Abstract
Type 1 diabetes (T1D) is caused by T cell mediated destruction of the insulin-producing β cells. CD4 T cell responses play a central role in β-cell destruction but the identity of the epitopes recognized by pathogenic CD4 T cells remains unknown. To address this we used a panel of diabetes triggering CD4 T cell clones isolated from non-obese diabetic (NOD) mice. Here we show that these pathogenic CD4 T cells target peptide ligands that are formed by covalent crosslinking of proinsulin peptides to other peptides present in β-cell secretory granules. These hybrid insulin peptides (HIPs) are highly antigenic for CD4 T cells and can be detected by mass spectrometry in β-cells. CD4 T cells from the residual pancreatic islets of two organ donors who had T1D also recognize HIPs. The discovery that autoreactive T cells target hybrid peptides may explain how immune tolerance is broken in T1D.
Introduction
Type 1 Diabetes (T1D) is an autoimmune disease mediated by T cells responding to self-antigens in the pancreatic β-cell. The most widely used animal model of T1D is the non-obese diabetic (NOD) mouse (1). Studies in the NOD model have shown that CD4 T cell responses to several β-cell proteins, most prominently proinsulin, have been implicated in diabetes (2). Despite these insights it is not clear how pathogenic T cells escape thymic deletion and how (pro)insulin becomes a target of the autoimmune T cell response. To address these questions we have used our Barbara Davis Center (BDC) panel of diabetes causing CD4 T cell clones (which includes the well-known BDC-2.5 clone) (3), in conjunction with proteomic analysis of β-cell extracts to identify the target antigens for these pathogenic CD4 T cell clones. Recently we reported two new autoantigens for CD4 T cells in autoimmune diabetes: chromogranin A (ChgA) (4) and islet amyloid polypeptide (IAPP) (5). Like insulin ChgA and IAPP are β-cell pro-hormonal secretory granule proteins. WE14, a naturally occurring peptide cleavage product of ChgA, was found to be antigenic in both the NOD mouse (4) and in T1D patients (6). However, because this peptide is not β-cell specific and only stimulates T cells weakly, we hypothesized that the natural ligand for pathogenic CD4 T cells may be a modified form of ChgA.
Post-translational modification (PTM) is a well-established property of antigens in many autoimmune diseases (7). A notable exception is T1D in which the investigation of modified peptides as antigenic epitopes has only just begun (8–11). Here we report a novel peptide fusion that occurs in islet β-cells and creates a highly immunogenic PTM in the form of hybrid insulin peptides (HIPs). The resultant peptides are very antigenic not only for diabetogenic CD4 T cell clones from the NOD mouse but also for CD4 T cell clones isolated from the residual islets of individuals with T1D. Collectively our data support the hypothesis that the autoimmune β-cell destruction that underlies T1D, in both NOD mice and humans, is driven by CD4 T cell responses against HIPs.
Results
Using mass spectrometric analysis we verified the presence of the peptide WE14 in chromatographic fractions of mouse β-cell extracts. However, the peptide distribution over individual fractions did not follow the antigen distribution of the natural ligand recognized by the WE14-responsive T cell clones, including BDC-2.5 (Fig. S1A, top). Conversely, the mouse insulin 1 C-peptide (Fig. S1A, bottom) does follow the antigen distribution profile (similar results are obtained with insulin 2 C-peptide). Furthermore, a broad panel of shorter C-peptide fragments (both insulin 1 and 2) could also be identified in peak antigenic fractions (Fig. S1B) and a large number of these peptides also correspond to the BDC-2.5 antigen distribution profile. The matching antigen/C-peptide distributions suggested that a C-peptide fragment is a component of the natural T cell ligand. The proposed WE14 / I-Ag7 binding register (4) in which the peptide WE14 fills only half of the MHC II binding groove (positions 5–9), leaves MHC II positions 1–4 unoccupied. The C-peptide fragments could fill these unoccupied positions, thereby adding MHC-anchor residues which would lead to an increased peptide-MHC binding affinity as well as providing additional residues for improved TCR recognition. This led us to hypothesize that peptide bonds within insulin C-peptide react with N-terminal amino groups of naturally occurring peptides such as WE14, resulting in the formation of hybrid insulin peptides (HIPs).
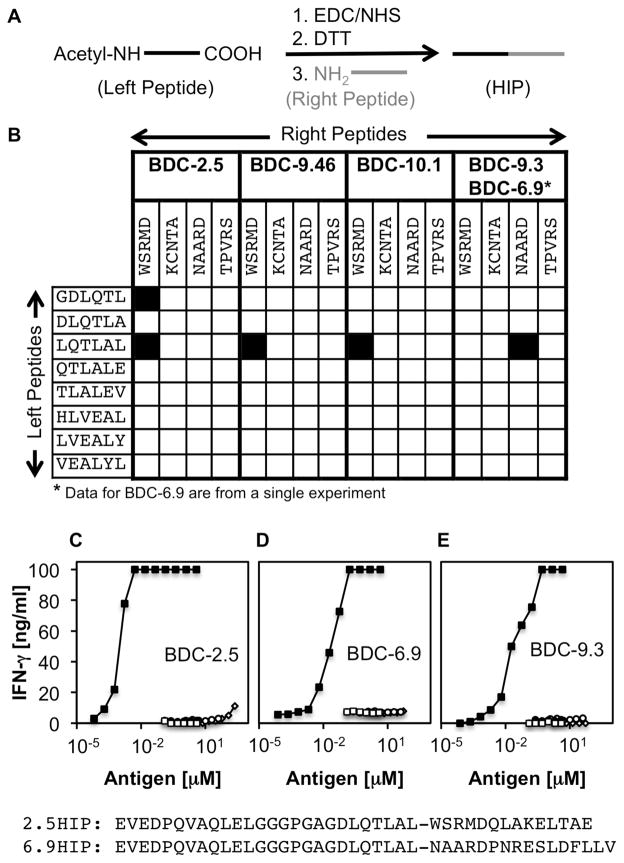
To investigate the possibility that BDC-2.5 and additional diabetogenic T cell clones from our panel are activated by HIPs, we utilized a chemical crosslinking strategy (outlined in Fig. 1A) to synthesize a HIP library for screening of our murine CD4 T cell clones (illustrated in Fig. 1B). The peptides in the library consist of insulin peptide sequences (left peptides) on the N-terminal side, covalently linked to peptide sequences from other secretory granule peptides (right peptides) on the C-terminal side. As left peptides we chose a selection of C-terminal amino acid sequences derived from proinsulin C-peptide and insulin B-chain fragments that we identified through mass spectrometry in high abundance in antigenic β-cell fractions (Tables S1 and S2). As right peptides we used N-terminal amino acid sequences from the ChgA peptide WE14 and the three natural cleavage products (12) of IAPP (Amylin, IAPP1 and IAPP2).
Fig. 1. Identification of HIP sequences that are recognized by pathogenic T cell clones.
(A) HIPs were synthesized upon chemical activation of the C-termini of acetylated left peptides with EDC/NHS, followed by quenching of residual EDC with DTT. Addition of a right peptide leads to covalent linkage of the right peptide N-terminal amine to the activated C-terminus of the left peptide. (B) A library of 32 HIPs was synthesized. Left peptides are C-terminal amino acid sequences of various mouse insulin C-peptide and B-Chain fragments. Right peptides reflect the N-terminal amino acid sequences of mouse WE14 (WSRMD), IAPP1 (TPVRS), Amylin (KCNTA) and IAPP2 (NAARD). T cell clones BDC-2.5, BDC-10.1, BDC-9.46 and BDC-9.3 were used to screen the peptide library. Black squares indicate positive T cell responses to individual HIPs. Data shown are from three separate experiments. To confirm antigenicity of HIPs, pure peptides (>95%) were obtained commercially for assay with the T cell clones: (C) WE14-reactive clones such as BDC-2.5 respond to 2.5HIP (solid squares) in the low nanomolar range whereas they are stimulated by unmodified WE14 only at high concentrations (100 μM) (open diamonds). IAPP-reactive clones, BDC-6.9 (D) and BDC-9.3 (E), respond to low nanomolar concentrations of the 6.9HIP (solid squares), but were not responsive to IAPP2 alone (open diamonds). None of the clones responds to Insulin 2 C-peptide (open circles) or the Insulin 2 C-peptide fragment ending with the amino acid sequence DLQTLAL (solid circles). Co-incubating the C-Peptide fragment with WE14 or IAPP2 without covalent linkage (open squares) did not increase antigenicity. Amino acid sequences for 2.5HIP and 6.9HIP are shown at the bottom of Fig. 1. Data are representative of at least three separate experiments.
As shown in Fig. 1B, the hybrid peptide LQTLAL-WSRMD, containing the C-peptide sequence LQTLAL on the left side and the WE14 sequence WSRMD on the right side, activates all three WE14-reactive clones (BDC-2.5, BDC-10.1 and BDC-9.46) from our BDC panel. The clones each express a distinct T cell receptor (TCR) and were isolated from different diabetic NOD mice, lending credence to the idea that HIPs are dominant targets for autoreactive T cells in autoimmune diabetes. To further test our hypothesis that antigenic HIPs play a key role in disease onset, we used a second subset of T cell clones (BDC-6.9 and BDC-9.3) that share the same TCR to screen the peptide library. Indirect evidence had previously indicated IAPP as the target antigen for BDC-6.9 and BDC-9.3, but IAPP specificity could not be verified since no IAPP peptide sequence could be identified that could activate these clones (Table S3). Upon screening of the HIP library with BDC-6.9 and BDC-9.3, we identified a single HIP sequence (LQTLAL-NAARD) that is recognized by both clones. This peptide contains on the left side the same C-peptide sequence LQTLAL in the HIP recognized by the WE14-reactive T cell clones and on the right side, the IAPP pro-peptide 2 (IAPP2) sequence NAARD. We concluded that the endogenous ligands for the two sets of pathogenic T cell clones are HIPs containing the C-peptide sequence LQTLAL on the left side and the natural cleavage products, WE14 or IAPP2, on the right side.
To confirm the antigenicity of the described peptides, we obtained commercially synthesized preparations of 2.5 and 6.9HIPs (HIPs antigenic for BDC-2.5 and BDC-6.9) spanning the full-length Ins2 C-peptide fragment ending in LQTLAL on the N-terminal ends and the entire WE14 or IAPP2 sequences on the C-terminal ends (see bottom of Fig.1). As illustrated with BDC-2.5 in Fig. 1C, the WE14-reactive T cell clones recognize the 2.5HIP at low nanomolar concentrations. As previously reported (4, 5), the unmodified peptide WE14 is poorly antigenic for WE14-reactive T cell clones, mandating high peptide concentrations (>10 μM) for T cell activation (Fig. 1C). Hence the HIP was >10,000-fold more potent than WE14 as a stimulator of BDC-2.5. The T cell clones from the second subset (BDC-6.9 and BDC-9.3) recognize the 6.9HIP, also at low nanomolar concentrations (Fig. 1D,E). BDC-6.9 and BDC-9.3 do not react to the unmodified IAPP2 peptide (Fig. 1D), and none of the clones respond to the full length Ins2 C-peptide or the C-peptide fragment ending with the amino acid sequence LQTLAL. T cell reactivity with WE14 or IAPP2 was not altered if these peptides were co-incubated without crosslinking with the C-peptide fragment ending in LQTLAL, indicating that the covalent attachment of the peptides is required to obtain full T cell activation.
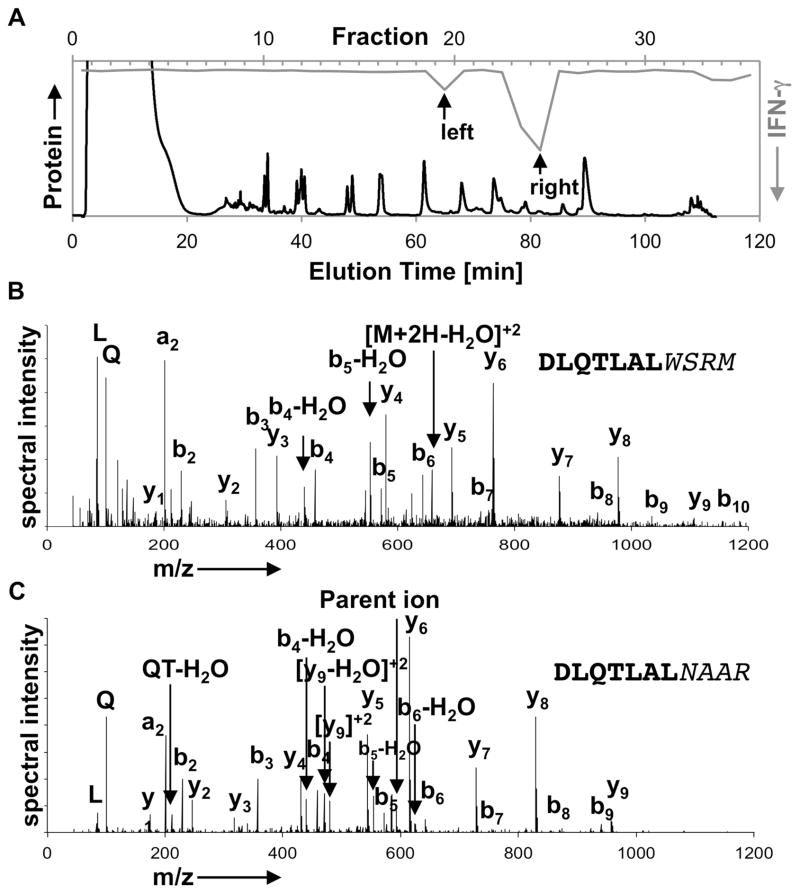
To validate the in vivo presence of HIPs in β-cells, we fractionated murine β-cell extracts by RP-HPLC and performed mass spectrometric analyses on antigenic fractions. As shown in Fig. 2A, the T cell clone BDC-2.5 responded to two chromatographic regions indicating that at least two distinct ligands (left/right peak) exist for this T cell clone. Following proteolytic digestion with flavastacin (AspN), which leads to the release of the core amino acid sequence of the hybrid peptide, and MS/MS analysis of the left antigen peak (Fig. 2A), we identified the peptide DLQTLAL-WSRM (Fig. 2B, Fig. S2). This spectrum provides physical evidence verifying the presence of the 2.5HIP in murine β-cells. Mice secrete two forms of insulin (Ins1/Ins2) with slightly different amino acid sequences, including differences in the C-peptide regions, and consequently, two distinct HIPs can be formed between Ins1/Ins2 C-peptide fragments (ending with the sequence DLQTLAL) and WE14. Because proteolytic processing of either HIP with AspN yields the identical core peptide (DLQTLAL-WSRM) for both HIPs, it is not possible to determine if the identified peptide originated from the Ins1 or Ins2 HIP. We have not identified a HIP in the antigen peak on the right. This lack of identification could be due to limited instrumental sensitivity, poor peptide ionization or the presence of yet another unknown HIP-sequence.
Fig. 2. Identification of HIPs in antigenic β-cell fractions.
(A) Size exclusion chromatography (SEC) fractions highly enriched for antigen were further fractionated by RP-HPLC (black line). IFN-γ T cell responses to individual fractions are shown for BDC-2.5 (grey line). (B) Following the proteolytic digest with AspN, the targeted MS/MS analysis of antigen-containing fractions reveals the spectrum of the HIP that contains the C-peptide amino acid sequence DLQTLAL and the WE14 sequence WSRM. (C) The corresponding 6.9HIP (DLQTLAL-NAAR) could also be identified in AspN digested fractions that contain the antigen recognized by BDC-6.9 and BDC-9.3. Data for chromatographic fractionation are representative of at least three separate experiments. Spectra shown are representative and were obtained from two separate experiments.
Purification of the endogenous ligand recognized by BDC-9.3 (Fig. S3), followed by mass spectrometric analysis, led to the identification of the corresponding 6.9HIP sequence DLQTLAL-NAAR (Fig. 2C, Fig. S4). Both the 2.5HIP and the 6.9HIP contain the C-peptide fragment ending with the amino acid sequence DLQTLAL, previously described to be a naturally occurring cleavage product of insulin C-peptide formed within the secretory granules of β-cells (13). This fragment may thus provide a preferred ligation site for the formation of HIPs, but additional insulin ligation sites may also exist.
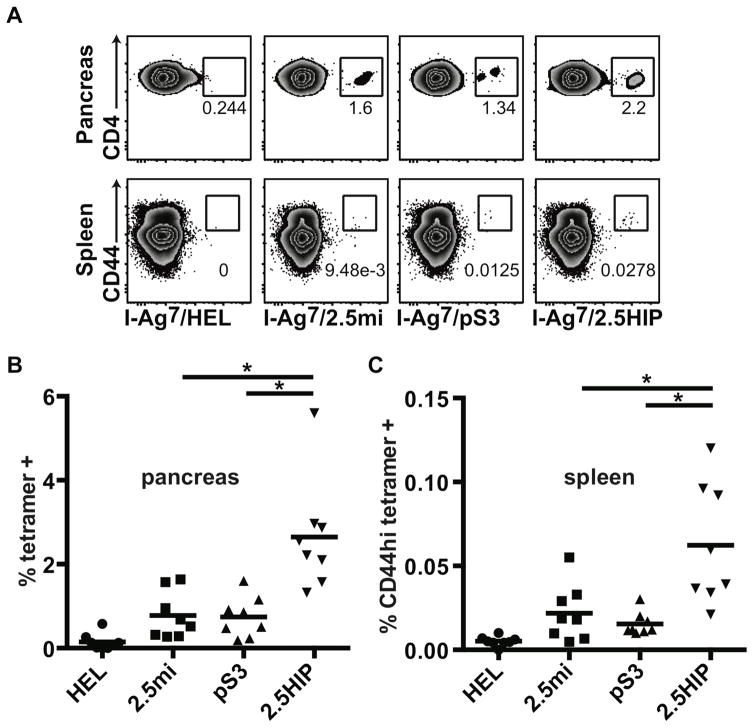
Identifying the autoreactive T cells involved in the pathogenic process is a major objective for T1D research because achieving this goal may enable us to monitor the disease process and devise antigen-specific methods to halt progression toward T1D and/or reverse disease. Peptide-tetramer reagents can be used to specifically identify pathogenic T cells (14). To determine if HIP-reactive T cells could be identified in diabetic NOD mice, we designed a peptide MHC class II (I-Ag7) tetramer containing the HIP sequences antigenic for BDC-2.5 and verified its specificity using CD4 T cell clones from the BDC panel. The 2.5HIP tetramer stained in an antigen-specific fashion all three 2.5HIP-reactive CD4 T cell clones (Fig. S5A). To examine the presence of HIP-reactive T cells in NOD mice, cell suspensions from spleens and pancreata of diabetic NOD mice were stained with the 2.5HIP tetramer (Fig. 3). Two additional tetramers constructed with other peptide sequences recognized by BDC-2.5 (peptide mimotopes), 2.5mi (15) and pS3 (4), were also used as positive controls. We found that 2.5HIP tetramer-positive cells were present in both the spleen and pancreas of NOD mice. In both tissues, there was increased binding by the HIP tetramer to CD4 T cells in comparison to the 2.5mi and the pS3 tetramers, a difference that was statistically significant in the pancreas. These data indicate that the 2.5HIP tetramer is more efficient in the detection of 2.5HIP-reactive CD4 T cells. CD44 expression on T cells in pancreas after collagenase digestion is difficult to assess but our data show that most of the 2.5HIP tetramer-positive cells in the spleen were CD44 high, indicating that a significant proportion of these cells were antigen-experienced and substantiating their involvement in the pathogenic process.
Fig. 3. Tetramer analysis of pancreas and spleen of diabetic NOD mice.
Single cell suspensions were prepared from pancreas and spleen of diabetic NOD female mice (n = 8) and stained with tetramers, antibodies and a live cell marker (7AAD). Gates were set on live leukocytes (7AAD-, CD45), CD4, dump- (CD8, CD11b, CD11c, CD19, F4/80) cells. (A) Tetramer staining in the pancreas and the spleen of a representative mouse. (B) Summary of CD4 tetramer-positive cells present in the pancreas. (C) Summary of CD4 CD44hi tetramer-positive cells present in the spleen. Each symbol represents a different mouse. Averages are indicated as a black horizontal bar. Data are from 4 independent experiments and were analyzed by two-tailed unpaired t-test. Statistical significance (*) was defined at a p value <0.05.
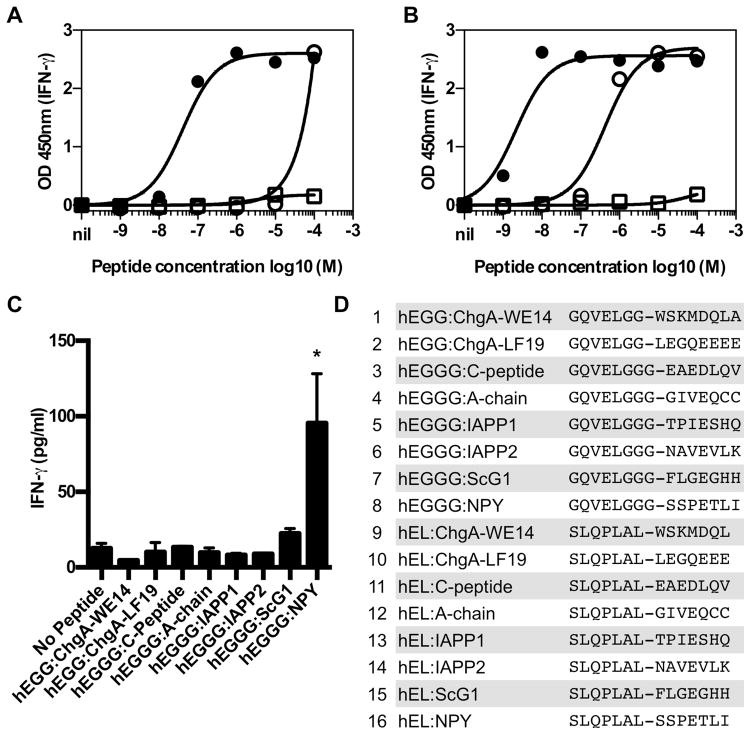
To determine if human CD4 T cells isolated from residual islets of individuals with T1D recognize HIPs, we screened a selection of HIPs (Fig. 4D) using CD4 T cells isolated from two patients. This selection of peptides was designed using the following strategy: for the right side of these HIPs we used corresponding human sequences of naturally occurring peptides that we identified in mouse β-cell extracts. DQ8 favors binding to peptides containing negatively charged anchor residues (E,D) in positions 1 and 9, and small anchor residues (A,S,G) in position 4 (16). The central C-peptide amino acid sequences ELGG or ELGGG, which form the left half of HIPs 1–8, satisfy these parameters for positions 1 and 4. For HIPs 9–16, we used the human version of the murine peptide sequence present in the 2.5 and 6.9 HIPs. Using CD4 T cell clones isolated from the first organ donor (19 year old male with 3 years of T1D (17), we screened a total of 16 HIPs (Fig. 4D). Two clones responded to the same HIP formed by the fusion of proinsulin C-peptide to IAPP2 (GQVELGGG-NAVEVLK) (Fig. 4A,B). Both clones responded weakly to proinsulin C-peptide (proinsulin 37–54), but the HIP was 1,000–10,000 times more potent as an agonist for the human clones than C-peptide. Risk of developing T1D is strongly associated with HLA alleles and HLA-DQ8 confers the highest risk of developing T1D of any single HLA allele (18). Both clones recognized HIP peptides presented by HLA-DQ8 (DQA1*03:01; DQB1*03:02) (Fig. S6). From a second organ donor (20 year-old male with 7 years of T1D), 17 CD4 T cell lines were grown from isolated residual islets, expanded and tested for reactivity against 8 of the HIPs (Fig. 4D, peptides 1–8) in context of Priess B cells (DR4+/+, DQ8+/+). CD4 line MG1 responded to the HIP formed by the fusion of proinsulin C-peptide to the pancreatic polypeptide, neuropeptide Y (GQVELGGG-SSPETLI) (Figure 4C). While pathogenicity of human T cells cannot be directly demonstrated, the presence of HIP-specific, CD4 T cells within the pancreatic islets of individuals with T1D supports the contention that these cells play a pathogenic role in human T1D.
Fig. 4. Human islet-infiltrating T cells respond to HIPs.
Human islet-infiltrating CD4 T cell clones A3.10 (A), A2.11 (B) and the cell line MG1 (C) respond to HIPs. (A and B) Clones from isolated islets of a 19 year-old male with 3 years of T1D were incubated with the indicated concentration of peptide and HLA-DQ2+, DQ8+ EBV transformed B-cell line overnight. Responses to peptide were detected by IFN-γ secretion measured by ELISA. Each point represents the mean and standard deviation of triplicate wells. Solid circles are the HIP (GQVELGGG-NAVEVLK), open circles are proinsulin 37–54 (LQVELGGGPGAGSLQ) open squares are another HIP incorporating the same region of IAPP2 (SLQPLAL-NAVEVLK). Clone A3.10 and A2.11 (Fig. S6) recognize HIPs presented by HLA-DQ8. (C) A CD4 T cell line grown from isolated islets of a 20 year-old male with 7 years of T1D (DR17, DR4, DQ2, DQ8) secreted IFN-γ when a HIP, formed by the fusion of proinsulin C-peptide to the pancreatic polypeptide, neuropeptide Y (GQVELGGG-SSPETLI), was presented by Priess B cells (DR4+/+, DQ8+/+); all peptides at 20μg/ml. *p=0.0395 as compared to the media control (paired Student’s t test). The mean and standard deviation of triplicates in shown in all graphs. For all data, one representative of at least two independent experiments is shown. (D) List of HIPs (>95% purity) screened.
This is the first report of autoantigens that consist of entirely new amino acid sequences formed through peptide bond formation between existing peptides. Importantly, CD4 T cells responding to these hybrid peptides play a role in the development of autoimmunity in the NOD mouse. Our data indicating that human CD4 T cells from T1D patients can respond to HIPs would suggest that HIP-reactive T cells also contribute to disease in humans. HIPs thus represent not only a novel form of PTM in autoimmune disease, but they also constitute a whole new family of autoantigens in T1D. Hybrid peptides that form through protein splicing have been previously noted in tumor antigen formation, a process that occurs through the proteosomal class I pathway, yielding antigens for CD8 T cells (19–21). The formation of such hybrid peptides involves the joining of two peptide sequences originating from within the same protein upon excision of an internal peptide fragment. Unlike this post-translational splicing of proteins in which the two joining peptides originate from the same protein, the HIPs described here form from two separate peptides not previously linked via an intramolecular peptide fragment. Hybrid peptides may well form in the secretory granules of other endocrine tissues and could be a source of self-antigens in other autoimmune disorders.
The identification of HIP-reactive CD4 T cells isolated from the islets of individuals with T1D, and the demonstration that several distinct pathogenic CD4 T cells from NOD mice target different HIPs that form in β-cells, strongly suggests that HIPs play a central role in the pathogenesis of T1D. We hypothesize that HIPs are key antigens for autoreactive T cells and are responsible for the loss of self-tolerance in human T1D. Identification of novel epitopes recognized by pathogenic CD4 T cells reveals new antigens for monitoring changes in human β-cell specific T cell responses and therefore may open new avenues for antigen-based approaches to predict, prevent, and/or cure T1D.
Supplementary Material
(A) IFN-γ response of BDC-2.5 to antigen in chromatographic fractions of β-cell extracts (black lines). Mass spectrometric analysis of fractions reveals the presence of WE14 and insulin 1 C-peptide in chromatographic fractions. The spectral intensity (grey lines) for insulin 1 C-peptide, but not WE14, follows the BDC-2.5 antigen distribution profile. (B) Various insulin 1 and 2 C-peptide fragments that follow the BDC-2.5 antigen distribution profile could be identified in antigenic fractions. This list is a selection of co-eluting insulin 2 C-peptide fragments that could be identified in antigenic fractions by mass spectrometry. Data are representative for at least three separate experiments.
(A) To confirm the identity of the natural peptide from RP-HPLC fractions analyzed in Fig. 2B, the synthetic peptide DLQTLAL-WSRM was obtained and analyzed on the same mass spectrometer. MS/MS product ion spectra for the natural and synthetic peptides were both normalized by dividing ion intensities by the intensity of the most abundant product ion in the spectrum. Normalized spectra were then overlaid. Representative of two independent experiments. (B) The observed m/z values of b- and y-ions in the MS/MS spectra of the peptide DLQTLAL-WSRM found in RP-HPLC fractions of purified β-cell antigen. The predicted m/z values are shown for comparison. The differences between the m/z of observed ions and the predicted values are reported in parts-per-million. Representative of two independent experiments.
(A) To confirm the identity of the natural peptide from RP-HPLC fractions analyzed in Fig. 2C, the synthetic peptide DLQTLAL-NAAR was obtained and analyzed on the same mass spectrometer. MS/MS product ion spectra for the natural and synthetic peptides were both normalized by dividing ion intensities by the intensity of the most abundant product ion in the spectrum. Normalized spectra were then overlaid. Representative of two independent experiments. (B) The observed m/z values of b- and y-ions in the MS/MS spectra of the peptide DLQTLAL-NAAR found in RP-HPLC fractions of purified β-cell antigen. The predicted m/z values are shown for comparison. The differences between the m/z of observed ions and the predicted values are reported in parts-per-million. Representative of two independent experiments.
The CD4 BDC T cell clones (1 x 10e5 cells) were stained with I-Ag7 tetramers, anti-CD4 and 7AAD before flow cytometry analysis. Gates were set on live (7AAD-), CD4+ cells. These data are representative of 3 similar experiments.
Fig. S6: Clones A2.11 and A3.10 recognize HIPs presented by HLA-DQ8
Only a monoclonal antibody specific for HLA-DQ inhibits A2.11’s (A) and A3.10’s (C) responses to HIP peptide. Mean and standard deviation of triplicates and one representative of at least two independent experiments is shown. Responses were compared by one-way ANOVA with the Giesser-Greenhouse correction. (*** p<0.0001, ** p<0.0013). Class II deficient T-2 cells transduced with the HLA molecules indicated were used to define the HLA-restriction. Clone A2.10 (B) and A3.10 (D) can only respond to HIP when presented by APC expressing HLA-DQ8. Mean and standard deviation of triplicates and one representative of at least two independent experiments is shown. Responses in the presence of different HLA-expressing APCs were compared by one-way ANOVA with the Giesser-Greenhouse correction. (*** p<0.0001 and ** p=0.0014).
Table S1: Insulin 1 peptides identified in antigenic size exclusion fractions
Table S2: Insulin 2 peptides identified in antigenic size exclusion fractions
Table S3: IAPP Peptide panel tested with BDC-6.9 and BDC-9.3
Acknowledgments
This research was supported by NIH grant 1K01DK094941 (TD) and American Diabetes Association Pathway to Stop Diabetes Grant 1-15-ACE-14 (TD); NIH grant 1R01DK081166 (KH); the Australian National Health and Medical Research Council (NHMRC) APP1061961) (SM); Juvenile Diabetes Research Foundation, Career Development Award 5-CDA2014210-AN (SM); Operational Infrastructure Support Program from the Victorian State Government of Australia (SM); Juvenile Diabetes Research Foundation grant 2-SRA-2015-68-Q-R (ACP); NIH DK104211, DK89572, DK72473 (ACP); Juvenile Diabetes Research Foundation SRA-2015-68-Q-R (ACP); Department of Veterans Affairs BX000666 (ACP); Vanderbilt Diabetes Research and Training Center DK20593 (ACP). We also thank the NIH tetramer core for providing tetramer reagents. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials section.
Footnotes
References and Notes
- 1.Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nature medicine. 1999 Jun;5:601. doi: 10.1038/9442. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama M, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005 May 12;435:220. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haskins K. Pathogenic T-cell clones in autoimmune diabetes: more lessons from the NOD mouse. Advances in immunology. 2005;87:123. doi: 10.1016/S0065-2776(05)87004-X. [DOI] [PubMed] [Google Scholar]
- 4.Stadinski BD, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nature immunology. 2010 Mar;11:225. doi: 10.1038/ni.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delong T, et al. Islet amyloid polypeptide is a target antigen for diabetogenic CD4+ T cells. Diabetes. 2011 Sep;60:2325. doi: 10.2337/db11-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb PA, et al. Chromogranin A is a T cell antigen in human type 1 diabetes. Journal of autoimmunity. 2013 Nov 12; doi: 10.1016/j.jaut.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle HA, Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Current opinion in immunology. 2012 Feb;24:112. doi: 10.1016/j.coi.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mannering SI, et al. The insulin A-chain epitope recognized by human T cells is posttranslationally modified. The Journal of experimental medicine. 2005 Nov 7;202:1191. doi: 10.1084/jem.20051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delong T, et al. Diabetogenic T-cell clones recognize an altered peptide of chromogranin A. Diabetes. 2012 Dec;61:3239. doi: 10.2337/db12-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Lummel M, et al. Posttranslational modification of HLA-DQ binding islet autoantigens in type 1 diabetes. Diabetes. 2014 Jan;63:237. doi: 10.2337/db12-1214. [DOI] [PubMed] [Google Scholar]
- 11.McGinty JW, et al. Recognition of posttranslationally modified GAD65 epitopes in subjects with type 1 diabetes. Diabetes. 2014 Sep;63:3033. doi: 10.2337/db13-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higham CE, et al. Processing of synthetic pro-islet amyloid polypeptide (proIAPP) ‘amylin’ by recombinant prohormone convertase enzymes, PC2 and PC3, in vitro. European journal of biochemistry / FEBS. 2000 Aug;267:4998. doi: 10.1046/j.1432-1327.2000.01548.x. [DOI] [PubMed] [Google Scholar]
- 13.Verchere CB, et al. Des-(27–31)C-peptide. A novel secretory product of the rat pancreatic beta cell produced by truncation of proinsulin connecting peptide in secretory granules. The Journal of biological chemistry. 1996 Nov 1;271:27475. doi: 10.1074/jbc.271.44.27475. [DOI] [PubMed] [Google Scholar]
- 14.Nepom GT. MHC Class II Tetramers. J Immunol. 2012 Mar 15;188:2477. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stratmann T, et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. The Journal of clinical investigation. 2003 Sep;112:902. doi: 10.1172/JCI18337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang KY, Unanue ER. Prediction of HLA-DQ8beta cell peptidome using a computational program and its relationship to autoreactive T cells. International immunology. 2009 Jun;21:705. doi: 10.1093/intimm/dxp039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pathiraja V, et al. Proinsulin-specific, HLA-DQ8, and HLA-DQ8-transdimer-restricted CD4+ T cells infiltrate islets in type 1 diabetes. Diabetes. 2015 Jan;64:172. doi: 10.2337/db14-0858. [DOI] [PubMed] [Google Scholar]
- 18.Todd JA, Bell JI, McDevitt HO. HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987 Oct 15–21;329:599. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 19.Warren EH, et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science. 2006 Sep 8;313:1444. doi: 10.1126/science.1130660. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004 Jan 15;427:252. doi: 10.1038/nature02240. [DOI] [PubMed] [Google Scholar]
- 21.Vigneron N, et al. An antigenic peptide produced by peptide splicing in the proteasome. Science. 2004 Apr 23;304:587. doi: 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- 22.Grabarek Z, Gergely J. Zero-length crosslinking procedure with the use of active esters. Analytical biochemistry. 1990 Feb 15;185:131. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 23.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988 Apr;37:413. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- 24.Ciantar JP, Mannering SI. An improved method for growing and analysing human antigen-specific CD4+ T-cell clones. Diabetes/metabolism research and reviews. 2011 Nov;27:906. doi: 10.1002/dmrr.1271. [DOI] [PubMed] [Google Scholar]
- 25.Mannering SI, et al. An efficient method for cloning human autoantigen-specific T cells. Journal of immunological methods. 2005 Mar;298:83. doi: 10.1016/j.jim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Balamurugan AN, et al. Suitability of human juvenile pancreatic islets for clinical use. Diabetologia. 2006 Aug;49:1845. doi: 10.1007/s00125-006-0318-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) IFN-γ response of BDC-2.5 to antigen in chromatographic fractions of β-cell extracts (black lines). Mass spectrometric analysis of fractions reveals the presence of WE14 and insulin 1 C-peptide in chromatographic fractions. The spectral intensity (grey lines) for insulin 1 C-peptide, but not WE14, follows the BDC-2.5 antigen distribution profile. (B) Various insulin 1 and 2 C-peptide fragments that follow the BDC-2.5 antigen distribution profile could be identified in antigenic fractions. This list is a selection of co-eluting insulin 2 C-peptide fragments that could be identified in antigenic fractions by mass spectrometry. Data are representative for at least three separate experiments.
(A) To confirm the identity of the natural peptide from RP-HPLC fractions analyzed in Fig. 2B, the synthetic peptide DLQTLAL-WSRM was obtained and analyzed on the same mass spectrometer. MS/MS product ion spectra for the natural and synthetic peptides were both normalized by dividing ion intensities by the intensity of the most abundant product ion in the spectrum. Normalized spectra were then overlaid. Representative of two independent experiments. (B) The observed m/z values of b- and y-ions in the MS/MS spectra of the peptide DLQTLAL-WSRM found in RP-HPLC fractions of purified β-cell antigen. The predicted m/z values are shown for comparison. The differences between the m/z of observed ions and the predicted values are reported in parts-per-million. Representative of two independent experiments.
(A) To confirm the identity of the natural peptide from RP-HPLC fractions analyzed in Fig. 2C, the synthetic peptide DLQTLAL-NAAR was obtained and analyzed on the same mass spectrometer. MS/MS product ion spectra for the natural and synthetic peptides were both normalized by dividing ion intensities by the intensity of the most abundant product ion in the spectrum. Normalized spectra were then overlaid. Representative of two independent experiments. (B) The observed m/z values of b- and y-ions in the MS/MS spectra of the peptide DLQTLAL-NAAR found in RP-HPLC fractions of purified β-cell antigen. The predicted m/z values are shown for comparison. The differences between the m/z of observed ions and the predicted values are reported in parts-per-million. Representative of two independent experiments.
The CD4 BDC T cell clones (1 x 10e5 cells) were stained with I-Ag7 tetramers, anti-CD4 and 7AAD before flow cytometry analysis. Gates were set on live (7AAD-), CD4+ cells. These data are representative of 3 similar experiments.
Fig. S6: Clones A2.11 and A3.10 recognize HIPs presented by HLA-DQ8
Only a monoclonal antibody specific for HLA-DQ inhibits A2.11’s (A) and A3.10’s (C) responses to HIP peptide. Mean and standard deviation of triplicates and one representative of at least two independent experiments is shown. Responses were compared by one-way ANOVA with the Giesser-Greenhouse correction. (*** p<0.0001, ** p<0.0013). Class II deficient T-2 cells transduced with the HLA molecules indicated were used to define the HLA-restriction. Clone A2.10 (B) and A3.10 (D) can only respond to HIP when presented by APC expressing HLA-DQ8. Mean and standard deviation of triplicates and one representative of at least two independent experiments is shown. Responses in the presence of different HLA-expressing APCs were compared by one-way ANOVA with the Giesser-Greenhouse correction. (*** p<0.0001 and ** p=0.0014).
Table S1: Insulin 1 peptides identified in antigenic size exclusion fractions
Table S2: Insulin 2 peptides identified in antigenic size exclusion fractions
Table S3: IAPP Peptide panel tested with BDC-6.9 and BDC-9.3