Abstract
Ubiquitin and some of its homologues target proteins to the proteasome for degradation. Other ubiquitin‐like domains are involved in cellular processes unrelated to the proteasome, and proteins containing these domains remain stable in the cell. We find that the 10 yeast ubiquitin‐like domains tested bind to the proteasome, and that all 11 identified domains can target proteins for degradation. Their apparent proteasome affinities are not directly related to their stabilities or functions. That is, ubiquitin‐like domains in proteins not part of the ubiquitin proteasome system may bind the proteasome more tightly than domains in proteins that are bona fide components. We propose that proteins with ubiquitin‐like domains have properties other than proteasome binding that confer stability. We show that one of these properties is the absence of accessible disordered regions that allow the proteasome to initiate degradation. In support of this model, we find that Mdy2 is degraded in yeast when a disordered region in the protein becomes exposed and that the attachment of a disordered region to Ubp6 leads to its degradation.
Keywords: degradation, proteasome, ubiquitin‐like domain
Subject Categories: Post-translational Modifications, Proteolysis & Proteomics
Introduction
Ubiquitin is a small single‐domain protein consisting of 76 amino acids that is conserved in all eukaryotes. It becomes attached to proteins post‐translationally, often in long polyubiquitin chains in which the first ubiquitin is modified with additional ubiquitin moieties. The first role discovered for polyubiquitin chains was to target proteins to the proteasome for degradation, but it is now clear that ubiquitin chains are also involved in the regulation of cellular processes via non‐proteolytic mechanisms ranging from membrane trafficking to DNA repair (Komander & Rape, 2012). It is not fully understood how cells use ubiquitin chains to specify distinct cellular processes.
Biochemically, ubiquitin chains function as transferable protein–protein interaction tags and are recognized by ubiquitin binding domains, sometimes within the same protein and sometimes in interacting proteins (Husnjak & Dikic, 2012). Individual ubiquitin moieties in polyubiquitin chains are connected to each other through different lysine residues, and chains with different linkage patterns are recognized with different affinities by ubiquitin receptors (Husnjak & Dikic, 2012; Komander & Rape, 2012). However, other properties of the modified proteins also affect their fate and provide additional information to the ubiquitin code (Prakash et al, 2004; Takeuchi et al, 2007; Zhao et al, 2010; Fishbain et al, 2011, 2015; Heinen et al, 2011; Inobe et al, 2011; van der Lee et al, 2014).
Cells also encode a number of ubiquitin homologues and in budding yeast (S. cerevisiae), at least eleven proteins show homology with ubiquitin. The sequence identity within these ubiquitin‐like (UbL) domains ranges between 7 and 53%, and they all share ubiquitin's β‐grasp fold (Grabbe & Dikic, 2009) (Fig 1). Although some UbL domains bind to the proteasome, it is thought that none of the proteins that carry UbL domains are degraded by the proteasome.
Figure 1. UbL domains.

- Phylogenetic relationship of UbL domains in S. cerevisiae and schematic representations of the corresponding UbL proteins. Sequences were clustered and mapped onto a N‐J tree by ClustalX2 (Larkin et al, 2007) and the PHYLIP package (Felsenstein, 1989). ASP, aspartyl protease domain; UBA, ubiquitin‐associated domain; USP, ubiquitin‐specific protease domain; DD, dimerization domain; RBD, Rad4‐binding domain; ST2, Sti1/Sti1 domain pair.
- Structural models of UbL domains generated by PyMOL (The PyMOL Molecular Graphics System, Version 0.99 Schrödinger, LLC). PDB IDs: Atg8 2ZPN, Atg12 3W1S, Dsk2 2BWF, Hub1 1M94, Mdy2 4GOC, Rad23 3M62, hHR23B 1P1A, Rub1 1BT0, Smt3 1EUV, Urm1 2PK0, ubiquitin 1UBQ.
Ubiquitin‐like proteins fall into two classes, ubiquitin‐like modifiers (ULMs) and ubiquitin‐like domain proteins (UDPs). Ubiquitin‐like modifiers are attached to proteins reversibly and post‐translationally, like ubiquitin (van der Veen & Ploegh, 2012). In yeast, the ULMs are Atg8, Hub1, Rub1 (Nedd8), Smt3 (SUMO), and Urm1, and these proteins are not thought to bind to the proteasome (Table 1). Rub1 may be an exception in that it can be incorporated into polyubiquitin chains, which in turn can be recognized by the proteasome (Singh et al, 2012). Ubiquitin‐like domain proteins are larger than ubiquitin and typically contain multiple domains, with only one homologous to ubiquitin. Four of the six UDPs in yeast are associated with the ubiquitin proteasome system (UPS) and interact directly with the proteasome through their UbL domains. Rad23, Dsk2, and Ddi1 contain N‐terminal UbLs, which are recognized by the proteasome subunits Rpn1, Rpn10, and Rpn13 (Elsasser et al, 2002; Funakoshi et al, 2002; Kaplun et al, 2005; Husnjak et al, 2008; Zhang et al, 2009; Gomez et al, 2011). Rad23, Dsk2, and Ddi1 also contain one or more ubiquitin‐associated (UBA) domains, which bind to polyubiquitin chains and allow the proteins to serve as diffusible proteasome substrate receptors (Saeki et al, 2002a; Elsasser et al, 2004; Kim et al, 2004; Verma et al, 2004; Elsasser & Finley, 2005; Kaplun et al, 2005; Zhang et al, 2009). The Ubp6 protein binds to the proteasome subunit Rpn1 through a UbL domain at its N‐terminus and through interactions of its catalytic domain (Leggett et al, 2002; Rosenzweig et al, 2012; Aufderheide et al, 2015; Bashore et al, 2015; Shi et al, 2016). Ubp6 modulates proteasome activity by trimming ubiquitin chains on proteasome substrates and by modulating proteasome activity allosterically (Crosas et al, 2006; Hanna et al, 2006; Koulich et al, 2008; Peth et al, 2009; Lee et al, 2010; Bashore et al, 2015). Neither the UbL‐UBA proteins nor Ubp6 are degraded by the proteasome. The remaining two UDPs are not associated with the UPS. Atg12 contains a C‐terminal UbL domain and participates in the autophagy pathway (Mizushima et al, 1998), whereas Mdy2 contains a central UbL domain and is involved in the biogenesis of tail‐anchored proteins in the endoplasmic reticulum (Wang et al, 2010; Chartron et al, 2012a).
Table 1.
Physical and sequence properties of UbL domains analyzed in this study as well as the function of the relevant proteins
| Protein | Class | Full length (aa) | UbL domain (aa) | Identity to Ub (%) | Identity to UbLRad23 (%) | Secondary structure identity to Ub (%SSE) | Function | K i relative to UbLRad23 (μM) |
|---|---|---|---|---|---|---|---|---|
| Rad23 | UDP | 398 | 1–77 | 22 | – | 83 | DNA excision repair; UPS | 0.45 ± 0.04 |
| hHR23B | UDP | 409 | 1–82 | 30 | 27 | 100 | DNA excision repair; UPS | 2.0 ± 0.2 |
| Dsk2 | UDP | 373 | 1–76 | 29 | 28 | 67 | Spindle pole body duplication; UPS | 3.3 ± 0.3 |
| Atg8 | ULM | 117 | 1–117 | 20 | 14 | 83 | Autophagy | 3.5 ± 0.2 |
| Hub1 | ULM | 73 | 1–73 | 22 | 18 | 83 | Pre‐mRNA splicing; morphogenesis | 9.8 ± 0.8 |
| ubiquitin | – | 76 | 1–76 | – | 22 | – | Ubiquitination | 16 ± 1 |
| Ddi1 | UDP | 428 | 1–80 | 20 | 19 | N/A | Mating type switching; UPS | 17 ± 2 |
| Ubp6 | UDP | 499 | 6–80 | 17 | 23 | N/A | Deubiquitination | 24 ± 3 |
| Urm1 | ULM | 99 | 1–99 | 28 | 6 | 83 | Sulfur carrier in tRNA modification | 30 ± 3 |
| Mdy2 | UDP | 212 | 74–152 | 22 | 34 | 83 | Biogenesis of TA proteins | 33 ± 3 |
| Rub1 | ULM | 77 | 1–77 | 53 | 22 | 100 | Cullin proteins neddylation | 34 ± 2 |
| Smt3 | ULM | 101 | 22–98 | 16 | 17 | 50 | Sumoylation | 65 ± 4 |
| Atg12 | UDP | 186 | 101–186 | 7 | 9 | 83 | Autophagy | N/A |
Ub, ubiquitin; UDP, ubiquitin‐like domain protein; ULM, ubiquitin‐like modifier; UPS, ubiquitin proteasome system; N/A, not available.
Here, we ask how the UbL domains specify different functions. The simplest model would be that domains in proteins not involved in the UPS do not bind to the proteasome. To test this model, we selected twelve ubiquitin‐like domains, eleven encoded by S. cerevisiae and one by H. sapiens, and examined whether they could target proteins to the yeast proteasome. We found that the yeast proteasome can bind all UbL domains tested, albeit with different affinities. The in vivo functions of the domains do not correlate with their apparent proteasome affinities; some UbL domains not involved in the UPS can bind the proteasome more tightly than bona fide interactors. We then asked why the proteasome does not degrade UbL proteins. We propose that proteins escape proteasomal degradation when they lack unstructured regions that allow the proteasome to initiate degradation. We tested this hypothesis on two ubiquitin domain proteins, Ubp6 and Mdy2. Ubp6 naturally lacks sites at which the proteasome can initiate degradation, but was depleted rapidly when a disordered region was introduced. The N‐terminal domain of Mdy2 is disordered but is buried by its binding partner Get4. Deleting Get4 exposed the disordered region and led to the degradation of Mdy2. We conclude that protein stability is determined not only by proteasome affinity, but also by the presence of sites at which the proteasome can engage its substrates and initiate degradation.
Results
UbL domains bind the proteasome
To investigate whether UbL domains bind to the proteasome, we tested whether the purified domains could displace a proteasome substrate and prevent its degradation. We constructed a model substrate with the UbL domain of S. cerevisiae Rad23 at its N‐terminus followed by a superfolder green fluorescent protein (GFP) domain (Pédelacq et al, 2006) and finally a previously characterized C‐terminal disordered region of 95 amino acids derived from S. cerevisiae cytochrome b 2 to allow the proteasome to initiate the degradation (Inobe et al, 2011) (UbLRad23‐GFP‐95; Fig 2A). We then expressed the protein in E. coli, isolated it, and presented it to purified yeast proteasome. The protein was degraded efficiently and at a rate comparable to that observed for model substrates targeted to the proteasome by ubiquitin tags under equivalent conditions (Fishbain et al, 2011; Kraut & Matouschek, 2012). The degradation followed Michaelis–Menten behavior and the measured K M value was independent of proteasome concentration, whereas V max values scaled linearly with it (Fig 2C). Thus, it was possible to characterize the degradation process using standard kinetic approaches.
Figure 2. UbL domains bind with proteasome.

- Sketch of UbLRad23‐GFP‐95 consisting of the UbL domain of S. cerevisiae Rad23, followed by superfolder GFP and a 95‐amino acid‐long tail derived from S. cerevisiae cytochrome b 2.
- In vitro degradation of UbLRad23‐GFP‐95 by purified S. cerevisiae proteasome. The graph plots the amount of substrate over time, estimated by fluorescence intensity monitored by plate reader (green) or electronic autoradiography of SDS–PAGE gel bands (red), normalized to the initial substrate amount as described in Materials and Methods.
- Michaelis–Menten analysis of UbLRad23‐GFP‐95 degradation by different concentrations of purified S. cerevisiae proteasome (green, 10 nM; red, 40 nM).
- UbLRad23 inhibits UbLRad23‐GFP‐95 degradation. The initial degradation rates of UbLRad23‐GFP‐95 at different concentrations of purified UbLRad23 domain are plotted and fitted to the equation describing competitive inhibition.
- The UbLDsk2 domain is a competitive inhibitor of UbLRad23‐GFP‐95 degradation. Lineweaver–Burk plot of UbLRad23‐GFP‐95 degradation with different concentrations of purified UbLDsk2 domain.
To investigate the binding of different UbL domains to the proteasome, we cloned and expressed the domains in E. coli and purified the corresponding proteins. We first characterized the UbL domains from Rad23 and Dsk2 because these two domains are known to bind to the proteasome (Schauber et al, 1998; Elsasser et al, 2002; Funakoshi et al, 2002; Saeki et al, 2002b). Adding increasing amounts of the purified UbL domains inhibited the degradation of UbLRad23‐GFP‐95 (Figs 2D and E, and EV1). Conversely, increasing amount of substrate overcame the inhibition by the UbL domains (Fig 2E). Thus, substrate and UbL domains competed for binding to the proteasome. The apparent equilibrium constants (K is) with which the competing UbL domains inhibited substrate degradation reflect the affinity of the domains to the proteasome (Table 1, Fig EV1). The measured K i values report only on binding to the sites at which the Rad23 UbL domain interacts with the proteasome. It is possible that the other UbL domains also bind to other sites on the proteasome (Gomez et al, 2011; Shi et al, 2016) but these interactions would not be detected by our assay, causing us to underestimate affinities in these cases.
Figure EV1. UbL domains inhibit UbLRad23‐GFP‐95 degradation.

-
A–LInitial degradation rates of UbLRad23‐GFP‐95 in the presence of different concentrations of purified UbL domains plotted against the competitor concentration and fitted to the equation describing competitive inhibition. The UbL domains were from yeast Rad23 (A), Dsk2 (C), Atg8 (D), Hub1 (E), ubiquitin (F), Ddi1 (G), Ubp6 (H), Urm1 (I), Mdy2 (J), Rub1 (K), Smt3 (L), and hHR23B (B). Graphs show one representative dataset of at least three independent experiments.
We then measured the ability of nine additional UbL domains, as well as monoubiquitin, to compete with Rad23's UbL domain for the proteasome (Table 1). The apparent K is fell into a range from 0.5 to 65 μM. UbL domains that are known to associate with the proteasome were distributed over the entire range of affinities, with monoubiquitin roughly in the middle.
UbL domains can target artificial substrates for proteasomal degradation in vitro
Next we examined whether all UbL domains that bound to the proteasome could also target proteins for degradation. We first addressed this question in vitro with model proteasome substrates consisting of a central E. coli dihydrofolate reductase (DHFR) domain with the relevant UbL domain fused to its N‐terminus and the 95‐amino acid tail described above fused to its C‐terminus, creating UbL‐DHFR‐95 (Fig 3A). We expressed these proteins by coupled in vitro transcription and translation and presented them to purified yeast proteasome. The UbL domain of Rad23 mediated efficient degradation. However, DHFR‐95 without the UbL domain and UbLRad23‐DHFR without the tail were stable (Fig 3A). Treatment with the proteasome inhibitor MG132 stabilized the UbLRad23‐DHFR‐95 protein, indicating that degradation was proteasome dependent (Fig 3A). Strikingly, all of the other UbL domains tested also mediated degradation (Fig 3B). As before, degradation depended on the presence of a disordered tail and was repressed by the proteasome inhibitor MG132. The rates and extent of degradation in the in vitro assays varied somewhat for the different UbL domains, but the variation did not seem to depend on whether the domains were naturally involved in proteasome‐dependent processes, nor did they correlate with the apparent inhibition constants (K is). For example, the yeast SUMO homolog Smt3 targeted DHFR for degradation as effectively as the Rad23 UbL domain but bound to the proteasome more weakly (Fig 3A and B, Table 1).
Figure 3. UbL domains target substrate to proteasome degradation in vitro .
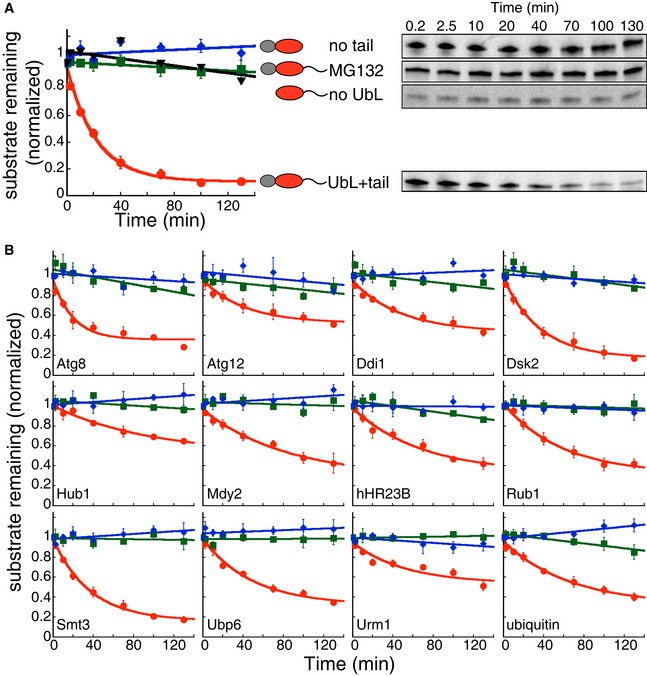
-
A, BIn vitro degradation of model proteins by purified S. cerevisiae proteasome. The model proteins (UbL‐DHFR‐95) consisted of an N‐terminal UbL domain, followed by an E. coli DHFR domain and a 95‐amino acid tail derived from S. cerevisiae cytochrome b 2. UbL‐DHFR‐95 proteins were degraded by proteasome (red solid circle) and stabilized by removing the UbL domain (no UbL, black triangles), by removing the 95‐amino acid tail (no tail, blue diamonds), or by proteasome inhibitor (MG132, green squares). (A) Degradation of model proteins containing the UbL domain of S. cerevisiae Rad23. (B) Degradation of model proteins containing other UbL domains or ubiquitin. The graphs plot the amount of substrate estimated by electronic autoradiography in SDS–PAGE gel bands over time as normalized to the initial substrate amount as described in Materials and Methods. Data points represent mean values determined from at least three repeat experiments; error bars indicate s.e.m.
Source data are available online for this figure.
UbL domains can target proteins for proteasome degradation in vivo
We next investigated whether UbL domains could mediate degradation by the proteasome in yeast. We created model substrates based on imidazoleglycerol‐phosphate dehydratase (His3) and yellow fluorescent protein (YFP). The abundance of His3 protein was estimated by its ability to support growth, and the abundance of yellow fluorescent protein (YFP) by measuring cellular fluorescence.
His3 catalyzes an essential step in histidine production in yeast, so that his3 mutant strains cannot grow in medium lacking histidine unless the mutation is complemented by a plasmid‐borne wild‐type HIS3 gene (Alifano et al, 1996). However, if the His3 protein expressed from the plasmid is degraded, then the complementation will fail. Proteasomal degradation of His3 protein can therefore modulate the yeast growth rate. We tested whether UbL domains could target His3 protein for degradation in yeast by attaching them to the N‐terminus of His3. The fusion proteins were expressed from a GAL1 promoter on a 2‐micron (multicopy) plasmid in a his3 mutant yeast strain (Fleming et al, 2002) (Appendix Table S1). We added 3‐amino‐1,2,4‐triazole (3‐AT), a competitive inhibitor of the His3 enzyme, to make the growth assay more sensitive (Hawkes et al, 1995). To allow the proteasome to initiate the degradation of any bound His3 fusion protein, we also fused a 51‐amino acid disordered tail (derived from subunit 9 of the F o component of the N. crassa ATPase, abbreviation Su9) to the C‐terminus of His3 (Fig 4A). Expression of His3 with an N‐terminal DHFR domain and the Su9 tail complemented his3 mutant cells, indicating that N‐ and C‐terminal modifications alone did not impede His3 activity (Fig 4B). However, fusing the Rad23 UbL domain to the N‐terminus of His3 with a Su9 tail prevented the complementation and inhibited cell growth (Fig 4B). His3 fusion protein could not be detected by Western blotting in cell lysate, unless the proteasome was inhibited by the proteasome inhibitor bortezomib (Fig EV2A), suggesting that the growth defect was caused by the depletion of UbL‐His3‐tail protein by the proteasome. Indeed, most of the yeast UbL domains we investigated prevented His3 from complementing the his3 mutation to some extent. The exceptions were the UbL domains of Ddi1 and Urm1, which affected growth similarly under selective and nonselective conditions, indicating that UbLDdi1 and UbLUrm1 failed to target His3 fusion proteins to degradation effectively. None of the UbL domains themselves disrupted His3 function as UbL‐His3 fusion proteins lacking the C‐terminal proteasome initiation tails were stable (Fig EV2B) and supported growth (Fig 2B). Expressing either UbL‐His3‐tail or UbL‐His3 constructs when histidine was present in the medium did not affect yeast growth. The simplest interpretation of our observations is that the different UbL domains target His3 to the proteasome. Furthermore, degradation by the proteasome required a disordered region at which the proteasome could initiate the degradation.
Figure 4. UbL domains target His3 substrates to proteasome degradation in yeast.

- Schematic representation of yeast growth assay. The model proteins consisted of an UbL domain and S. cerevisiae imidazoleglycerol‐phosphate dehydratase (His3), followed by stop codon (no tail) or a 51‐amino acid tail derived from subunit 9 of the F o component of the Neurospora crassa ATPase (Su9) at the C‐terminus. In his3 mutant cells, the absence of His3 protein caused a growth defect when grown on selective media. Stable His3 proteins escaped proteasome degradation and rescued the his3 mutant cells from the growth defect.
- UbL domains mediated the degradation of His3 fusion proteins with a Su9 tail and affected yeast growth under selective condition (+3‐AT, −his). Replacing UbL domains with DHFR domains rescued the growth defect. Cells expressing His3 fusion proteins in late log phase were serially diluted and stamped on selective plates. The plates were incubated at 30°C for 3 days for imaging.
Figure EV2. UbL domain‐mediated His3 protein degradation depends on proteasome, but not ubiquitination.
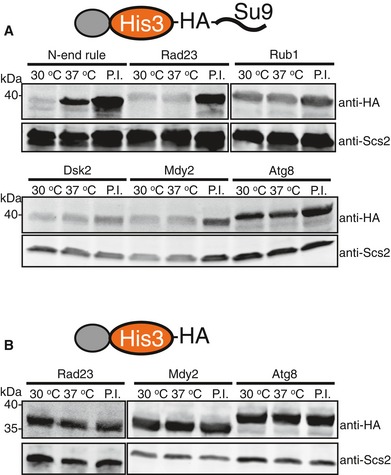
- In the E1 temperature‐sensitive strain uba1‐204, the cellular abundance of UbL‐His3‐tail proteins was similar at the permissive (30°C) and restrictive (37°C) temperatures. In contrast, a His3 fusion protein containing a classic N‐end rule degron was stabilized at the restrictive temperature (37°C), showing that its degradation depended on the ubiquitination machinery. Bortezomib stabilized both the UbL‐His3‐tail and N‐degron‐His3‐tail proteins.
- UbL‐His3 proteins without initiation region were stable and not affected by the shift to the restrictive temperature or treatment with bortezomib.
The his3 complementation experiments report on cellular protein abundance indirectly through the His3 protein's enzymatic activity and its effect on cell metabolism and growth. The abundance of fluorescent proteins can be estimated directly from the total cell fluorescence measured by flow cytometry (Yen et al, 2008; Sharon et al, 2012). Therefore, we repeated the degradation experiments with yellow fluorescent protein (YFP) as the reporter protein. We fused either one of the eleven UbL domains or a DHFR domain to the N‐terminus of YFP and a Su9 tail to its C‐terminus, and then expressed the fusion proteins (UbL‐YFP‐tail) from the constitutive promoter pACT1 on a CEN plasmid derived from pYCplac33 (Gietz & Sugino, 1988). The same plasmid also expressed the red fluorescent protein dsRed Express2 (Strack et al, 2008) from a second pACT1 promoter as a normalization control for variation in plasmid copy number, global protein expression levels, and cell size. We again first investigated the yeast Rad23 UbL domain, measuring the fluorescence of yeast cells expressing the fusion protein UbLRad23‐YFP‐tail together with dsRed (Fig 5A). Replacing the UbL domain with DHFR increased the yellow fluorescence roughly 9‐fold, while the red fluorescence remained unchanged (Fig 5A). Inhibiting the proteasome with bortezomib increased yellow fluorescence without changing red fluorescence (Figs 5A and EV3B). Degradation required a C‐terminal tail; removing the tail increased YFP fluorescence 9‐fold and bortezomib did not affect the fluorescence of cells expressing UbLRad23‐YPF protein without the tail (Fig EV3B and C).
Figure 5. UbL domains target fluorescence substrates to proteasome degradation in yeast.
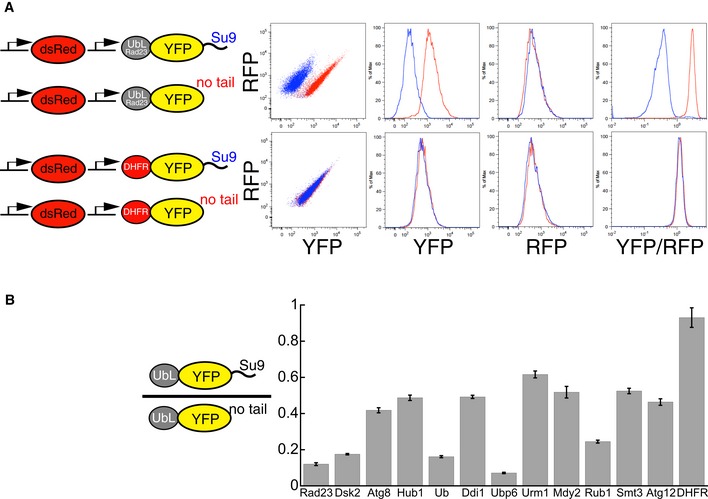
- Fluorescence‐based degradation assay in S. cerevisiae. The substrate proteins consisted of a UbL domain, followed by a yellow fluorescent protein (YFP) domain and a disordered tail (51 amino acids derived from subunit 9 of the F o component of the Neurospora crassa ATPase, Su9). Control substrates lacked the disordered tail. Substrates and a red fluorescent protein were expressed from consecutive constitutive promoters (pACT1) on the same CEN plasmid (YCplac33). Fluorescence profiles of cells expressing different substrates monitored by flow cytometry. UbLRad23 targeted YFP‐Su9 protein for degradation. Removing the Su9 tail or replacing UbLRad23 with a DHFR domain stabilized YFP protein in cells.
- UbL domains targeted YFP substrates to degradation in the presence of an initiation region. The Y‐axis plots the abundance of the UbL‐YFP‐tail proteins normalized by a translation control, which is the same UbL‐YFP protein without the tail. The ratio is equal to one for domains that are not recognized by the proteasome, and smaller than one if the UbL is recognized by the proteasome and the UbL‐YFP‐tail protein is degraded. YFP fluorescence was also corrected for plasmid copy number using RFP fluorescence. The median of YFP/RFP ratio for each construct was calculated from 10,000 cells collected in one flow cytometry run and reflected the abundance of YFP substrates in yeast cells as described in Materials and Methods. Data points represent the mean values determined from at least three repeat experiments; error bars indicate s.e.m.
Figure EV3. UbL domain‐mediated YFP protein degradation depends on proteasome, but not ubiquitination.

- Schematic representation of UbL‐YFP constructs. The fluorescent substrates consisted of a UbL domain, followed by a yellow fluorescent protein (YFP) domain and a tail (51 amino acids derived from subunit 9 of the F o component of the Neurospora crassa ATPase, Su9) or a stop codon at the C‐terminus. dsRed Express2 (expression control) and UbL‐YFP‐Su9 (substrate) were expressed from the consecutive constitutive ACT1 promoters on the same CEN plasmid (YCplac33).
- Stabilization of UbL‐YFP substrates with tails (Su9, gray) or without tails (no tail, red) by proteasome inhibitor (bortezomib) treatment. The extent of recovery was calculated as the fraction of the YFP/RFP ratios of cells expressing each construct after bortezomib treatment divided by the YFP/RFP ratios for cells with the same construct after DMSO treatment.
- Levels of UbL‐YFP proteins with tails (Su9, gray) or without tails (no tail, red) in the E1 temperature‐sensitive strain uba1‐204 at the permissive temperature (30°C; UbL‐YFP‐Su9, dark gray; UbL‐YFP, dark red) and the restrictive temperature (37°C; UbL‐YFP‐Su9, light gray; UbL‐YFP, light red). The median of YFP/RFP value for each construct was calculated from 10,000 cells collected in one flow cytometry run and was used to indicate the concentration of YFP substrate in yeast cells as described in Materials and Methods.
We then investigated the 10 other yeast UbL domains. Here, we compared the fluorescence of cells expressing the UbL‐YFP fusion with and without the proteasome initiation tail to minimize the effects of the N‐terminal UbL domain on protein expression (Fig 5B). The tail itself had little effect on YFP fluorescence as cells expressing DHFR‐YFP fusions with or without the tail showed similar fluorescence (Fig 5A and B). All UbL domains reduced cellular YFP fluorescence at least twofold (Fig 5B), and inhibiting the proteasome with bortezomib increased YFP fluorescence (Fig EV3B), suggesting that all the UbL domains targeted YFP for degradation by the proteasome. The proteasome inhibitor affected neither the fluorescence of cells expressing UbL‐YFP fusions without proteasome initiation tails nor the fluorescence of cells expressing DHFR‐YFP fusions (Fig EV3B). Therefore, both the N‐terminal UbL domain and the C‐terminal tail were required for degradation by the proteasome.
Degradation of the His3 and YFP fusion proteins depended on the UbL domain but was not due to ubiquitination. S. cerevisiae encodes a single ubiquitin‐activating enzyme (McGrath et al, 1991), Uba1, and its temperature‐sensitive allele uba1‐204 makes it possible to reduce protein ubiquitination effectively by shifting cells to the restrictive temperature (Ghaboosi & Deshaies, 2007). When expressed in uba1‐204 cells, the steady‐state levels of all the UbL fusion proteins with or without proteasome initiation tail were similar at the restrictive and permissive temperatures, while a protein containing a classic N‐end rule degron was stabilized at the restrictive temperature (Figs EV2 and EV3C). Thus, the UbL domains did not serve as ubiquitination signals but rather targeted proteins to the proteasome directly.
The UbL domains share the β‐grasp fold of ubiquitin (Fig 1). The proteasome recognizes ubiquitin through a hydrophobic patch on its surface centered around the residues L8, I44, and V70 (Beal et al, 1996; Sloper‐Mould et al, 2001), and we asked whether the UbL domains are recognized in a similar manner. We mutated the residues corresponding to the ubiquitin hydrophobic patch in the UbL domains of Rad23, Dsk2, Rub1, and Mdy2 to glutamic acid (Fig EV4A), and tested whether the mutations prevented proteasomal degradation. We found that the triple Glu mutations stabilized UbL‐His3‐tail proteins, as judged by Western blotting against an HA‐tag inserted between the His3 domain and the Su9 tail (Fig EV4C). The mutations also allowed the proteins to complement the his3 mutation under selective growth conditions. Thus, proteasome targeting mediated by UbL domains was disturbed by the mutations and the UbL domains of Rad23, Dsk2, Rub1, and Mdy2 appeared to bind the proteasome through the same interaction surface as ubiquitin in the assays described here.
Figure EV4. Four UbL domains bind to the proteasome via similar residues as ubiquitin.

- The proteasome‐interacting residues on ubiquitin and the UbL domains of yeast Rad23, Mdy2, Rub1, and Dsk2 that were mutated to glutamic acid residues (3E mutations) in this study are shown in red. Structural models of ubiquitin and UbL domains generated by PyMol (version 0.99). PDB IDs: Dsk2 2BWF; Mdy2 4GOC; Rad23 3M62; Rub1 1BT0; ubiquitin 1UBQ.
- 3E mutations rescued the growth defect of his3 strains expressing UbL‐His3‐tail constructs under selective conditions (+3‐AT, −his). Cells expressing the His3 fusion proteins in late log phase were serially diluted and stamped on selective plates. The plates were incubated at 30°C for 3 days for imaging.
- 3E mutations stabilized UbL‐His3‐tail proteins in yeast cells. Protein levels were determined by Western blotting for the HA‐tag and for Scs2 protein as a loading control in SDS–PAGE gels of S. cerevisiae protein extracts.
Source data are available online for this figure.
In summary, the eleven yeast UbL domains we tested were able to target proteins to the proteasome for degradation in at least one of the cell‐based assays and degradation depended on the presence of a disordered tail, presumably to allow the proteasome to initiate the degradation.
Degradation of natural UbL proteins Mdy2 and Ubp6
The observation that all UbL domains we tested could target artificial proteins for degradation was unexpected because only a subset of UbL domains is thought to function in the UPS physiologically. Degradation of the model proteins in yeast required the presence of an unstructured region in the protein in addition to the UbL domain to allow the proteasome to initiate degradation (Fishbain et al, 2011). Perhaps stable natural UbL proteins also escape degradation because they lack sites at which the proteasome can engage them (Prakash et al, 2004, 2009; Takeuchi et al, 2007; Zhao et al, 2010; Fishbain et al, 2011, 2015; van der Lee et al, 2014). We tested this hypothesis on two UDPs, Mdy2 and Ubp6.
Mdy2 (also known as Get5) catalyzes a step in the guided entry of tail‐anchored protein insertion (GET) pathway through which tail‐anchored proteins are inserted into the ER membrane (Jonikas et al, 2009). It is a 212‐amino acid protein with a central UbL domain flanked by an N‐terminal domain (NTD) and a C‐terminal dimerization domain (Chang et al, 2010; Chartron et al, 2010, 2012a,b; Simon et al, 2013) (Fig 6A). Mdy2 expressed by coupled in vitro transcription and translation was degraded by purified yeast proteasome (Fig 6B left panel). Degradation was inhibited by the addition of an excess of purified UbLMdy2 domain, indicating that the UbL domain of Mdy2 was required for its degradation (Fig 6B right panel). Degradation also required the N‐terminal domain, since deleting the first 73 amino acids of Mdy2 stabilized it against degradation (Fig 6B left panel). The N‐terminal domain is largely disordered (Chang et al, 2010; Chartron et al, 2010) and could be required to allow the proteasome to initiate the degradation. Indeed, attaching an unrelated 95‐amino acid tail to the C‐terminus of Mdy2 restored proteasomal degradation of Mdy2 lacking its N‐terminal domain (Fig 6B left panel). In yeast, Get4 binds to Mdy2's N‐terminal domain and buries it (Chang et al, 2010; Chartron et al, 2010). Adding purified Get4 to Mdy2 in the degradation assays protected Mdy2 from proteolysis (Fig 6B right panel). Get4 did not, however, prevent proteasome binding; fusing a 95‐amino acid tail to the C‐terminus of Mdy2 allowed it to be degraded even in the presence of Get4 (Fig 6B right panel). Thus, Get4 protected Mdy2 from proteasome degradation in vitro by preventing the proteasome from initiating degradation.
Figure 6. Degradation of Mdy2 proteins in vitro and in vivo .

- Schematic representation of S. cerevisiae Mdy2. Structural models of the N‐terminal domain (NTD) of Mdy2 (amino acids 1–59, green), with its binding partner Get4 (red, PDB ID: 2WPV), the central UbL domain of Mdy2 (amino acids 74–148, gray, PDB ID: 4GOC), and the C‐terminal dimerization domain (DD) of Mdy2 (amino acids 152–212, blue, PDB ID: 2LNZ).
- In vitro degradation of Mdy2 proteins by purified S. cerevisiae proteasome. Mdy2 (red solid circles) was degraded by the proteasome; removing the NTD (Mdy2ΔN, black solid circles) stabilized the protein but attaching a C‐terminal tail to the truncated protein (Mdy2ΔN‐95, blue solid circles) restored degradation. Mdy2 (red solid circles) was also stabilized by the addition of its binding partner Get4 (red open squares, dashed line) or UbLMdy2 as a competitor for proteasome binding (red open diamonds, dotted line). When Get4 binding buried the NTD, fusion of a tail to the C‐terminus of Mdy2 (green open squares, dotted line) restored degradation. The graph plots the amount of substrate remaining estimated by electronic autoradiography in SDS–PAGE gel bands over time as normalized to the initial amount of substrate as described in Materials and Methods. Data points represent the mean values determined from at least three repeat experiments; error bars indicate s.e.m.
- Degradation of Mdy2 in vivo. Steady‐state levels of overexpressed C‐terminally HA‐tagged full‐length Mdy2 and a mutant in which the NTD was deleted (Mdy2ΔN) in S. cerevisiae, in the absence of Get4 or in the presence of endogenous Get4 or overexpressed N‐terminally Flag‐tagged Get4. Protein levels were determined by Western blotting for the HA‐ or Flag‐tag, or for the protein Scs2 as a loading control, of SDS‐PAGE gels of S. cerevisiae protein extracts. The proteasome was inhibited by bortezomib as indicated.
- Steady‐state levels of endogenous Mdy2 in Get4 wild‐type (GET4) or deletion (get4Δ) S. cerevisiae strains in the presence or absence of the proteasome inhibitor bortezomib. Protein levels were determined by Western blotting for Mdy2, or Scs2 as the loading control, of SDS–PAGE gels of S. cerevisiae protein extracts.
Source data are available online for this figure.
Next, we studied Mdy2 degradation in yeast cells by expressing Mdy2 under the control of a constitutive promoter (pTPI1) (Partow et al, 2010), and Get4 under the control of an inducible promoter (pGAL1) on the same CEN plasmid. Mdy2 was tagged with a C‐terminal HA epitope and Get4 with an N‐terminal Flag epitope, so that their abundance in yeast cells could be estimated by Western blotting. Plasmid‐expressed Mdy2 was degraded by the proteasome, as demonstrated by the increase in steady‐state levels when the proteasome was inhibited with bortezomib (Fig 6C). Overexpressing Get4 at the same time protected Mdy2 from degradation and proteasome inhibition no longer increased Mdy2 steady‐state levels (Fig 6C). Mdy2 that lacked the NTD was not degraded by the proteasome as neither proteasome inhibition nor Get4 overexpression affected its steady‐state levels (Fig 6C).
Up to this point, we analyzed overexpressed Mdy2 and it was possible that endogenous Mdy2 was protected by other mechanisms such as proteins binding to its UbL domain. However, we found that Mdy2 at physiological levels was also susceptible to proteasomal degradation (Fig 6D). Deletion of GET4 (yeast strain get4Δ, Appendix Table S1) led to the depletion of Mdy2 from cells in a proteasome‐dependent manner. Thus, endogenous Mdy2 can be degraded by the proteasome but appears to be protected by the binding of Get4 to a disordered region where the proteasome can initiate the degradation.
Ubp6 binds to the proteasome subunit Rpn1 through an N‐terminal UbL domain and through interactions of its catalytic (ubiquitin‐specific protease, USP) domain with Rpn1 (Leggett et al, 2002; Rosenzweig et al, 2012; Aufderheide et al, 2015; Bashore et al, 2015). By analogy to Mdy2, it seemed possible that Ubp6 escaped degradation because it lacked a region at which the proteasome can initiate the degradation. Indeed, Ubp6 expressed by coupled in vitro transcription and translation was not degraded by purified yeast proteasome unless a 95‐amino acid disordered tail was fused to its C‐terminus (Fig 7A). Deleting the UbL domain inhibited, but did not completely abolish the degradation of Ubp6 with the tail (Fig 7A). Similarly, fusing the 51‐amino acid Su9 tail to the C‐terminus of Ubp6 allowed the proteasome to degrade the resulting fusion proteins when overexpressed in yeast (Fig 7B). The proteasome inhibitor bortezomib increased the steady‐state levels of overexpressed Ubp6 fusions with a tail but not without a tail. Degradation of overexpressed Ubp6 with a tail was not eliminated by the deletion of the UbL domain, presumably because the truncated protein was still able to bind to the proteasome. Degradation of Ubp6 was not simply caused by overexpression. Attaching a short disordered tail (an 11‐aa linker followed by the 8‐aa Flag epitope) to endogenous Ubp6 also destabilized the protein, and a longer disordered tail (54 aa) caused complete depletion of Ubp6 (Fig 7C). These results suggested that Ubp6 binds to the proteasome through a two‐site interaction, yet avoids degradation by the proteasome because it lacks an initiation region.
Figure 7. Degradation of Ubp6 proteins in vivo and in vitro .

- In vitro degradation of Ubp6 proteins by purified S. cerevisiae proteasome. Full‐length Ubp6 (Ubp6, black open circles) and a UbL deletion mutant (Ubp6ΔN, green open circles) escaped degradation. Attachment of a C‐terminal 95‐amino acid tail derived from S. cerevisiae cytochrome b 2 allowed the proteasome to degrade the Ubp6 proteins (Ubp6‐95, black solid circles and Ubp6ΔN‐95, green solid circles). The graph plots the amount of substrate estimated by electronic autoradiography in SDS–PAGE gel bands over time as normalized to the initial substrate amount as described in Materials and Methods. Data points represent the mean values determined from at least three repeat experiments; error bars indicate s.e.m.
- Degradation of Ubp6 proteins in vivo. Steady‐state levels of N‐terminally HA‐tagged Ubp6 mutants in S. cerevisiae. N‐terminally HA‐tagged Ubp6 and Ubp6∆N with or without a Su9‐tail attached to their C‐termini were overexpressed in yeast. Protein levels were determined by Western blotting for the HA‐tag, or the protein Scs2 as a loading control, of SDS–PAGE gels of S. cerevisiae protein extracts. The proteasome was inhibited by bortezomib where indicated.
- Steady‐state levels of endogenous Ubp6 and Ubp6 derivatives with short (19 amino acid) or long (54 amino acid) tails at their C‐termini. Protein levels were determined by Western blotting for Ubp6, or Scs2 as a loading control, of SDS–PAGE gels of S. cerevisiae protein extracts. The proteasome was inhibited by bortezomib as indicated.
Source data are available online for this figure.
Discussion
Ubiquitin and UbL domains mediate protein–protein interactions in the UPS and other cellular processes, but it is not clear how the cell distinguishes between the different signals and assigns them to the appropriate processes. The simplest explanation would be that the proteasome does not recognize ubiquitin signals that direct proteins to processes other than degradation. However, this mechanism seems to be only part of the explanation. For example, polyubiquitin chains formed through both Lys48 and Lys63 are recognized by the proteasome (Saeki et al, 2009; Komander & Rape, 2012), yet Lys63‐linked chains are thought to function mostly in membrane trafficking and DNA repair (Erpapazoglou et al, 2014). One likely basis for targeting specificity is that ubiquitin receptors compete with each other for substrates so that even though the proteasome may be able to recognize a protein, the receptors involved in a competing pathway bind them more effectively because of higher affinity or local concentration (Hicke et al, 2005; Grabbe & Dikic, 2009; Dores et al, 2010; Nathan et al, 2013). Degradation by the proteasome requires a second signal in the target protein in the form of a disordered region at the which the proteasome can initiate degradation (Inobe & Matouschek, 2014). In yeast, the proteins Cdc34, which autoubiquitinates, and Rad23, which contains a UbL domain, escape degradation when overexpressed, but are rapidly degraded by proteasome when disordered tails are fused to their C‐termini (Fishbain et al, 2011, 2015). More broadly, cellular proteins that contain disordered regions at which the proteasome can initiate degradation have on average shorter half‐lives than proteins that lack appropriate initiation regions (van der Lee et al, 2014; Fishbain et al, 2015).
We find here that UbL domains in proteins associated with proteasome‐independent processes are recognized by the proteasome with apparent affinities similar to those of bona fide proteasome‐interacting domains. All the UbL domains we tested can bind to the proteasome and target model proteins for degradation in vitro and in yeast. However, in the more stringent proteasome‐targeting assay, the degradation of the stable YFP protein, the four highest scoring UbL domains were the ones previously known to bind to the proteasome.
Several factors may complicate the comparison of in vitro and in vivo observations quantitatively. In vivo, UbL domains could bind the proteasome indirectly. For example, the UbL domains of Rad23, Dsk2, and Ddi1 are thought to bind to their own and each others' UBA domains (Saeki et al, 2002a; Heessen et al, 2005; Kang et al, 2006; Heinen et al, 2011). Similarly, other binding partners and receptors for individual UbL domains may modulate their interaction with the proteasome, positively or negatively, as Sgt2 is thought to do for Mdy2 (Chartron et al, 2012a), Ufo1 for Ddi1 (Ivantsiv et al, 2006), UbL‐UBA proteins for Rub1 (Singh et al, 2012), Snu66 for Hub1 (Mishra et al, 2011), and Shp1 for Atg8 (Krick et al, 2010), as may subcellular localization. Under some circumstances, the K is may underestimate the strengths of the UbL–proteasome interactions. We measured K is in kinetic competition experiments with the UbL domain of Rad23 so that any interactions that are completely independent of the UbLRad23 binding site would not be detected. Indeed, it was recently reported that the UbL domain of Ubp6 can bind to a site on Rpn1 not recognized by UbLRad23 (Shi et al, 2016).
Nevertheless and importantly, even weak proteasome interactions can be physiologically relevant. The UbL domain of Ddi1 has one of the lower proteasome affinities in our competition assay, in agreement with earlier studies (Saeki et al, 2002a; Gomez et al, 2011; Rosenzweig et al, 2012), and it targets the model proteins we tested only poorly for proteasomal degradation in vivo. However, Ddi1 functions as an extrinsic substrate receptor for the proteasome and is required for the degradation of Ho endonuclease during the regulation of mating type switching in yeast (Kaplun et al, 2005). As part of the same process, the UbL domain of Ddi1 also mediates Ddi1's interaction with the ubiquitin ligase Ufo1 through several UIM domains in Ufo1 (Ivantsiv et al, 2006). It has been proposed that the UbL domain switches between the two binding partners as it mediates Ho degradation and the lower apparent proteasome affinity may be a reflection of this mechanism (Voloshin et al, 2012).
The UbL domain in Mdy2 appears to bind the proteasome even less tightly than the UbL of Ddi1 (K i = 33 ± 3 μM for UbLMdy2 vs. K i = 17 ± 2 μM for UbLDdi1), but it can target model proteins as well as Mdy2 itself to the proteasome for degradation. Indeed, even endogenous Mdy2 can be degraded by the proteasome. The first 73 amino acids of Mdy2 are disordered in structures of Mdy2 alone and the length and amino acid composition of this region are in the range observed in sequences that allow the proteasome to initiate degradation efficiently. In yeast, the Get4 protein binds tightly to this region and buries it entirely (Chang et al, 2010; Chartron et al, 2010). There are no other disordered regions in Mdy2, so that the proteasome is unable to initiate degradation when it binds to Mdy2. The UbL domain of Mdy2 also interacts with Sgt2, another component of the GET pathway (Chartron et al, 2012a). Why would the cell use a UbL domain that interacts with both its binding partner in the GET pathway and the proteasome? The arrangement may keep Mdy2 abundance in a specific range. Mdy2 that accumulated above the amount of Get4 would be subject to degradation by the proteasome. An upper concentration threshold might prevent competition with interaction partners, such as Sgt2, that are shared with other pathways.
At the same time, tight proteasome interaction does not necessarily lead to degradation. Ubp6 binds the proteasome with high affinity (Leggett et al, 2002; Rosenzweig et al, 2012; Aufderheide et al, 2015; Bashore et al, 2015; Shi et al, 2016) but escapes proteolysis because it lacks a site at which the proteasome is able to initiate degradation. We have found previously that the UbL‐UBA protein Rad23 also escapes the degradation because it lacks effective proteasomal initiation sites and it is likely that the other two UbL‐UBA proteins in yeast, Dsk2 and Ddi1, are protected by a similar mechanism (Fishbain et al, 2011, 2015). The last remaining UbL domain protein in yeast, Atg12, contains a C‐terminal UbL domain, and its N‐terminal half is predicted to be disordered. We were not able to purify Atg12's UbL domain and therefore do not know whether it is able to bind to the proteasome directly.
Of the eleven yeast UbL proteins, five (Atg8, Hub1, Rub1, Smt3, and Urm1) are ubiquitin‐like modifiers that become attached to various macromolecules. Two of these, Atg8 and Hub1, bind the proteasome more tightly than the UbL of Ddi1 in our in vitro assay. Atg8 (K i = 3.3 ± 0.3 μM) competed as effectively for proteasome binding as the UbL of the proteasomal substrate receptor Dsk2 (K i = 3.5 ± 0.2 μM) and Hub1 competed only slightly less effectively (K i = 9.8 ± 0.8 μM). Atg8 becomes attached to phosphatidylethanolamine in the autophagy pathway (Ichimura et al, 2000), and Hub1 is thought to bind to proteins non‐covalently (Lüders et al, 2003; Yashiroda & Tanaka, 2004; Mishra et al, 2011). The interaction of Hub1 with its binding partner Snu66 leaves the likely proteasome interaction surface free (Mishra et al, 2011) so that Hub1 might still be able to bind to the proteasome when in complex with Snu66. Snu66 is predicted to contain long disordered regions yet may escape the degradation because of competition with other binding partners or its localization. It is also possible that ubiquitin‐like modifiers destabilize target proteins under some circumstances. In human cells, the ubiquitin‐like modifier FAT10 is incorporated into proteasome degradation signals (Hipp et al, 2005). Future work will reveal how common this mechanism is and whether ubiquitin‐like modifiers also target proteins to degradation in yeast.
In summary, we find that 10 of the 11 ubiquitin‐like domains encoded in the yeast genome are able to bind to the proteasome and most of them are able to target proteins for degradation. Thus, protein stability is not solely controlled by proteasome affinity as determined by the UbL domains but also by the presence or absence of sites at which the proteasome can engage its substrates to initiate the degradation.
Materials and Methods
Substrate proteins
Protein substrates were derived from E. coli DHFR, N. crassa ATPase F o component subunit 9, S. cerevisiae cytochrome b 2, UbL proteins, His3, and Homo sapiens hHR23B. Fluorescent proteins were gifts from B. S. Glick (University of Chicago). The coding sequences were cloned into the plasmid pGEM‐3Zf (+) (Promega) for expression in vitro and in E. coli for protein purification as described previously (Fishbain et al, 2011, 2015), or into the yeast CEN plasmid YCplac33 or 2‐micron plasmid pYES2 for in vivo experiments. In model proteins, N‐terminal UbL domains were connected to E. coli DHFR, S. cerevisiae His3, GFP, or YFP by the linker sequence (VDGGSGGGS) as described previously (Kraut & Matouschek, 2011). C‐terminal tails derived from cytochrome b 2 or subunit 9 were attached to DHFR and the other proteins through a two‐amino acid linker (PR), and lysine residues in the cytochrome b 2 tails were replaced by arginine. Model proteins without a tail or with cytochrome b 2 tails had a hexahistidine tag at the very C‐terminus. Isolated individual UbL domains all ended with the linker VDGGSGGGS followed by a C‐terminal hexahistidine tag for purification.
Protein expression and purification
Yeast proteasome was purified from S. cerevisiae strain YYS40 (Appendix Table S1) by immunoaffinity chromatography using anti‐FLAG antibodies (M2 agarose affinity beads, Sigma) as described previously with modifications (Saeki et al, 2005). Proteasome preparations were analyzed by SDS–PAGE and compared to published compositions (Lander et al, 2012). Each proteasome preparation was checked for activity by testing the degradation of the proteasome substrate UbLRad23‐DHFR‐95 and for contamination by proteases by testing the stability of proteins that lacked a proteasome‐binding tag (DHFR‐95) as described previously (Fishbain et al, 2011). For in vitro degradation experiments, radioactive substrates were expressed from a T7 promoter by a coupled in vitro transcription–translation reaction using the RTS 100 E. coli HY Kit (Roche) or TNT® Coupled Reticulocyte Lysate Systems (Promega) containing [35S]‐methionine following the manufacturer's protocol. After synthesis, the substrates were either partially purified by high‐speed centrifugation followed by precipitation in two volumes of saturated (NH4)2SO4 or affinity‐purified using Talon magnetic beads (Clontech) as described previously (Kraut & Matouschek, 2011).
Proteasomal degradation assays
Degradation assays with radiolabeled substrates were performed as described previously (Kraut & Matouschek, 2011). Briefly, the assays were carried out at 30°C by adding radiolabeled substrates to 40 nM of purified yeast proteasome in a reaction buffer containing a creatine phosphate and creatine kinase ATP‐regenerating system. Samples were removed at designated times, added to SDS–PAGE sample buffer to stop the reaction, and analyzed by SDS–PAGE. Protein amounts were determined by electronic autoradiography (Instant Imager, Packard); 100 μM MG132 was used to inhibit the proteolytic activity of proteasome where indicated. The degradation assays monitoring fluorescence intensity were performed in 384‐well plates in a plate reader (Infinite M1000 PRO, Tecan). Assays were carried out at 30°C by adding fluorescent substrates to 40 nM of purified yeast proteasome in a reaction buffer containing a creatine phosphate and creatine kinase ATP‐regenerating system. Fluorescence intensity was read every 30 s (excitation: 485 nm, 5 nm bandwidth; emission: 525 nm, 10 nm bandwidth) for a total reaction time of one hour. Protein amounts were corrected based on fluorescence intensity of the reaction in each well using standard curves describing the relationship between fluorescence intensity and protein concentration. Each assay was repeated at least three times. Initial degradation rates are given by the slope of the decay curves at time zero and were calculated as the product of the amplitude and the rate constant of the decay curve determined by nonlinear fitting to a single exponential decay to a constant offset using the software package Kaleidagraph (version 4.1, Synergy Software).
Yeast transformation and strain construction
His3 and fluorescent substrates were expressed in S. cerevisiae from a CEN plasmid (YCplac33) or a 2‐micron plasmid (pYES2) with a URA3 selection marker. The plasmids were transformed into S. cerevisiae strains (Appendix Table S1) using the Frozen‐EZ Yeast Transformation II Kit (Zymo Research). To attach a tail to endogenous Ubp6, linearized DNA encoding the tail sequence followed by a LEU2 selection cassette and flanked by sequences homologous to the insertion site was transformed into the host strain UBP6‐GFP (Appendix Table S1). The constructs were confirmed by Sanger sequencing.
In vivo protein abundance determination
HA‐tagged Mdy2 protein derivatives were expressed from the constitutive triosephosphate isomerase (TPI1) promoter and Flag‐tagged Get4 from the inducible GAL1 promoter. Both proteins were expressed from the CEN plasmid YCplac33 in S. cerevisiae strains pdr5Δ or get4Δ (Appendix Table S1). HA‐tagged Ubp6 protein derivatives were expressed under the control of the inducible GAL1 promoter on the CEN plasmid YCplac33 in S. cerevisiae strain pdr5Δ. HA‐tagged UbL‐His3 proteins were expressed from the inducible GAL1 promoter on the 2‐micron plasmid pYES2 in S. cerevisiae strain pdr5Δ. Cells with overexpressed or endogenous Mdy2 and Ubp6 proteins were grown to mid–log phase with glucose or galactose as a carbon source and lysed by vortexing with glass beads (BioSpec Products). Protein extracts were prepared and analyzed by Western blotting using standard protocols as described (Fishbain et al, 2015). HA‐tagged Mdy2 or Ubp6 proteins were detected with a monoclonal anti‐HA antibody (1:500, Roche Life Science, #12158167001); Flag‐tagged Get4 was detected by a monoclonal anti‐Flag antibody (Cell Signaling, #8146). Scs2p was detected by an anti‐Scs2 polyclonal antibody (1:1,000, gift from J. Brickner, Northwestern University). Endogenous Mdy2 was detected by an anti‐Mdy2 polyclonal antibody (Liou et al, 2007) (1:10,000, gift from C. Wang, Institute of Molecular Biology, Academia Sinica). Endogenous Ubp6 was detected by an anti‐Ubp6 antibody (Leggett et al, 2002) (1:500, gift from D. Finley, Harvard Medical School). Alexa‐680‐labeled goat anti‐rabbit antibody (1:20,000, Invitrogen, #A21109) and Alexa‐800‐labeled goat anti‐mouse antibody (1:20,000, Rockland Immunochemicals, #610‐132‐121) were used as secondary antibodies. Protein amounts were estimated by direct infrared fluorescence imaging (Odyssey, LI‐COR Biosciences). 100 μM bortezomib was used to inhibit proteasome activity where indicated.
Flow cytometry
Yeast cells were grown at 30°C to early log phase before harvesting for analysis. Fluorescence signals in dsRed and YFP channels were collected on an LSR Fortessa (BD Biosciences) flow cytometer and analyzed by FlowJo to calculate the median YFP over RFP fluorescence ratio of 10,000 cells in each population. Assays were repeated at least three times; 100 μM bortezomib was used to inhibit proteasome activity where indicated.
Yeast growth assay
His3 proteins were expressed under the control of the inducible GAL1 promoter on a 2‐micron plasmid (pYES2) in S. cerevisiae strain pdr5Δ (Appendix Table S1). Cells were grown with galactose as a carbon source at 30°C to late log phase, serially diluted (OD600 from 10−1 to 10−6), and stamped on synthetic media plates. The plates were incubated at 30°C for 3 days before imaging.
Author contributions
HY, GK, and CMY performed experiments, analyzed data, and co‐wrote the paper. AM directed experiments, analyzed data, and co‐wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Drs. Suzanne Elsasser, Yuan Shi, and Dan Finley (Harvard Medical School), Kylie Walters (National Cancer Institute), Chung Wang (Academia Sinica), Ben S. Glick (University of Chicago), Jason Brickner (Northwestern University), Dan Kraut (Villanova University), and Dennis Wylie and Richard Salinas (The University of Texas at Austin) for advice and reagents. We are also grateful to Matouschek laboratory members for helpful comments on the manuscript. We acknowledge the use of the instruments of the Microscopy and Imaging Facility of the Institute for Cell and Molecular Biology at The University of Texas at Austin. This work was supported by the US National Institutes of Health (U54GM105816, R21CA191664, R21CA196456), the Cancer Prevention and Research Institute of Texas (CPRIT) (RP140328), and the Welch Foundation (F‐1817).
The EMBO Journal (2016) 35: 1522–1536
References
- Alifano P, Fani R, Liò P, Lazcano A, Bazzicalupo M, Carlomagno MS, Bruni CB (1996) Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol Rev 60: 44–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Förster F (2015) Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci USA 112: 8626–8631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A (2015) Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol 22: 712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C (1996) Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci USA 93: 861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y‐W, Chuang Y‐C, Ho Y‐C, Cheng M‐Y, Sun Y‐J, Hsiao C‐D, Wang C (2010) Crystal structure of Get4‐Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem 285: 9962–9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Suloway CJM, Zaslaver M, Clemons WM (2010) Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA 107: 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, VanderVelde DG, Clemons WM (2012a) Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Rep 2: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, VanderVelde DG, Rao M, Clemons WM (2012b) Get5 carboxyl‐terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J Biol Chem 287: 8310–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Dores MR, Schnell JD, Maldonado‐Baez L, Wendland B, Hicke L (2010) The function of yeast epsin and Ede1 ubiquitin‐binding domains during receptor internalization. Traffic 11: 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, Feng MT, Tübing F, Dittmar GAG, Finley D (2002) Proteasome subunit Rpn1 binds ubiquitin‐like protein domains. Nat Cell Biol 4: 725–730 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chandler‐Militello D, Müller B, Hanna J, Finley D (2004) Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem 279: 26817–26822 [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D (2005) Delivery of ubiquitinated substrates to protein‐unfolding machines. Nat Cell Biol 7: 742–749 [DOI] [PubMed] [Google Scholar]
- Erpapazoglou Z, Walker O, Haguenauer‐Tsapis R (2014) Versatile roles of K63‐linked ubiquitin chains in trafficking. Cells 3: 1027–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP ‐ Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fishbain S, Prakash S, Herrig A, Elsasser S, Matouschek A (2011) Rad23 escapes degradation because it lacks a proteasome initiation region. Nat Commun 2: 192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, Matouschek A (2015) Sequence composition of disordered regions fine‐tunes protein half‐life. Nat Struct Mol Biol 22: 214–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK (2002) Complementary whole‐genome technologies reveal the cellular response to proteasome inhibition by PS‐341. Proc Natl Acad Sci USA 99: 1461–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H (2002) Budding yeast Dsk2p is a polyubiquitin‐binding protein that can interact with the proteasome. Proc Natl Acad Sci USA 99: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaboosi N, Deshaies RJ (2007) A conditional yeast E1 mutant blocks the ubiquitin‐proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol Biol Cell 18: 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A (1988) New yeast‐Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six‐base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA‐UBL domain protein Ddi1. BMC Biol 9: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabbe C, Dikic I (2009) Functional roles of ubiquitin‐like domain (ULD) and ubiquitin‐binding domain (UBD) containing proteins. Chem Rev 109: 1481–1494 [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D (2006) Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: 99–111 [DOI] [PubMed] [Google Scholar]
- Hawkes TR, Thomas PG, Edwards LS, Rayner SJ, Wilkinson KW, Rice DW (1995) Purification and characterization of the imidazoleglycerol‐phosphate dehydratase of Saccharomyces cerevisiae from recombinant Escherichia coli. Biochem. J. 306: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heessen S, Masucci MG, Dantuma NP (2005) The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell 18: 225–235 [DOI] [PubMed] [Google Scholar]
- Heinen C, Acs K, Hoogstraten D, Dantuma NP (2011) C‐terminal UBA domains protect ubiquitin receptors by preventing initiation of protein degradation. Nat Commun 2: 191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin‐binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hipp MS, Kalveram B, Raasi S, Groettrup M, Schmidtke G (2005) FAT10, a Ubiquitin‐Independent Signal for Proteasomal Degradation. Mol Cell Biol 25: 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Dikic I (2012) Ubiquitin‐binding proteins: decoders of ubiquitin‐mediated cellular functions. Annu Rev Biochem 81: 291–322 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin‐like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Inobe T, Fishbain S, Prakash S, Matouschek A (2011) Defining the geometry of the two‐component proteasome degron. Nat Chem Biol 7: 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inobe T, Matouschek A (2014) Paradigms of protein degradation by the proteasome. Curr Opin Struct Biol 24C: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivantsiv Y, Kaplun L, Tzirkin‐Goldin R, Shabek N, Raveh D (2006) Unique role for the UbL‐UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol Cell Biol 26: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse DM, Crosas B, Finley D, Church GM (2004) Localization to the proteasome is sufficient for degradation. J Biol Chem 279: 21415–21420 [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, Schuldiner M (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Vossler RA, Diaz‐Martinez LA, Winter NS, Clarke DJ, Walters KJ (2006) UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol 356: 1027–1035 [DOI] [PubMed] [Google Scholar]
- Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D (2005) The DNA damage‐inducible UbL‐UbA protein Ddi1 participates in Mec1‐mediated degradation of Ho endonuclease. Mol Cell Biol 25: 5355–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H (2004) Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15: 3357–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Koulich E, Li X, DeMartino GN (2008) Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol Biol Cell 19: 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut DA, Matouschek A (2011) Proteasomal degradation from internal sites favors partial proteolysis via remote domain stabilization. ACS Chem Biol 6: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut DA, Matouschek A (2012) Sequence‐ and species‐dependence of proteasomal processivity. ACS Chem Biol 7: 1444–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen E‐L, Thumm M (2010) Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin‐like Atg8. J Cell Biol 190: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A (2012) Complete subunit architecture of the proteasome regulatory particle. Nature 482: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown N, Chenna R, Mcgettigan P, Mcwilliam H, Valentin F, Wallace I, Wilm A, Lopez R, Thompson J, Gibson T, Higgins D (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee B‐H, Lee MJ, Park S, Oh D‐C, Elsasser S, Chen P‐C, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D (2010) Enhancement of proteasome activity by a small‐molecule inhibitor of USP14. Nature 467: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Lang B, Kruse K, Gsponer J, Sánchez de Groot N, Huynen MA, Matouschek A, Fuxreiter M, Babu MM (2014) Intrinsically disordered segments affect protein half‐life in the cell and during evolution. Cell Rep 8: 1832–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507 [DOI] [PubMed] [Google Scholar]
- Liou S‐T, Cheng M‐Y, Wang C (2007) SGT2 and MDY2 interact with molecular chaperone YDJ1 in Saccharomyces cerevisiae . Cell Stress Chaperones 12: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J, Pyrowolakis G, Jentsch S (2003) The ubiquitin‐like protein HUB1 forms SDS‐resistant complexes with cellular proteins in the absence of ATP. EMBO Rep 4: 1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JP, Jentsch S, Varshavsky A (1991) UBA 1: an essential yeast gene encoding ubiquitin‐activating enzyme. EMBO J 10: 227–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Ammon T, Popowicz GM, Krajewski M, Nagel RJ, Ares M, Holak TA, Jentsch S (2011) Role of the ubiquitin‐like protein Hub1 in splice‐site usage and alternative splicing. Nature 474: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y (1998) A protein conjugation system essential for autophagy. Nature 395: 395–398 [DOI] [PubMed] [Google Scholar]
- Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL (2013) Why do cellular proteins linked to K63‐polyubiquitin chains not associate with proteasomes? EMBO J 32: 552–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjørn S, Nielsen J, Maury J (2010) Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast 27: 955–964 [DOI] [PubMed] [Google Scholar]
- Pédelacq J‐D, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2006) Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24: 79–88 [DOI] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL (2009) Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36: 794–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A (2004) An unstructured initiation site is required for efficient proteasome‐mediated degradation. Nat Struct Mol Biol 11: 830–837 [DOI] [PubMed] [Google Scholar]
- Prakash S, Inobe T, Hatch AJ, Matouschek A (2009) Substrate selection by the proteasome during degradation of protein complexes. Nat Chem Biol 5: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem 287: 14659–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Saitoh A, Toh‐e A, Yokosawa H (2002a) Ubiquitin‐like proteins and Rpn10 play cooperative roles in ubiquitin‐dependent proteolysis. Biochem Biophys Res Commun 293: 986–992 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Sone T, Toh‐e A, Yokosawa H (2002b) Identification of ubiquitin‐like protein‐binding subunits of the 26S proteasome. Biochem Biophys Res Commun 296: 813–819 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Isono E, Toh‐E A (2005) Preparation of ubiquitinated substrates by the PY motif‐insertion method for monitoring 26S proteasome activity. Meth. Enzymol. 399: 215–227 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh‐e A, Tanaka K (2009) Lysine 63‐linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J 28: 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 391: 715–718 [DOI] [PubMed] [Google Scholar]
- Sharon E, Kalma Y, Sharp A, Raveh‐Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E (2012) Inferring gene regulatory logic from high‐throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee B‐H, Shi Y, Zhang N, dePoot SAH , Tuebing F, Sun S, Vannoy J, Tarasov SG, Engen JR, Finley D, Walters KJ (2016) Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351: aad9421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AC, Simpson PJ, Goldstone RM, Krysztofinska EM, Murray JW, High S, Isaacson RL (2013) Structure of the Sgt2/Get5 complex provides insights into GET‐mediated targeting of tail‐anchored membrane proteins. Proc Natl Acad Sci USA 110: 1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Zerath S, Kleifeld O, Scheffner M, Glickman MH, Fushman D (2012) Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1‐ubiquitin chains by components of the ubiquitin‐proteasome system. Mol Cell Proteomics 11: 1595–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper‐Mould KE, Jemc JC, Pickart CM, Hicke L (2001) Distinct functional surface regions on ubiquitin. J Biol Chem 276: 30483–30489 [DOI] [PubMed] [Google Scholar]
- Strack RL, Strongin DE, Bhattacharyya D, Tao W, Berman A, Broxmeyer HE, Keenan RJ, Glick BS (2008) A noncytotoxic DsRed variant for whole‐cell labeling. Nat Methods 5: 955–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J, Chen H, Coffino P (2007) Proteasome substrate degradation requires association plus extended peptide. EMBO J 26: 123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen AG, Ploegh HL (2012) Ubiquitin‐like proteins. Annu Rev Biochem 81: 323–357 [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ (2004) Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin‐proteasome system. Cell 118: 99–110 [DOI] [PubMed] [Google Scholar]
- Voloshin O, Bakhrat A, Herrmann S, Raveh D (2012) Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL‐UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS ONE 7: e39210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Brown EC, Mak G, Zhuang J, Denic V (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell 40: 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda H, Tanaka K (2004) Hub1 is an essential ubiquitin‐like protein without functioning as a typical modifier in fission yeast. Genes Cells 9: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Yen H‐CS, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008) Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D (2009) Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain‐length sensor. Mol Cell 36: 1018–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang N‐Y, Zurawel A, Hansen KC, Liu C‐W (2010) Degradation of some polyubiquitinated proteins requires an intrinsic proteasomal binding element in the substrates. J Biol Chem 285: 4771–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 3
Source Data for Figure 6
Source Data for Figure 7


