||||||||
| Absent | 202 | 42 | 20.8 | 96 | 47.5 | 64 | 31.7 | 0.07 | 0.03 |
| Present | 44 | 16 | 36.4 | 19 | 43.2 | 9 | 20.5 | ||
| Lymphovascular invasion | |||||||||
| Absent | 177 | 34 | 19.2 | 92 | 52.0 | 51 | 28.8 | 0.01 | 0.01 |
| Present | 69 | 24 | 34.8 | 23 | 33.3 | 22 | 31.9 | ||
| p53 status | |||||||||
| Wild type | 207 | 48 | 23.2 | 98 | 47.3 | 61 | 29.5 | 0.90 | 0.83 |
| Mutant | 39 | 10 | 25.6 | 17 | 43.6 | 12 | 30.8 | ||
| EGFR statusb | |||||||||
| Wild type | 133 | 30 | 22.6 | 62 | 46.6 | 41 | 30.8 | 0.92 | 0.76 |
| Mutant | 111 | 27 | 24.3 | 52 | 46.8 | 32 | 28.8 | ||
| KRAS status | |||||||||
| Wild type | 214 | 54 | 25.2 | 99 | 46.3 | 61 | 28.5 | 0.25 | 0.13 |
| Mutant | 32 | 4 | 12.5 | 16 | 50.0 | 12 | 37.5 | ||
| Adjuvant chemotherapy | |||||||||
| Received | 47 | 14 | 29.8 | 27 | 57.4 | 6 | 12.8 | 0.02 | 0.26 |
| Not received | 199 | 44 | 22.1 | 88 | 44.2 | 67 | 33.7 | ||
| Chemotherapy | |||||||||
| Received | 16 | 3 | 18.8 | 9 | 56.2 | 4 | 5.5 | 0.80 | 0.55 |
| Not received | 11 | 4 | 36.4 | 4 | 36.4 | 3 | 27.31 | ||
NA, not available; AC, adenocarcinoma.
P values were calculated by chi‐square test. P < 0.05 are shown in bold.
Performance status at surgery and EGFR mutation status remains unknown in some cases.
Figure 1.
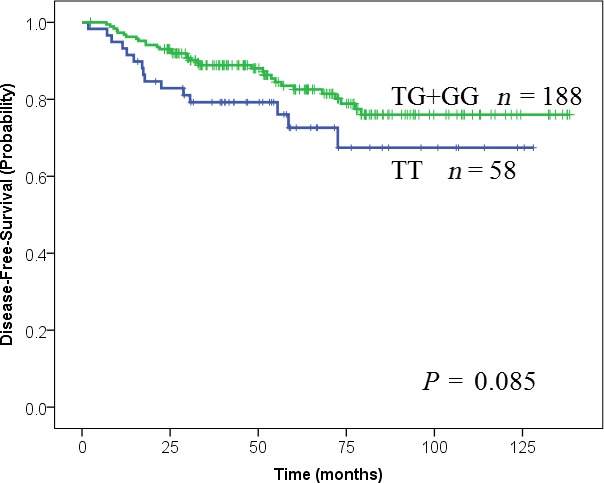
Kaplan–Meier estimates of disease‐free survival in patients with stage I lung adenocarcinoma. MDM2 c.309T>G (T/T, blue; T/G + G/G, green). The P value was calculated using the log‐rank test.
Figure 2.
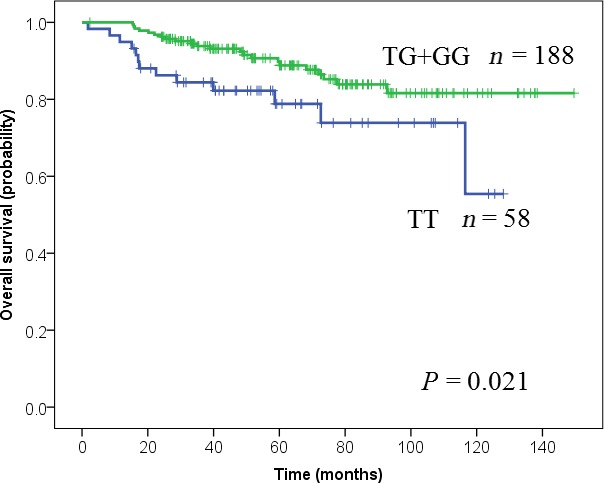
Kaplan–Meier estimates of overall survival in patients with stage I lung adenocarcinoma. MDM2 c.309T>G (T/T, blue; T/G + G/G, green). The P value was calculated using the log‐rank test.
Table 4.
Univariate and multivariate analysis in disease‐free survival and overall survival of stage I AC patients
| DFS | OS | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| Variables | HR | 95% CI | P a | HR | 95% CI | P a | HR | 95% CI | P a | HR | 95% CI | P a |
| Sex | ||||||||||||
| Women | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| Men | 3.32 | 1.77–6.21 | <0.0001 | 2.11 | 0.90–4.98 | 0.087 | 7.39 | 3.05–17.9 | <0.0001 | 5.69 | 1.78–18.2 | 0.003 |
| Age | ||||||||||||
| <70 | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| ≥70 | 2.75 | 1.50–5.03 | 0.001 | 1.95 | 1.04–3.66 | 0.039 | 3.07 | 1.50–6.27 | 0.002 | 2.39 | 1.12–5.09 | 0.024 |
| Smoking history | ||||||||||||
| Never smoker | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| Smoker | 3.4 | 1.84–6.29 | <0.0001 | 0.919 | 0.60–1.42 | 0.22 | 4.73 | 2.21–10.1 | <0.0001 | 1.14 | 0.69–1.91 | 0.089 |
| p53 Arg72Pro | ||||||||||||
| Arg/Arg + Arg/Pro | 1.0 | 1.0 | ||||||||||
| Pro/Pro | 0.82 | 0.37–1.83 | 0.62 | 0.80 | 0.31–2.07 | 0.65 | ||||||
| MDM2 c.309T>G | ||||||||||||
| T/G + G/G | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| T/T | 1.71 | 0.92–3.15 | 0.09 | 1.45 | 0.76–2.76 | 0.254 | 2.20 | 1.10–4.36 | 0.025 | 2.23 | 1.07–4.65 | 0.033 |
| Performance status | ||||||||||||
| 0 | 1.0 | 1.0 | ||||||||||
| 1,2 | 0.94 | 0.29–3.05 | 0.92 | 1.34 | 0.41–4.43 | 0.63 | ||||||
| Differentiation | ||||||||||||
| Well | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| Moderate and poorly | 3.94 | 2.08–7.48 | <0.0001 | 0.79 | 0.54–1.16 | 0.22 | 4.76 | 2.16–10.5 | <0.0001 | 0.79 | 0.49–3.31 | 0.34 |
| T factor | ||||||||||||
| T1 | 1.0 | 1.0 | 1.0 | |||||||||
| T2 | 2.93 | 1.64–5.23 | <0.0001 | 1.76 | 0.84–3.72 | 0.14 | 2.62 | 1.34–5.11 | 0.005 | 1.27 | 0.49–3.31 | 0.62 |
| Lymphovascular invasion | ||||||||||||
| Negative | 1.0 | 1.0 | 1.0 | |||||||||
| Positive | 4.36 | 2.43–7.82 | <0.0001 | 1.48 | 1.04–2.10 | 0.03 | 6.10 | 3.00–12.4 | <0.0001 | 2.50 | 1.03–6.11 | 0.044 |
| Pleural invasion | ||||||||||||
| Negative | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||
| Positive | 2.49 | 1.35–4.60 | 0.004 | 1.19 | 0.542–2.61 | 0.66 | 3.00 | 1.51–5.96 | 0.002 | 1.13 | 0.44–2.90 | 0.80 |
| EGFR gene mutation | ||||||||||||
| Mutant | 1.0 | 1.0 | 1.0 | |||||||||
| Wild type | 2.02 | 1.08–3.78 | 0.027 | 2.94 | 1.34–6.48 | 0.007 | 1.46 | 0.60–3.56 | 0.41 | |||
| KRAS gene mutation | ||||||||||||
| Wild type | 1.0 | 1.0 | ||||||||||
| Mutant | 1.13 | 0.51–2.53 | 0.76 | 1.31 | 0.54–3.16 | 0.55 | ||||||
| p53 gene mutation | ||||||||||||
| Wild type | 1.0 | 1.0 | ||||||||||
| Mutant | 1.43 | 0.69–2.97 | 0.33 | 1.75 | 0.79–3.84 | 0.17 | ||||||
| Adjuvant chemotherapy | ||||||||||||
| Not received | 1.0 | 1.0 | ||||||||||
| Received | 1.56 | 0.79–3.07 | 0.2 | 1.25 | 0.54–2.89 | 0.6 | ||||||
HR and 95% CI are shown as the values of the latter compared to the former (HR = 1.0). P < 0.05 are shown in bold. DFS, disease‐free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval.
P values were calculated by Cox regression analysis.
Stratified analyses of the prognostic effects of the MDM2 genotypes
We further evaluated the associations between the prognostic effects of c.309T>G of the MDM2 gene and p53 status using stratified analyses (Fig. 3). Stronger relationships were observed among p53 wild‐type group (HR = 3.69) and p53 Arg72Pro Arg/Arg + Arg/Pro group (HR = 2.99). Further stratified analyses of patients with a p53 wild‐type tumor and Arg/Arg + Arg/Pro genotype of p53 Arg72Pro showed a higher (HR = 4.39), but these results are underpowered due to small sample size.
Figure 3.
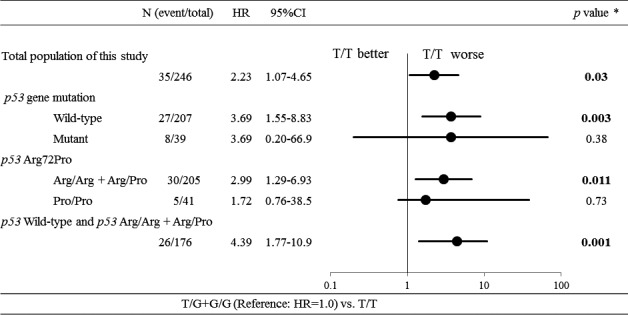
Subset analysis of overall survival in stage I lung adenocarcinoma. The forest plot shows the multivariate Cox regression for each subgroup. P values correspond to hazard ratios adjusted by multivariate regression. The P values <0.05 are shown in bold.
Discussion
Because disruption of p53 tumor suppressor function is important in cancer development, we hypothesized that the 309G allele of the MDM2 gene would be associated with worse survival outcomes among surgically treated lung cancer patients. Unexpectedly, our study demonstrated that the T/T genotype of c.309T>G was a significant independent unfavorable prognostic factor, and the associated tumors tended to show pleural invasion or LVI among stage I lung AC patients.
So far, seven studies have analyzed the association between c.309T>G of the MDM2 gene and lung cancer prognosis, but the results were contradictory (Table 5). Our present results are consistent with the two reports from Taiwan and China 17, 20. Heist et al. 15. investigated the impact of MDM2 gene polymorphism in early‐stage (stage I or II) NSCLC patients in the United States and reported that the G/G genotype was associated with worse OS. These findings might seem contrary to our results. However, when analyzed in detail, Heist's subgroup analyses (Table 5) and ours had similar results. First, Heist showed that the G/G genotype was associated with worse OS only in patients with stage IB/II NSCLC and squamous cell carcinoma (SQ) histology 15. Similarly, we showed that the T/T genotype was associated with better OS in patients with SQ histology (although it did not reach statistical significance). Next, Heist showed that, the G/G genotype was associated with better OS in patients with stage IA NSCLC or those with AC histology (although not significant) 15. This is consistent with our results (Table 2), since we showed that the T/T genotype was associated with worse OS in patients with stage I NSCLC or those with AC histology. We believe that the difference in statistical power between the two studies may be due to the difference of study population. Recently, SNP285 has been reported to act as an antagonist to SNP309 only observed in Caucasians 10, furthermore, Ryan et al. 12. showed that neither SNP309 nor SNP285 were associated with lung survival. Therefore, this point is still a matter of debate. SNP285 per se could not explain the discrepancies between Heist's study and ours. Han et al.[ 16 and Liu et al. 19. investigated stage III or IV NSCLC patients and reported disparate findings. Survival outcome of advanced lung cancer depends strongly on tumor size, lymph node metastasis, or therapeutic regimen. Therefore, known genetic factors might have less influence on cancer prognoses if study subjects have only advanced‐stage NSCLC. Our results support the previous study from Asia (Taiwan) focusing on stage I NSCLC17 and which reported a tendency for the T/T group to be a poor prognostic factor compared to the G/G group (P = 0.05).
Table 5.
Comparison of previous reports concerning c.309T>G and lung cancer prognosis
| Author | Year | Country | Ethnic group | Smoker (%) | Stage | Histology | Subgroup | N | MDM2 c.309T>G SNP309 | HR | 95% CI | P | Correlation | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heist | 2007 | States | Caucasian | 93% | Stage I, II | NSCLC | 383 | T//T vs. G/G | 1.57 | 1.03–2.40 | 0.04 | G/G risk | 15 | |
| AC | 186 | T/T vs. G/G | 0.93 | 0.48–1.80 | 0.82 | NS | ||||||||
| SQ | 110 | T/T vs. G/G | 3.77 | 1.88–7.57 | 0.0002 | G/G risk | ||||||||
| Stage IA | 200 | T/T vs. G/G | 0.65 | 0.33–1.27 | 0.2 | NS | ||||||||
| Stage IB or II | 183 | T/T vs. G/G | 3.19 | 1.80–5.65 | <0.0001 | G/G risk | ||||||||
| Noncurrent smoker | 233 | T/T vs. G/G | 1.17 | 0.66–2.08 | 0.58 | NS | ||||||||
| Current smoker | 150 | T/T vs. G/G | 2.93 | 1.52–5.67 | 0.001 | G/G risk | ||||||||
| Han | 2008 | Korea | Asian | 75% | Stage IIIB or IV | NSCLC | 148 | T/T vs. T/G + G/G | 1.74 | 1.05–2.89 | 0.032 | T/G + G/G risk | 16 | |
| Chua | 2010 | Singapore | Asian | 0% | Lung cancer | 123 | T/T vs. T/G vs. G/G | NA | 0.27(log‐rank) | N.S. | 18 | |||
| Chien | 2010 | Taiwan | Asian | 51% | Stage I‐III | NSCLC | 198 | T/T vs. G/G | 0.62 | 0.41–0.95 | 0.003 | T/T risk | 17 | |
| Stage I | 127 | T/T vs. G/G | 0.47 | 0.22–1.01 | 0.05 | NS | ||||||||
| Stage I with wild‐type p53 | 99 | T/T vs. G/G | 0.34 | 0.15–0.80 | 0.01 | T/T risk | ||||||||
| Stage I with p53 mutation | 28 | T/T vs. G/G | 5.02 | 0.31–28.20 | 0.35 | NS | ||||||||
| Stage II, III | 179 | T/T vs. G/G | 0.82 | 0.48–1.39 | 0.45 | NS | ||||||||
| Liu | 2011 | China | Asian | 59% | Stage III or IV | NSCLC | 199 | T/T vs. G/G | 1.05 | 0.66–1.67 | 0.91(log‐rank) | NS | 19 | |
| Dong | 2011 | China | Asian | 72% | Stage I‐IV | NSCLC | 561 | G/G vs. T/T + T/G | 1.37 | 1.06–1.78 | 0.017 | T/T + T/G risk | 20 | |
| Ryan | 2012 | States | Caucasian and African‐American | 92% | Stage I‐IV | NSCLC | 197 | T/T vs. G/G | 0.8 | 0.51–1.24 | 0.31 | NS | 12 | |
| This study | 2015 | Japan | Asian | 45% | Stage I | AC | 179 | T/G + G/G vs. T/T | 2.23 | 1.07–4.65 | 0.033 | T/T risk |
HR, hazard ratio; CI, confidence interval; Ref., Reference; NSCLC, non‐small‐cell lung cancer; NS, not significant; NA, not available; AC, adenocarcinoma; SQ, squamous cell carcinoma.
The T/T genotype was associated with poor survival in patients with aggressive bladder cancer 27, in line with our observations. The cited authors concluded that p53 mutational status was of prognostic value, but, in this study, the p53 mutation levels did not differ significantly by MDM2 genotype (Table 3). Any prognostic utility of the SNP309 marker in gastric cancer, renal cell carcinoma, and breast cancer, remains controversial 28, 29, 30. Furthermore, of early‐stage cancers, only lung cancer has been analyzed 15, 17.
Regarding the p53 Arg72Pro, it has been reported that the Arg/Arg variant encodes a highly proapoptotic protein, whereas the Pro/Pro variant has the opposite effect 31. We analyzed the associations between p53 status (p53 mutation and p53 Arg72Pro) and c.309T>G of the MDM2 gene, and consequently found that the T/T genotype was associated with worse OS among p53 wild‐type group (HR = 3.69) and the p53 Arg72Pro31 Arg/Arg + Arg/Pro group (HR = 2.99) (Fig. 3), although these results are underpowered due to small sample size. Our findings are consistent with those of Chien et al. about p53 mutation status 17. p53 function is considered normal (not inactivated) in patients with the T/T genotype, Arg/Arg + Arg/Pro, or p53 wild‐type group compared to the 309G allele carrier, Pro/Pro, or p53 mutant group. Furthermore, among patients in the abovementioned groups, the T/T genotype was associated with worse OS (HR = 4.39). These results suggest a positive interaction between the T/T genotype, p53Arg72Pro RR + RP, and p53 wild type in increasing the risk of death.
The tumorigenic functions of MDM2 in both p53‐dependent and ‐independent pathways are complicated, and analysis of one polymorphic variant may not address all the MDM2 functions. The precise mechanism underlying the worse OS with the T/T genotype being associated with p53 status remains unknown 32. However, tumors of MDM2 T/T patients tended to be positive in LVI and pleural invasion (Table 3), which have been reported to be worse prognostic factors associated with tumor proliferation and aggressiveness 33, 34. These results indicate that the tumors of T/T patients in the stage I period might have overall malignant potential, although p53 tumor suppressor function is normal. Based on our results, tumors that develop under normal p53 might have a malignant potential rather than tumors that develop under abnormal p53, and genotyping of c.309T>G might simply be a selection tool for malignant potential for stage I lung AC.
Throughout this study, we found that c.309T>G was a predictive factor of postoperative survival among p‐stage I lung AC patients in a Japanese population. Analysis of MDM2 polymorphism has several advantages over somatic cell mutations analysis. First, the MDM2 309T>G polymorphism can be used to predict which individuals are at an increased risk of death after surgery. Second, the assessment of an individual's polymorphism status does not require an extraction of tumor‐specific DNA. In this study, the EGFR, KRAS, and p53 mutations were not independently associated with prognosis as previously reported 35. Although our findings need to be validated in prospective studies, c.309T>G would be a useful prognostic marker that is detectable at any stage of diagnosis or treatment and influences the therapeutic strategies. Furthermore, we had already established the Duplex SmartAmp method22 to detect c.309T>G with a single drop (5 μL) of blood within 40 min from sample collection. If we can make this method more practical, we will detect this SNP more easily and quickly in any clinical situation.
This study possesses several limitations. First, we could gather data on OS but not on cancer‐specific survival because the sources of survival data did not indicate the cause of death, although it would be useful to know the cause of death especially for early‐stage cancer patients. Another limitation of our study is its retrospective nature, although blood sample collection was performed preoperatively, and the database was run prospectively. Therefore, patient populations might be biased. Finally, the sample size and number of events in this study might be too small to draw meaningful conclusions associated with p53 status. Further prospective studies with a larger, more homogeneous study population would be desirable to abrogate these limitations.
In conclusion, to the best of our knowledge, this is the first study to analyze the effects of c.309T>G in the MDM2 gene together with p53 Arg72Pro as well as mutations in the EGFR. KRAS and p53 genes on the prognoses of lung cancer patients. We conclude that the T/T genotype of c.309T>G affects OS in surgically resected stage I lung AC patients and represents an independent prognostic factor in a Japanese population. Further studies are warranted to clarify the biological importance of these findings and the usefulness of the MDM2 309T>G polymorphism as a predictive marker for therapy selection and outcome prediction in NSCLC.
Conflicts of Interest
None declared.
Acknowledgments
The authors thank Dr. Hisayoshi Nakazawa for his fruitful discussion concerning genetic polymorphisms of the MDM2 gene. In addition, our thanks go to Mr. Ryosuke Konuma, Ms. Nao Kobayashi, Mr. Masaki Shinohara, and Ms. Yuriha Iwata (Gunma University) for their technical assistance.
Cancer Medicine 2016; 5(8):1791–1801
References
- 1. Jemal, A. , Bray F., Center M. M., Ferlay J., Ward E., and Forman D.. 2011. Global cancer statistics. CA Cancer J. Clin. 61:69–90. [DOI] [PubMed] [Google Scholar]
- 2. Travis, W. D. , Brambilla E., Müller‐Hermelink K., and Harris C. C. 2004. Pathology & genetics tumours of the lung, pleura, thymus and heart. IARC Press, Lyon. [Google Scholar]
- 3. Sawabata, N. , Miyaoka E., Asamura H., Nakanishi Y., Eguchi K., Mori M., et al. 2011. Japanese lung cancer registry study of 11,663 surgical cases in 2004: demographic and prognosis changes over decade. J. Thorac. Oncol. 6:1229–1235. [DOI] [PubMed] [Google Scholar]
- 4. Herbst, R. S. , Heymach J. V., and Lippman S. M.. 2008. Lung cancer. N. Engl. J. Med. 359:1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin, S. , and Levine A. J.. 2001. The p53 functional circuit. J. Cell Sci. 114(Pt 23):4139–4140. [DOI] [PubMed] [Google Scholar]
- 6. Thut, C. J. , Goodrich J. A., and Tjian R.. 1997. Repression of p53‐mediated transcription by MDM2: a dual mechanism. Genes Dev. 11:1974–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Momand, J. , Zambetti G. P., Olson D. C., D. George, and A. J. Levine. 1992. The mdm‐2 oncogene product forms a complex with the p53 protein and inhibits p53‐mediated transactivation. Cell 69:1237–1245. [DOI] [PubMed] [Google Scholar]
- 8. Bond, G. L. , Hu W. W., Bond E. E., Robins H., Lutzker S. G., Arva N. C., et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119:591–602. [DOI] [PubMed] [Google Scholar]
- 9. Bond, G. L. , Hu W., and Levine A.. 2005. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 65:5481–5484. [DOI] [PubMed] [Google Scholar]
- 10. Knappskog, S. , and Lonning P. E.. 2011. MDM2 promoter SNP285 and SNP309; phylogeny and impact on cancer risk. Oncotarget 2:251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pine, S. R. , Mechanic L. E., Bowman E. D., Welsh J. A., Chanock S. C., Shields P. G., et al. 2006. MDM2 SNP309 and SNP354 are not associated with lung cancer risk. Cancer Epidemiol. Biomarkers Prev. 15:1559–1561. [DOI] [PubMed] [Google Scholar]
- 12. Ryan, B. M. , Calhoun K. M., Pine S. R., Bowman E. D., Robles A. I., Ambs S., et al. 2012. MDM2 SNP285 does not antagonize the effect of SNP309 in lung cancer. Int. J. Cancer 131:2710–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilkening, S. , Bermejo J. L., and Hemminki K.. 2007. MDM2 SNP309 and cancer risk: a combined analysis. Carcinogenesis 28:2262–2267. [DOI] [PubMed] [Google Scholar]
- 14. Enokida, Y. , Shimizu K, Kakegawa S, Atsumi J., Y. Takase , Miyamae Y., et al. 2014. Single‐nucleotide polymorphism (c.309T>G) in the MDM2 gene and lung cancer risk. Biomed. Rep. 2:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heist, R. S. , Zhou W., Chirieac L. R., Cogan‐Drew T., Liu G., Su L., et al. 2007. MDM2 polymorphism, survival, and histology in early‐stage non‐small‐cell lung cancer. J. Clin. Oncol. 25:2243–2247. [DOI] [PubMed] [Google Scholar]
- 16. Han, J. Y. , Lee G. K., Jang D. H., Lee S. Y., and Lee J. S.. 2008. Association of p53 codon 72 polymorphism and MDM2 SNP309 with clinical outcome of advanced nonsmall cell lung cancer. Cancer 113:799–807. [DOI] [PubMed] [Google Scholar]
- 17. Chien, W. P. , Wong R. H., Cheng Y. W., Chen C. Y., and Lee H.. 2010. Associations of MDM2 SNP309, transcriptional activity, mRNA expression, and survival in stage I non‐small‐cell lung cancer patients with wild‐type p53 tumors. Ann. Surg. Oncol. 17:1194–1202. [DOI] [PubMed] [Google Scholar]
- 18. Chua, H. W. , Ng D., Choo S., Lum S. S., Li H., Soh L. Y., et al. 2010. Effect of MDM2 SNP309 and p53 codon 72 polymorphisms on lung cancer risk and survival among non‐smoking Chinese women in Singapore. BMC Cancer 10:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu, L. , Wu C., Wang Y., Zhong R., Duan S., Wei S., et al. 2011. Combined effect of genetic polymorphisms in P53, P73, and MDM2 on non‐small cell lung cancer survival. J. Thorac. Oncol. 6:1793–1800. [DOI] [PubMed] [Google Scholar]
- 20. Dong, J. , Ren B., Hu Z., Chen J., Hu L., Dai J., et al. 2011. MDM2 SNP309 contributes to non‐small cell lung cancer survival in Chinese. Mol. Carcinog. 50:433–438. [DOI] [PubMed] [Google Scholar]
- 21. Sobin, L. , Gospodrowicz M., and Wittekind C.. 2009. TNM classification of malignant tumours, 7th ed. Wiley‐Liss, New York. [Google Scholar]
- 22. Enokida, Y. , Shimizu K., Atsumi J., Lezhava A., Y. Tanaka , Kimura Y. et al. 2013. Rapid detection of SNP (c.309T>G) in the MDM2 gene by the Duplex SmartAmp method. PLoS One 8:e60151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ara, S. , Lee P. S., Hansen M. F., and Saya H.. 1990. Codon 72 polymorphism of the TP53 gene. Nucleic Acids Res. 18:4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Araki, T. , Shimizu K., Nakamura T., Baba M., Kawai Y., Nakamura K., et al. 2011. Clinical screening assay for EGFR exon 19 mutations using PNA‐clamp smart amplification process version 2 in lung adenocarcinoma. Oncol. Rep. 26:1213–1219. [DOI] [PubMed] [Google Scholar]
- 25. Miyamae, Y. , Shimizu K., Mitani Y., Araki T., Kawai Y., Baba M., et al. 2010. Mutation detection of epidermal growth factor receptor and KRAS genes using the smart amplification process version 2 from formalin‐fixed, paraffin‐embedded lung cancer tissue. J. Mol. Diagn. 12:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang, C. C. , Cheng Y. W., Chen M. C., Lin Y. S., Chou M. C., and Lee H.. 2006. Different p53 mutation patterns in colorectal tumors from smokers and nonsmokers. Environ. Mol. Mutagen. 47:527–532. [DOI] [PubMed] [Google Scholar]
- 27. Sanchez‐Carbayo, M. , Socci N. D., Kirchoff T., Erill N., Offit K., Bochner B. H., et al. 2007. A polymorphism in HDM2 (SNP309) associates with early onset in superficial tumors, TP53 mutations, and poor outcome in invasive bladder cancer. Clin. Cancer Res. 13:3215–3220. [DOI] [PubMed] [Google Scholar]
- 28. Ohmiya, N. , Taguchi A., Mabuchi N., Itoh A., Hirooka Y., Niwa Y., et al. 2006. MDM2 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. J. Clin. Oncol. 24:4434–4440. [DOI] [PubMed] [Google Scholar]
- 29. Hirata, H. , Hinoda Y., Kikuno N., Kawamoto K., Suehiro Y., Tanaka Y., et al. 2007. MDM2 SNP309 polymorphism as risk factor for susceptibility and poor prognosis in renal cell carcinoma. Clin. Cancer Res. 13:4123–4129. [DOI] [PubMed] [Google Scholar]
- 30. Boersma, B. J. , Howe T. M., Goodman J. E., H. G. Yfantis , Lee D. H., Chanock S. J., et al. 2006. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J. Natl. Cancer Inst. 98:911–919. [DOI] [PubMed] [Google Scholar]
- 31. Dumont, P. , Leu J. I., Della Pietra A. C., George D. L., and Murphy M.. 2003. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33:357–365. [DOI] [PubMed] [Google Scholar]
- 32. Robles, A. I. , Linke S. P., and Harris C. C.. 2002. The p53 network in lung carcinogenesis. Oncogene 21:6898–6907. [DOI] [PubMed] [Google Scholar]
- 33. Takise, A. , Kodama T., Shimosato Y., Watanabe S., and Suemasu K.. 1988. Histopathologic prognostic factors in adenocarcinomas of the peripheral lung less than 2 cm in diameter. Cancer 61:2083–2088. [DOI] [PubMed] [Google Scholar]
- 34. Nentwich, M. F. , Bohn B. A., Uzunoglu F. G., M. Reeh , Quaas A., Grob T. J., et al. 2013. Lymphatic invasion predicts survival in patients with early node‐negative non‐small cell lung cancer. J. Thorac. Cardiovasc. Surg. 146:781–787. [DOI] [PubMed] [Google Scholar]
- 35. Kosaka, T. , Yatabe Y., Onozato R., Kuwano H., and T. Mitsudomi . 2009. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J. Thorac. Oncol. 4:22–29. [DOI] [PubMed] [Google Scholar]


