Abstract
Fatty acid and fat synthesis in liver is a highly regulated metabolic pathway critical for energy distribution. Having common features at their promoter regions, lipogenic genes are coordinately regulated at the transcription level. Transcription factors, such as USF, SREBP-1c, LXR and ChREBP play critical roles in this process. Recently, insights have been gained into how various signaling pathways regulate these transcription factors. After feeding, high blood glucose and insulin induce lipogenic genes through several pathways, including DNA-PK, aPKC and Akt-mTOR. Various transcription factors and coregulators undergo specific modifications, such as phosphorylation, acetylation, or ubiquitination, which affect their function, stability, or localization. Dysregulation of lipogenesis can contribute to hepatosteatosis, which is associated with obesity and insulin resistance.
Glucose from excess dietary carbohydrate undergoes glycolysis in liver, and is eventually converted into fatty acids (FA) to be esterified to TAG for VLDL secretion. The process of converting glucose to fatty acids, de novo lipogenesis (DNL), is tightly controlled by hormones and nutritional status1 (Box 1). In fasting, DNL is very low due to the increased glucagon and cAMP levels. After eating, blood glucose and insulin levels increase and stimulates insulin signaling leading to activation of specific kinases, including PI3K and its multiple downstream kinases, Akt, aPKC or mTORC, as well as phosphatases such as PPI and PP2. If the diet is rich in carbohydrate, as blood glucose and insulin levels are elevated to a greater extent, fatty acid and fat synthesis is induced even at a higher degree. Many of the enzymes involved in FA and TAG production, are regulated during the fasting-feeding cycle1 (Box 2). Activities of these enzymes are maintained low during fasting and are increased after feeding2. Lipogenic enzymes can be regulated by multiple mechanisms. Allosteric control and post-translational modification, such as phosphorylation-dephosphorylation, mediate rapid regulation. For example, ACC is activated through dephosphorylation by PP1. PFK-2 is also activated by dephosphorylation by PP1; this generates fructose-2,6-bisphosphate, which in turn is a potent allosteric activator of PFK-1, a critical regulatory enzyme in glycolysis (Box 2).
Box 1. Insulin and glucagon regulate blood glucose levels and lipid metabolism.
Glucose and lipid metabolism is regulated together to balance energy use and storage for maintenance of blood glucose concentrations within a narrow range. Glucagon and insulin, which are secreted from pancreatic islets, have opposing roles in the regulation of glucose and lipid metabolism. Following fasting, low glucose levels stimulate glucagon secretion from islet α cells, which functions mainly in the liver to increase hepatic glucose production by increasing glycogenolysis (the enzymatic breakdown of glycogen), and gluconeogenesis (the synthesis of glucose mainly from lactate and amino acids) involving PKA-cAMP signaling pathway. In contrast, high glucose levels, for example after ingestion of carbohydrates, trigger secretion of insulin from pancreatic β cells, which stimulates glucose uptake and utilization, and promotes glycogen and fatty acid synthesis in the liver. Fatty acids generated from de novo lipogenesis (DNL), along with those taken up from circulation, are then used for sequential esterification of glycerol backbone to produce triacylglycerols (TAGs) in the liver. TAGs are secreted into circulation as VLDL. VLDL secretion is followed by the LPL-mediated mobilization of TAGs to FAs that are taken up by adipose tissue for long-term storage after re-esterification. DNL and fat synthesis are executed through series enzymes that are regulated for metabolic homeostasis to adapt to changing nutritional and hormonal conditions.
Box 2. Metabolic pathways for Fatty acid and TAG synthesis.
Enzymes involved include: 1) glycolytic enzymes, such as glucokinase (GK), Phosphofructokinase-1, and −2 (PFK-1 and −2), and liver pyruvate kinase (L-PK), to provide the carbon source for FA and TAG synthesis, 2) enzymes for FA synthetic pathway, such as ATP-citrate lyase (ACLY), acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), stearoyl-CoA desaturase (SCD) and elongase of long chain fatty acids family 6 (ELOV6), 3) enzymes for the production of NADPH used in fatty acid synthesis, including oxidative branch of the pentose-phosphate pathway, such as glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), as well as malic enzyme (ME), and 4) enzymes involved in esterification for TAG production, such as mitochondrial glycerol-3-phosphate acyltransferase (mGPAT), 1-acylglycerol-3-phosphate acyltransferase (AGPAT), phosphatidate phosphatase (PAP), diacylglycerol acyltransferase (DGAT).
In the longer term, many of the enzymes that are involved in DNL and TAG synthesis are primarily regulated at the transcriptional level in a coordinate fashion. Transcriptional activation of lipogenic genes after a meal may be via complex mechanisms involving multiple transcription factors in response to insulin and glucose signaling. Thus, insulin can activate specific kinases and phosphatases that may modify transcription factors, such as USF, SREBP-1c, ChREBP and LXR, or their expression levels (Table 1). In addition, specific glucose metabolites that increase after a meal may also affect function or localization of certain transcription factors. These transcription factors are then recruited to the promoter regions of lipogenic genes, where they may form complexes with other transcription factors or coactivators for transcriptional regulation. In this review, we summarize the studies of major transcription factors and signaling pathways for induction of lipogenesis after a meal. Silencing of lipogenic genes upon fasting has been previously reviewed 3, 4.
Table 1.
Transcription factors, lipogenic targets, and the types of modification that activate or inhibit lipogenesis.
| TF | Binding site | Target genes | Activation | Inactivation |
|---|---|---|---|---|
| USF-1 | E-box 5′-CANNTG-3′ |
SREBP-1c, FAS, ACC, ACLY, SCD, mGPAT | P-S262 by DNA-PK AC-K237 by P/CAF |
deAC-K237 by HDAC9 |
| SREBP-1c | SRE 5′-TCACNCCAC-3′ |
SREBP-1c, FAS, ACC, ACLY, SCD, mGPAT, PK, GK |
P-S117, P-S63 and P-T426 by MAPK AC-K289 and K309 by P300 |
P-S31, P-S314 by PKA P-S372 by AMPK deAC-K289, deAC-K309 by SIRT |
| ChREBP | ChoRE 5′-CAYGNGN5CNCRTG-3′ |
L-PK, FAS, ACC, ACLY, SCD | deP-S196, deP-T666 by PP2A AC-K672 by P300 O-GlcNAcylation or O-GlcNAc |
P-S196, P-T666 by PKA P140, P196 by PKA P-586 by AMPK |
| LXR | LXRE 5′-AGGTCAN4AGGTCA-3′ |
SREBP-1c, ChREBP, LXR, FAS, ACC | O-GlcNAcylation or O-GlcNAc | P-S195, P-S196, P-S290, P-S291 by PKA |
P, phosphorylation; deP, dephosphorylation; AC, acetylation; deAC, deacetylation
Finally, we also discuss the role of chromatin remodeling and circadian rhythm in the regulation of lipogenic gene transcription, as well as the contribution of lipogenesis to hepatosteatosis and insulin resistance.
USF is critical for lipogenic gene activation by insulin
USF-1 and USF-2 are basic helix-loop-helix leucine zipper (bHLH-LZ) transcription factors and form homo- or heterodimers to bind an E-box, 5’-CATGTG-3’ at target promoters for transcriptional activation. Originally, USF had been identified to bind an E-box at the L-PK promoter that can be activated upon feeding when both glucose and insulin levels are elevated 5, 6, later USF was also found to be binding the promoter of fatty acid synthase which is the key enzymes for lipogenesis7. The requirement of USF in lipogenesis has been demonstrated in vivo in USF-1 or USF-2 knockout mice that show significantly impaired lipogenic gene activation upon high carbohydrate feeding8. However, USF ablation did not affect insulin resistance, probably because USF affected lipogenesis but not glucose metabolism8-10. Mice overexpressing USF in liver may also help in understanding the role of USF in hepatosteatosis and insulin resistance. In this regard, quantitative trait mapping studies have identified USF-1 as a candidate gene for familial combined hyperlipidemia11.
USF has been shown to be required for FAS promoter activation by insulin, via binding to the −65 E-box (Figure 1)12. Thus, mutation of the −65 E-box abolishes the response of the FAS promoter to insulin in cultured cells. Overexpression of dominant negative USF-1 or USF-2 mutants that lack the DNA binding domain impairs the insulin stimulation of the FAS promoter activity13. The FAS promoter has been examined in an in vivo context employing transgenic mice carrying various 5’-deletions of the FAS promoter region fused to the chloramphenicol acetyltransferase (CAT) reporter gene, as well as mutation of the −65 E-box14. CAT reporter activity measurements and chromatin immunoprecipitation (ChIP) of livers from these transgenic mice showed that USF-1 and USF-2 are bound to the E-boxes at −332 and −65 of the FAS promoter15. Deletion or mutation of the −65 E-box abolishes feeding or insulin dependent activation of the FAS promoter, providing in vivo evidence of the requirement of the −65 E-box in response to feeding and insulin treatment. Additionally, presence of the −332 E-box increases the promoter activation further, showing the significance of this second −332 E-box in the FAS promoter activation15.
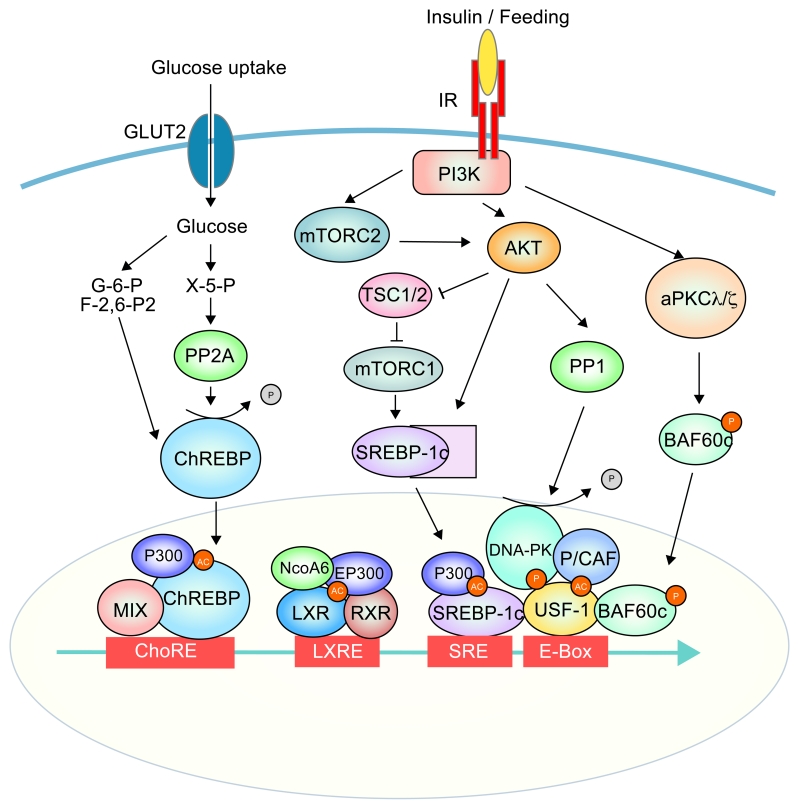
Figure 1.
Transcription factors and coregulators and the signalling pathways for hepatic lipogenic gene activation by insulin and glucose
Signaling and transcription network for activation of hepatic lipogenesis. Insulin signaling and glucose uptake trigger activation of series of kinases downstsream of PI3K, such as Akt, aPKC (PKC λ/⎛), mTORC1/2, DNA-PK, as well as phosphatases, such as PP1 and PP2. These kinases and phosphotases modify transcription factors by phosphorylation or dephosphorylation, respectively. Activated transcription factors, including USF-1, SREBP-1c, ChREBP and LXR, are bound to lipogenic gene promoter regions at E-box, SRE, ChoRE and LXRE, respectively, for transcriptional activation. USF-1 also recruits lipoBAF complex leading to chromatin remodeling for transcription.
Phosphorylation and acetylation of USF-1
USF levels do not change during fasting-feeding and USFs are bound to the E-box at the promoter regions of FAS and other lipogenic enzymes, such as mGPAT, in both conditions16-20. This suggests that posttranslational modification(s) of USF may occur during fasting-feeding cycle. Indeed, mass spectrometry (MS) analysis employing liver nuclear extracts from fasted and fed mice revealed feeding-specific S262 phosphorylation and K237 acetylation of USF-121. S262A, S262D phosphorylation mutants, as well as K237R and K237A acetylation mutants of USF-1 were used to test the importance of these posttranslational modifications. Greatly higher activation of the FAS promoter was observed using S262D and K237A mutants that mimic hyperphosphorylation and hyperacetylation, respectively, whereas the S262A and K237R mutants that mimic dephosphorylation and deacetylation, respectively, could not effectively activate FAS promoter. These studies clearly demonstrate the importance of S262 phosphorylation and K237 acetylation for USF function in transcriptional activation upon feeding or insulin treatment (Figure 2).
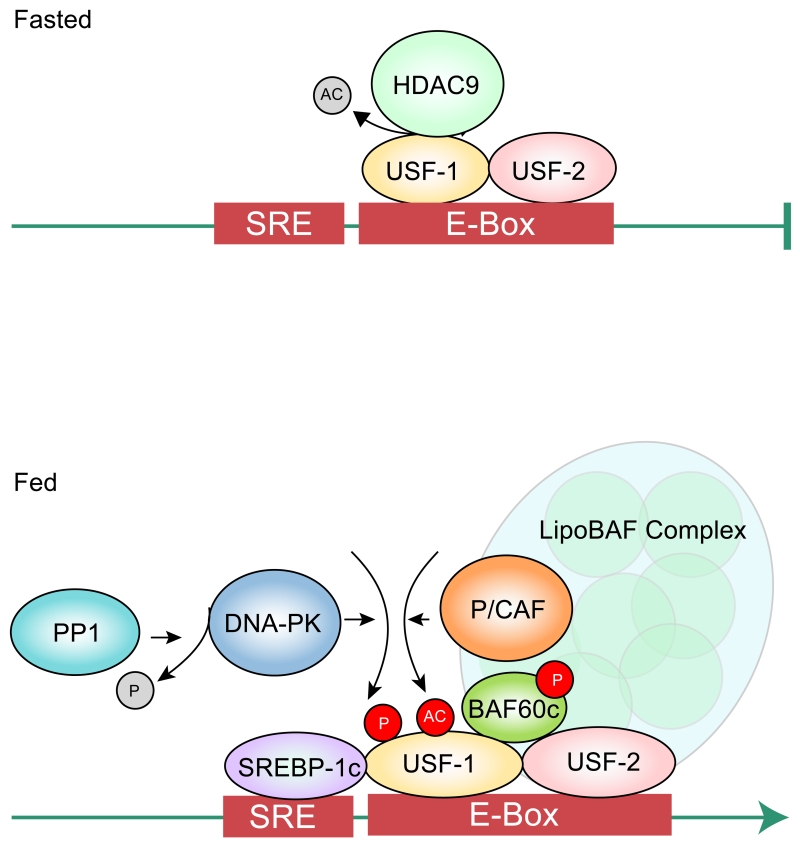
Figure 2.
Modifications of USF during the fasting and feeding transition.
In fasting, USF is deacetylated by HDAC9. Upon feeding, USF is phosphorylated by DNA-PK and subsequently acetylated by P/CAF. Phosphorylated acetylated USF interacts with BAF60c for recruitment of the LipoBAF Complex.
In defining signaling pathways and upstream components for posttranslational modification of USF, various USF-1 interacting proteins that are specific to fasting or feeding have been identified by tandem affinity purification followed by MS analysis. The enzymes identified include DNA-PK, a kinase known to be involved in DNA damage repair, and its regulatory subunits Ku70 and Ku80, PP1 that can dephosphorylate and activate DNA-PK, as well as PARP-1, Topoisomerase IIβ, and P/CAF21. It has been shown that upon feeding or insulin treatment, PP1 translocates to the nucleus for activation of DNA-PK. USF-1 can be phosphorylated at S262 by DNA-PK in vitro, as well as in vivo upon feeding or insulin treatment. Decreased lipogenic gene activation in DNA-PK deficient SCID mice during feeding with impaired hepatic lipogenesis and lower TAG levels demonstrates the critical role of DNA-PK in lipogenic gene activation22. Moreover, S262 phosphorylation of USF-1 allows interaction and recruitment of P/CAF, which in turn acetylates USF-1 at K23721. In contrast, in fasting, USF-1 preferentially interacts with histone deacetylase 9 (HDAC9) that can deacetylate USF-1, preventing the recruitment of various factors21. ChIP detected S262 phosphorylated and K237 acetylated USF-1 on FAS and other lipogenic promoters only in fed condition, whereas dephosphorylated and deacetylated USF-1 is found on these promoters in fasted condition. In this regard, USF-1 has also been implicated in suppression of gluconeogenesis. It has also been reported that USF-1 is activated by AMPK in a DNA-PK dependent manner, stimulating hepatic transcription of tyrosine phosphatase inhibitor, SHP. This resulted in suppression of transcription of PEPCK and glucose-6-phosphatase catalytic subunit (G6PC), thereby reducing glucose production23. These studies suggest that USF that is phosphorylated by DNA-PK may participate in coordinating changes in lipogenesis and gluconeogenesis during fasting-feeding cycle.
Interestingly, in addition to DNA-PK and P/CAF that phosphorylate and acetylate USF-1, respectively, other DNA break/repair components including PARP-1 and Topoisomerase, were identified to be present in the USF-1 complex21. Indeed, transient DNA breaks were observed during feeding-induced FAS gene activation21. Parenthetically, involvement of PARP-1 and topoisomerases in ligand-dependent function of nuclear hormone receptors has been described previously24-27. These authors proposed that transient DNA break may serve to release torsion stress generated from RNA polymerase II (Pol II)-dependent transcription.
USF recruits SREBP-1c to lipogenic promoters
SREBPs belong to bHLH-LZ transcription factor family that bind as dimers to SRE, 5’-TCACNCCAC-3’, of the target genes of lipid metabolism. Among the three isoforms, SREBP-1a, 1c, and 2, SREBP-1c is primarily responsible for the expression of lipogenic genes, although there is some functional overlap between SREBPs. A crucial role of SREBP-1c in the transcriptional activation of lipogenic genes has been shown in vivo by hepatic overexpression of SREBP-1c in transgenic mice that leads to increased hepatic lipid accumulation and insulin resistance28. Interestingly, insulin resistant obese mice with hepatosteatosis show constitutively elevated SREBP-1c expression29, 30. Conversely, SREBP-1 knockout mice showed defects in induction of lipogenic enzymes by feeding31. However, induction of lipogenic genes was diminished by approximately 50%, but was not completely eliminated by SREBP-1c ablation32. Thus, SREBP-1c activity alone could not account for full stimulation of liver TAG synthesis in response to a high-carbohydrate diet. Although it is possible that other SREBP isoforms might compensate SREBP-1c, the partial effect of SREBP-1c ablation on hepatic lipogenesis is more likely due to contribution of other transcription factor(s), such as USF, ChREBP and LXR, that are required or enhance lipogenic gene induction.
Although SREBPs are known to function by binding to SRE, because of an atypical tyrosine residue that replaces a conserved arginine present in the basic regions, SREBPs can also bind to E-boxes, at least in vitro33, 34. It has been reported that SREBP binds to the −65 E-box of the FAS promoter for activation, while others reported SREBP binding to the nearby −150 SRE35, 36. However, ChIP using transgenic mice carrying FAS promoter-CAT reporter with a mutation at −150 SRE or −65 E-box firmly established in vivo that SREBP binds to the −150 SRE, but not the −65 E-box, for the FAS promoter activation35-37.
More importantly, although binding of SRBEP-1c to the −150 SRE is critical for the feeding/insulin response, SREBP-1c itself cannot bind the SRE, upon mutation of the USF binding site of −65 E-box in vivo. These observations suggest a potential interaction of the two transcription factors and the critical role of USF for transcriptional activation of lipogenic genes35. In this regard, SREBPs have been shown to function together with other transcription factors, such as Sp1 and NF-Y, for transcriptional activation38-41. For example, although the physical interaction has not been examined, SREBP-1 has been reported to cooperate with Sp1 at adjacent sites to activate FAS and ACC promoters42, 43. The requirement of USF for the SREBP-1c function was also revealed by USF knockout mice that showed severely delayed FAS induction during feeding, even when the level of the mature form of SREBP remained unchanged8. Indeed, interaction between USF and SREBP-1 has been shown both in vitro and in vivo36. The basic HLH domain of USF interacts directly with the basic HLH domain of N-terminal region of SREBP-1. In addition, through cotransfection of USF-1 and SREBP-1c, synergistic activation of the FAS promoter by these two transcription factors has been shown. Furthermore, functional domain mapping using USF and SREBP deletion constructs indicated that the activation domains of both proteins are required for the functional synergy. As described above, USF-1 undergoes phosphorylation and acetylation upon feeding. Co-IP studies using USF-1 mutants containing S262D and S262A showed that S262 phosphorylation increases its interaction with SREBP-1c21. This indicates that posttranslational modifications of USF-1 are critical for recruitment of SREBP-1c to bind the nearby SRE for the synergistic activation of lipogenic genes by USF and SREBP-1c during feeding or insulin treatment21. Similar synergistic effect of USF and SREBP-1c was observed for mGPAT promoter also44. Numerous lipogenic genes, including SREBP-1c, ACC, ACLY, and mGPAT, contain E-box and SRE at their promoter regions, suggesting a common mechanism of recruitment of SREBP-1c by USF in the induction of lipogenic genes.
Regulation of SREBP-1c expression and its cleavage
As mentioned above, during the fasting-feeding transition, expression of SREBP-1c is similar to that of other lipogenic enzymes, and is probably controlled by a common transcriptional regulatory mechanism. It can be predicted that, by directly interacting with SREBP-1c, USF-1 recruits SREBP-1c to bind SRE at the SREBP-1c promoter region for transcriptional activation21. In accord with this notion, expression of SREBP-1c is induced by SREBP-1c itself and USF binding to the SREBP-1c promoter region has been detected by ChIP35, 45. USF phosphorylation by DNA-PK enhances SREBP-1c binding to its own promoter region, as observed with other lipogenic genes. In addition, as described below in more detail, LXRα can bind and activate lipogenic promoters, including SREBP-1c32, 46, to induce lipogenesis.
SREBP-1c, as all SREBPs, resides at the ER membrane as a larger precursor complexed with SCAP and INSIG. After proteolytic cleavage, the mature N-terminal SREBP-1c translocates into nucleus where it binds to SRE to activate transcription (Figure 1). Insulin activates SREBP-1c cleavage through two mechanisms involving Akt and mTORC1. Although specific phosphorylation site(s) are not known, Akt has been shown to directly phosphorylate SREBP-1c increasing the affinity of SCAP-SREBP-1c complex for Sar1/Sec23-24 complex of COPII-coated vesicles, resulting in facilitation of SREBP-1c transport to the Golgi where SREBP-1c can undergo cleavage47. Processing of SREBP-1c upon insulin stimulation has also been reported to depend on mTORC1 and its downstream target kinase, p70 S6K48-50. In addition, mTORC1-independent Akt activation decreased Insig-2a mRNA and thus the Insig-2a pool at the ER membrane, releasing SREBP-1c/SCAP to be transported into Golgi51, 52.
SREBPs undergo posttranslational modifications as well. SREBPs were reported to be phosphorylated by MAPK family, p38, ERK and JNK, at different sites53. Mice overexpressing SREBP-1a containing S63, S117, and T426 phosphorylation site mutants were protected from fatty liver development54. Since these phosphorylation sites are in mature form of SREBPs and are conserved in SREBP-1c, MAPK may play a role in SREBP-1c function, either by DNA binding or transactivation, for lipogenic gene activation. In addition, SREBP-1 was reported to be phosphorylated by PKA (SREBP-1c at S314), resulting in attenuation of SREBP-1 binding to SRE at the lipogenic promoter regions55. The N-terminal region of SREBP-1c was also reported be phosphorylated by GSK3, a kinase inactivated upon its phosphorylation by Akt, to negatively regulate SREBP-1c56.
SREBP-1c may undergo acetylation-deacetylation. SREBP-1c was reported to be acetylated by p300/CBP in conditions of high glucose and insulin at K289 and K309, which can be deacetylated by SIRT1. Deacetylation of SREBP-1c by SIRT1 inhibited binding to its target lipogenic promoters30. Interestingly, SREBP-1c deacetylation by SIRT1 has also been reported to promote ubiquitination and proteasomal degradation of SREBP-1c, implicating dual effects of SREBP-1c acetylation30, 57. In addition, during nutritional deprivation, PKA mediated SREBP-1c phosphorylation increases its interaction of SREBP-1c with PIASy, the small ubiquitin-related modifier (SUMO) E3 ligase, thus augmenting sumoylation of SREBP-1c at K98, leading to SREBP-1c degradation58. These studies further illustrate how different posttranslational modifications of SREBP-1c operate together in regulating lipogenic gene transcription.
Liver X receptor (LXR) activates lipogenic genes
LXRs belong to the nuclear hormone receptor superfamily of sterol-regulated transcription factors that are activated by the binding of physiological ligands, such as oxysterols59, 60 and certain intermediates in the cholesterol biosynthetic pathway, such as desmosterol. LXR target genes include ABC transporters, such as ABCA1, ABCG1, ABCG5, ABCG861 and apolipoprotein E62, which play a central role in liver cholesterol metabolism. Two isoforms of LXR, LXRα and LXRβ, bind to LXR response element (LXRE), 5’-AGGTCAN4AGGTCA-3’, at the target promoter regions, with the obligate partner, 9-cis retinoic acid receptor (RXR). Of the two LXR isoforms, LXRα is abundantly found in lipogenic tissues, such as liver. Mice ablated of both LXRα and LXRβ showed defective hepatic lipid metabolism decreasing lipogenesis by 80% and were resistant to obesity, demonstrating the role of LXR in hepatic lipogenesis46, 63, 64. Although they remained obese, LXRα and LXRβ deficient-ob/ob mice also had reduced hepatosteatosis.
It has been shown that LXRα can increase lipogenic gene transcription by binding and activating the SREBP-1c46, 65, 66. ChREBP may also be a target of LXR and as it was reported to have two LXRE at the ChREBP promoter regions, and LXR agonist treatment increased ChREBP expression67, 68. However, ChREBP expression was reported to be unaltered in LXRα/β ablated mice, contradicting ChREBP as a target of LXR69. Regardless, LXRE are also found in the promoter regions of several lipogenic genes, such as FAS, ACC and SCD, as well as LXRα itself. In fact, activation of FAS promoter that contains LXRE at −700 bp region has been reported70. Therefore, LXR may activate lipogenic gene transcription directly, as well as indirectly through induction of SREBP-1c and ChREBP70, 71.
It is generally believed that LXR functions in insulin-mediated lipogenesis66, but physiologically relevant ligand(s) of LXR for lipogenic activation have not been well established. It has been reported that glucose and glucose-6-phosphate can directly bind to LXR68 to stimulate transcription of its target genes. It has also been reported that LXR interacts with O-GlcNAc transferase and is modified by O-linked ß-N-acetylglucosamine (O-GlcNAc) for glucose sensing to induce LXR target genes72-74. Under hyperglycemic and hypoinsulinemic condition, LXR maintain their ability to upregulate the expression of glycolytic and lipogenic enzymes, including GK, SREBP-1c and ChREBPα and β 70. However, others reported that LXR is not required for induction of ChREBP or glucose-regulated genes, which suggests that glucose and its metabolites cannot be physiological ligands for LXR or that the posttranslational modification of LXR may not present a glucose sensing mechanism. Regardless, upon agonist binding, the conformation of the LXR–RXR complex may be altered, leading to the release of nuclear receptor corepressors, such as silencing mediator of retinoic acid and thyroid hormone receptor (SMRT, also known as NCoR2) and NCoR1, and recruitment of nuclear receptor coactivators, such as E1A-associated protein p300 (EP300) and activating signal cointegrator 2 (ASC2, also known as NCOA6)75, 76.
In fasting, LXRα can be phosphorylated by PKA at S195/S196 and S290/S291, which prevents dimerization with RXR and binding to LXRE, and recruits corepressor NcoR1, rather than coactivator SCR-177. Although the specific site(s) are not known, AMPK has also been reported to phosphorylate LXRα at threonine residue(s), resulting in the inhibition and attenuation of TAG synthesis, directly opposing its phosphorylation at serine residues by S6K1 activated through mTORC1, described in more detail below78.
ChREBP mediates lipogenic gene activation by Glucose
ChREBP was purified and identified by DNA affinity purification using the L-PK promoter sequence that responds to glucose, of which other E-box binding transcription factors, such as USF and MLTF, were previously implicated. ChREBP is the bona fide transcription factor that is primarily responsive to glucose. ChREBP has bHLH-Zip domain and forms heterodimeric complex with another bZIP protein Max-like protein X (MlX) to bind ChoRE composed of two E-boxes separated by five base pairs, 5’-CAYGNGN5CNCRTG-3’, at the promoter region of target genes79-81. Well-accepted targets of ChREBP include not only enzymes in glucose metabolism, such as L-pyruvate kinase (L-PK), G6PC, GLUT4, GPDH, and GKRP79, 82, but also lipogenic enzymes, such as ACLY, FAS, ACC, and SCD183, 84. Recently it has been reported that an isoform of full length of ChREBP (also named ChREBPα), named ChREBPβ, which derived from an alternative promoter, shorter than the conventional isoform of ChREBP has higher transcriptional activity 85, 86. Because ChREBP regulates both glucose and lipid metabolism, ChREBP causes complex metabolic changes in loss- and gain-of function studies87. ChREBP ablated mice not only show reduced glycolysis but also about a 60% reduction in lipogenesis and these mice exhibited insulin resistance84. However, liver-specific ChREBP deletion by adenoviral shRNAs improves hepatosteatosis, but also improves insulin resistance in ob/ob mice88. Conversely, adenoviral overexpression of ChREBP in liver increases hepatic TAG levels but blunts HFD-induced insulin resistance89. Overall, these studies clearly demonstrate the involvement of ChREBP in lipogenic gene transcription, but might dissociate hepatosteatosis from insulin resistance. In this regard, ChREBP expression in liver biopsies from patients with nonalcoholic steatohepatitis was increased when steatosis was found to be greater than 50%, but decreased in the presence of severe insulin resistance89.
ChREBP undergoes posttranslational modification. In fasting, ChREBP can be phosphorylated by PKA at S196 and T666, causing prevention of nuclear entry and decrease its DNA binding, respectively90, 91. In addition, ChREBP phosphorylation at S586 by AMPK also decreases its DNA binding activity92. ChREBP phosphorylated by PKA at S140 and S196 binds 14-3-3 with higher affinity to retain ChREBP in the cytosolic compartment93, 94. In high glucose conditions, ChREBP is dephosphorylated to become an active form and to translocate into nucleus to increase ChREBP binding to its response element91. In addition to phosphorylation-dephosphorylation mechanism, ChREBP may be regulated by Xylulose 5-phosphate, an intermediate of the pentose phosphate shunt activated in high glucose conditions, and has been reported to specifically activate protein phosphatase 2A (PP2A) for ChREBP dephosphorylation90. It has also been reported that, although underlying mechanism is not clear, other phosphometabolites that are generated during glucose metabolism: Glucose 6-phosphate and fructose-2,6-bisphophate, promote nuclear translocation of ChREBP95, 96 . Furthermore, acetylation of ChREBP at K672 by p300 has also been shown to increase its recruitment to the target gene promoter regions97. Intramolecular interaction has been proposed to be a mode of controlling ChREBP activity. ChREBP contains both a low glucose inhibitory domain (LID) and a glucose response activation conserved element (GRACE). LID inhibits the ChREBP transactivation activity conferred by GRACE, and this inhibition is lifted under high glucose conditions93, 98, 99. Therefore, ChREBP would be modified in a manner that relieves the repressive intramolecular interaction in high glucose condition.
In addition, similar to SREBP-1c, transcription of ChREBP itself is induced by glucose and exhibits a feed-forward auto-regulation by ChREBP/MlX binding to its own ChoRE82. LXR has been shown to directly bind to the promoter region to transcriptionally activate ChREBP67,68, although others reported that LXR is not required for induction of ChREBP or other glucose regulated genes, as stated above. Protein stability may also be a regulatory mode of the ChREBP levels. ChREBP has been shown to undergo O-GlcNAcylation in high glucose condition for its stabilization, potentially affecting its phosphorylation 100, 101. Treating cells with PUGNAc, an inhibitor of O-GlcNAcase or overexpression of O-glcNActransferase increased ChREBP function and lipogenic gene expression in high glucose condition, whereas treating cells with DON, an inhibitor of glutamine fructose amindotransferase that decreases O-GlcNAc modification or overexpression of O-GlcNAcase blocked lipogenic gene expression in vitro and in vivo100, 101.
Chromatin remodeling for lipogenic gene transcription
Dynamic chromatin structure plays an essential role in the control of gene transcription. While low endonuclease accessibility of the FAS and mGPAT promoter regions is observed in serum starvation, the endonuclease accessibility is remarkably increased upon insulin treatment. These observations demonstrate altered chromatin structure of lipogenic promoter regions during fasting-feeding cycle102. Two highly conserved mechanisms underlie the alteration of chromosome structure: 1) Posttranslational modification of histones, and 2) ATP-dependent chromosome remodeling. Modifications of histones include acetylation, methylation, and phosphorylation that primarily occur at N-terminal tails. Many of these enzymes such as p300/CBP and P/CAF that are involved in lipogenic gene transcription were originally described as proteins for general transcription and p300/CBP is found to be associated with the Pol II holoenzyme. In this regard, as is observed in most transcriptionally active genes, acetylation of H3 at K14 and phosphorylation of S10 at H3 were found at the FAS promoter region along with Pol II, in response to feeding and insulin102.
Since methylation status of histones has been shown to be important for transcription, especially that of H3 tail region, efforts have been made to characterize the effects of methylation-demethylation for transcriptional regulation. Upon feeding or insulin treatment, activation marks, such as H3K4me2/3 and H3K79me3, are predicted at lipogenic promoter regions. Conversely, the repressive mark, H4K20me3, may be detected in fasting. In this regard, histone demethylase LSD1 functions on H3K4 and H3K9 for repression and activation, respectively. LSD1 has recently been reported to be required for activation of the FAS promoter by SREBP-1. Transfection of LSD1 mutant lacking demethylase domain or treating cells with LSD1 inhibitor, trans-2-phyneycyclopropylamine, caused a decrease in lipogenic gene expression103. Histone modifications at the lipogenic genes in the liver during feeding or insulin treatment need further investigation.
In addition to histone posttranslational modifications, SWI/SNF complex carries out ATP-dependent chromatin remodeling. Within the mammalian SWI/SNF complex, the BAF (Brg1/Brm-associated factor) complex is composed of a core ATPase, Brg1/BAF190 or Brm and other BAF subunits, such as BAF155, BAF170, and BAF250, which act as modulators, as well as BAF57 and BAF60, which may allow interaction of the BAF complex with transcriptional machinery. Recent studies have revealed that, by direct binding, USF recruits BAF60c, one of the three isoforms of BAF60, which, in turn, recruits other BAF subunits, such as Brg1/BAF190, BAF155, and BAF250, to form the lipoBAF complex for chromatin remodeling required for activation of the lipogenic program upon feeding/insulin treatment (Figure 2)102. Thus knockdown of BAF60c resulted in a decrease in hepatic lipogenesis, whereas overexpression of BAF60c caused lipogenic induction in vivo102. In addition, reduced endonuclease accessibility of chromatin was detected upon knockdown of BAF60c at the lipogenic gene promoter region.
Upon feeding or insulin treatment, BAF60c was found to be phosphorylated at S247 by aPKC, PKCλ and PKCζ . Thus, S247 phosphorylation is abolished by siRNA mediated knockdown of aPKC or by overexpression of dominant negative aPKC, an evidence that aPKC is the bona fide kinase for the S247 phosphorylation of BAF60c. Phosphorylated BAF60c then translocates from the cytosol to nucleus. Phosphorylated BAF60c directly interacts with phosphorylated/acetylated USF-1 to allow recruitment of lipoBAF complex to lipogenic promoter regions for remodeling of chromatin in a manner conducive for transcription. Furthermore, USF-1 phosphorylation by DNA-PK and subsequent acetylation by P/CAF and BAF60c phosphorylation by aPKC are both required for their direct interaction for chromatin remodeling of the lipogenic genes. In fact, in addition to Akt, insulin-mediated activation of atypical PKC, PKCλ and PKCζ, has been shown to increase SREBP-1c expression and lipogenesis104. Liver specific knockout of PKCλ leads to a marked decrease in SREBP-1c expression105. BAF60c phosphorylation may be the underlying mechanism for this observation. Interestingly, it has been reported that, during fasting, BAF60a, by interacting with PGC1α, recruits SWI/SNF complex to various fatty acid oxidative genes, including Acaa1b and Acox1106. This suggests isoform specific function of BAF60s for chromatin remodeling during different metabolic states. It would be interesting to examine whether BAF60a is posttranslationally modified during fasting to increase its interaction with PGC1α.
Role of mTOR pathway in lipogenic gene activation
During the energy abundance of the fed state, lipogenesis may also be activated by mTOR, a pathway known to control cell growth and metabolism in response to nutrients, growth factors, or energy status107. TOR exists as distinct complexes, TORC1 and TORC2, which function independently. mTORC1 has been shown to increase SREBP-1c expression upon Akt-mediated phosphorylation of TSC1/2, which inhibits mTORC1 activity. Although mTORC1 has been shown to be essential108, ablation of TSC1, a negative regulator of mTORC1, could not elevate SREBP-1c expression50, 52. The Akt-mediated suppression of Insig2a may explain this discrepancy. Upon insulin signaling, mTORC2 has also been shown to induce SREBP-1c expression through phosphorylation and activation of S473 Akt109. Thus, liver-specific ablation of an mTORC2 component, Rictor, impaired hepatic lipogenesis. However, constitutively active Akt2 could not fully restore lipogenesis in mTORC2-deficient hepatocytes, suggesting an involvement of Akt independent mechanism110. Moreover, whether mTOR pathway activates lipogenic genes only indirectly via induction of SREBP-1c expression as reported or also by directly affecting transcription factor function has not been examined.
Lipogenic genes are regulated by circadian rhythm
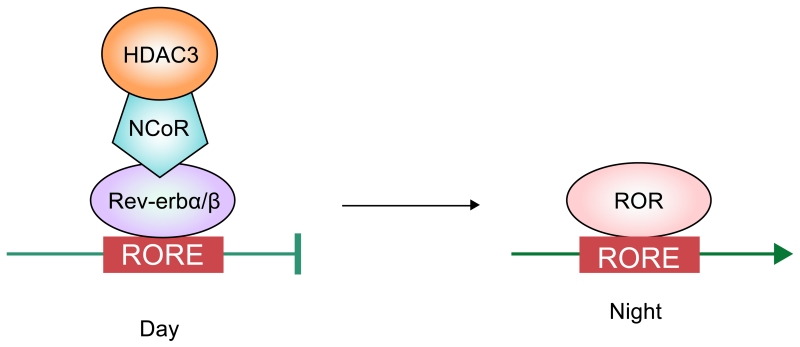
In addition to being regulated during fasting-feeding, lipogenesis fluctuates in day-night cycle of circadian rhythm (Box 3)111. Food intake can affect circadian rhythm in return112-114. Circadian regulated lipogenesis in liver has been reported to function through Reverse-erb α/β (Rev-erbα/β) and retinoic acid receptor related orphan receptors (RORα/β/γ). They represent orphan nuclear receptors that can bind as a monomer their response element, RORE 5’-AGGTCA-3’ ‘half site’ with a 5’ AT-rich extension115. By recruitment of specific repressors or coactivators, Rev-erbα/β and ROR can repress and activate gene transcription, respectively (Figure 3). In rodents, during the light period when they are inactive, as Rev-erbα/β expression level rises, it recruits HDAC3, preventing lipogenic gene activation116. Conversely, during dark time of active period in rodents, Rev-erbα/β levels are low and lipogenic genes are activated. Two groups of researchers reported that deletion of Rev-erbα in mice alters glucose and lipid metabolism, leading to elevation of circulating TAG levels and exacerbated hepatosteatosis117-120. Furthermore, it has been shown that Rev-erb recruits HDAC3 via NCoR121. ChIP-sequencing has revealed that HDAC3 is enriched in >100 lipid biosynthetic genes, including FAS, SCD1, ACLY during light time, but less during the dark period, inversely correlating with histone acetylation and RNA Pol II recruitment120. Histone deacetylation by HDAC3 alone, however, was insufficient for lipogenic gene suppression, but required interaction with NCoR. Thus, liver-specific ablation of NcoR in mice causes metabolic and transcriptional alterations similar to hepatic ablation of HDAC3122. Although both Rev-erbα/β and RORs are under the control of circadian rhythm machinery, the effect of ROR on the activation of lipogenic genes during circadian rhythm has not been examined. Regardless, stagger mice (RORa sg/sg), a natural mutant mice containing a RORa deletion showed protection from hepatosteatosis and insulin resistance, demonstrating its activating role in lipogenic gene transcription123. Interestingly, PPARδ which is subjected to circadian fluctuation was reported to control diurnal expression of hepatic lipogenic genes in the dark cycle. The authors proposed that PPARδ regulated hepatic metabolic program increases phosphatidylcholine (18:0/18:1) production in liver, which then can increase muscle fatty acid uptake for oxidation113.
Box 3. Regulation of circadian rhythm.
The core mammalian clock is composed of a heterodimer of the transcription factors circadian locomotor output cycles protein kaput (CLOCK) and brain and muscle ARNT-like 1 (BMAL1), which activate the transcription of themselves and period circadian clock (PER) and cryptochrome (CRY) genes via E box sequences within their promoters. PER and CRY proteins form dimers and directly interact with the CLOCK–BMAL1 heterodimers, thus suppressing their activity. The peak expression of the CLOCK– BMAL1 complex is 12 hours out of phase with peak PER and CRY expression to have the 24 hour pace maker. Both expression of retinoic acid receptor-related orphan receptor (ROR) and Rev-erb are regulated by Clock/BMAL and PER/CRY loop, and undergoes circadian oscillations. Conversely, ROR and Rev-erbα/β feedback to activate or suppress transcription of CLOCK and BMAL1, respectively132.
Figure 3.
Transcription factors and coregulators involved in the regulation of lipogenesis by circadian rhythm.
During the light time, Rev-erbα/β binds to RORE and recruits HDAC3 and NCoR to inhibit lipogenesis in rodents. ROR competitively binds to the same region as Rev-erbα/β at night to activate lipogenesis.
It has recently been reported that BMAL1, a core component of circadian transcription complex (Box 3), is required to drive the feeding/insulin-dependent activation of lipogenic genes in liver via stabilizing Rictor, an essential component of mTORC2 that can directly phosphorylate Akt at S473124. Since BMAL1 targets include phosphatases and ubiquitin-specific proteases, BMAL1 was suggested to inhibit phosphorylation-dependent ubiquitination and degradation of Rictor in promoting its stability. Thus, mTORC2 activity may be under the control of circadian rhythm and, as mentioned above, regulation of mTORC2 by Rictor may, in turn, affect SREBP-1c and other lipogenic gene transcription. It has also been reported that circadian clock deficiency can cause persistent activation of IRE1α pathway for ER stress, resulting in aberrant activation of SREBPs and hepatic lipid metabolism125. The role of ER stress in lipogenesis will not be addressed in detail in this review since it has been discussed in great detail previously 126
Relationship between lipogenesis and insulin resistance
Dysregulation of lipid metabolism often leads to adverse consequences, such as obesity, excess hepatic TAG accumulation, i.e., hepatosteatosis, and insulin resistance. The relationship of lipogenesis and hepatosteatosis with insulin resistance, however, is complex and needs better understanding at the molecular level. Hepatosteatosis can result from combination of excessive lipogenesis through insulin signaling. Various mouse models in which a component of insulin signaling pathway has been ablated (liver-specific insulin receptor knockout, liver-specific knockout of two p85 subunits of PI3K, ablation of p110α subunit of PI3K, and liver-specific knockout of Akt2) all exhibited hypolipidemia with reduced hepatic lipogenic gene expression. In liver-specific deletion of PTEN, a negative regulator of insulin signaling that acts upstream of Akt, development of fatty liver and a marked increase in FAS expression are observed. Under overnutrition conditions, PTEN deficient mice also exhibit increased hepatic lipogenesis, as well as the development of hepatosteatosis and hypertriglyceridemia. Furthermore, due to the reduced suppression of gluconeogenesis resulting from impaired insulin signaling resulting, these mice also exhibit hyperglycemia and insulin resistance. These phenotypes demonstrate the contribution of insulin pathway for lipogenesis and development of hepatosteatosis.
In insulin-resistant states of obesity and type 2 diabetes, hepatic lipid production is increased, paradoxically, in concert with increased hepatic glucose production. Resistance to the specific effects of insulin in the suppression of hepatic glucose production, but not lipogenesis, is referred to as selective insulin resistance. How does lipogenesis persist in insulin resistance? It has been proposed that mTORC1, which lies downstream of Akt, mediates SREBP-1c induction despite the impaired signaling during insulin resistant conditions. Because mTORC1 can function independently of Akt, it does not phosphorylate and inhibit FoxO1, the critical transcription factor for gluconeogenic gene activation, as Akt normally does, and thus promotes lipogenic gene transcription while gluconeogenic gene transcription is still active48. Inhibition of mTORC1 blocked insulin-induced upregulation of SREBP-1c and other lipogenic genes, independently of known mTORC1 downstream target S6K. However, mTORC1 inhibition did not affect insulin-mediated suppression of gluconeogenic genes. It is also possible that lipogenic genes are activated in insulin resistance by insulin independent mechanisms. In this regard, mTOR pathway is regulated by nutritional conditions. For example, mTORC signaling can be suppressed through Raptor upon amino acid or energy deprivation in a TSC1/2 independent manner127. In addition, mTOR can also be inactivated by AMPK-mediated phosphorylation of Raptor or TSC1/2128. Thus, mTOR pathway provides a potential mechanism for a persistent hepatic lipogenesis even in insulin resistance. Moreover, high glucose levels in insulin resistance may activate ChREBP for robust induction of, not only glycolytic, but also lipogenic genes. Expression levels of ChREBP are positively correlated with hepatosteatosis and adenoviral overexpression of ChREBP in mouse liver increases hepatic TAG levels. Therefore, activation of lipogenic genes by ChREBP even when insulin signaling is blunted may partly explain sustained lipogenesis during insulin resistance. Paradoxically, ChREBP expression levels are negatively correlated to insulin resistance in humans89. Furthermore, high fat diet-induced insulin resistance is blunted by ChREBP overexpression, because ChREBP decreases expression of gluconeogenic genes and reduces hepatic glycogen storage. Therefore, ChREBP knockout mice fed high carbohydrate diet display mitigated hepatosteatosis, but develop severe insulin resistance84. It has recently been reported that liver–specific ablation of three FoxOs prevents the induction of glucose–6–phosphatase and the repression of glucokinase during fasting, thus increasing lipogenesis at the expense of glucose production129. Conversely, overexpression of FoxO1 increased fasting glucose and reduced triglyceride level with impaired glucose tolerance130. Overall, selective insulin resistance may be the result of nutrient fluxes to the liver from the diet, from hepatic metabolism or from extrahepatic tissues129, 131.
Perspectives
Hepatic lipogenesis is regulated in complex fashion responding to hormones and nutrients. Lipogenic enzymes as well as transcription factors for lipogenic genes, such as USF, SREBP-1c, LXR, ChREBP, are coordinately activated upon insulin and glucose signaling involving multiple downstream molecules, including various kinases and phosphatases. These transcription factors are posttranslationally modified in response to insulin and glucose for activation of lipogenic gene transcription. Overnutrition or dysregulation of carbohydrate and lipid metabolism can result in accumulation of excess TAG in the liver, i.e., hepatosteatosis. Although critical transcription factors for hepatic lipogenesis have been identified and studied, more questions remain to explain molecular details of transcriptional regulation of lipogenesis. 1. How do multiple transcription factors work together to activate lipogenic genes in a concerted manner? 2. What enzymes and posttranslational events are involved in histone modification and chromatin remodeling in response to insulin and glucose? 3. Are there other modes of epigenetic regulation, such as DNA modification, involved in lipogenic gene transcription? The work discussed here shows that various transcription factors and coregulators and the signaling pathways involved in lipogenesis may provide future therapeutic targets for selective suppression of lipogenesis to combat insulin resistance and fatty liver disease.
Online summary.
Lipogenic genes are coordinately regulated at the transcription level during fasting-feeding cycle, as well as by circadian rhythm. Tightly regulated modifications of lipogenic transcription factors and coregulators by hormones and nutrients by various kinases-phosphatases and acetyltransferases-deacetylases involving multiple signaling pathways regulate lipogenic gene transcription. Detailed understanding in lipogenic gene transcription may provide future targets for the selective suppression of lipogenesis during insulin resistance or hepatosteatosis.
Glossary terms
- Glycolysis
a series enzyme-catalyzed reactions that converts glucose into pyruvate.
- Hyper- or Hypolipidemia
abnormally elevated or lower levels of any or all lipids and/or lipoproteins in the blood.
Biographies
Biographies
Hei Sook Sul is professor and Doris Calloway Chair in Human Nutrition in the Dept of Nutritional Sciences and Toxicology, University of California, Berkeley. Her research focuses on regulation of adipogenesis and lipid metabolism.
Yuhui Wang received her Ph.D. from Tokyo University in 2003. Since then, she has been a research specialist at the University of California, Berkeley. Her research is on mechanisms for adipocyte differentiation and lipogenic gene transcription.
Jose Viscarra is a postdoctoral fellow in the laboratory of Hei Sook Sul at the University of California, Berkeley. He completed his Ph.D. in 2013 from the laboratory of Rudy Ortiz at the University of California, Merced. His current research focuses on the regulation of hepatic lipogenic gene transcription.
Sun-Joong Kim received his Ph.D. from Korea University in 2012. His research focuses on the effects of energetic stress on adipose TAG metabolism.
References
- 1.Czech MP, Tencerova M, Pedersen DJ, Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia. 2013;56:949–964. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rui L. Energy Metabolism in the Liver. Comprehensive Physiology. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemper JK, Choi SE, Kim DH. Sirtuin 1 deacetylase: a key regulator of hepatic lipid metabolism. Vitam Horm. 2013;91:385–404. doi: 10.1016/B978-0-12-407766-9.00016-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viollet B, et al. Activation of AMP-activated protein kinase in the liver: a new strategy for the management of metabolic hepatic disorders. J Physiol. 2006;574:41–53. doi: 10.1113/jphysiol.2006.108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Z, Thompson KS, Towle HC. Carbohydrate regulation of the rat L-type pyruvate kinase gene requires two nuclear factors: LF-A1 and a member of the c-myc family. J Biol Chem. 1993;268:12787–95. [PubMed] [Google Scholar]
- 6.Diaz Guerra MJ, et al. Functional characterization of the L-type pyruvate kinase gene glucose response complex. Mol Cell Biol. 1993;13:7725–33. doi: 10.1128/mcb.13.12.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D, Sul HS. Upstream stimulatory factors bind to insulin response sequence of the fatty acid synthase promoter. USF1 is regulated. J Biol Chem. 1995;270:28716–22. doi: 10.1074/jbc.270.48.28716. [DOI] [PubMed] [Google Scholar]
- 8.Casado M, Vallet VS, Kahn A, Vaulont S. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J Biol Chem. 1999;274:2009–13. doi: 10.1074/jbc.274.4.2009. [DOI] [PubMed] [Google Scholar]
- 9.Vallet VS, et al. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J Biol Chem. 1998;273:20175–9. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 10.Vallet VS, et al. Glucose-dependent liver gene expression in upstream stimulatory factor 2 −/− mice. J Biol Chem. 1997;272:21944–9. doi: 10.1074/jbc.272.35.21944. [DOI] [PubMed] [Google Scholar]
- 11.Pajukanta P, et al. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1) Nat Genet. 2004;36:371–6. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- 12.Wong RHF, Sul HS. Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Curr Opin Pharmacol. 2010;10:684–691. doi: 10.1016/j.coph.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Sul HS. Upstream stimulatory factor binding to the E-box at −65 is required for insulin regulation of the fatty acid synthase promoter. J Biol Chem. 1997;272:26367–74. doi: 10.1074/jbc.272.42.26367. [DOI] [PubMed] [Google Scholar]
- 14.Soncini M, Yet S-F, Moon Y, Chun J-Y, Sul HS. Hormonal and Nutritional Control of the Fatty Acid Synthase Promoter in Transgenic Mice. J Biol Chem. 1995;270:30339–30343. doi: 10.1074/jbc.270.51.30339. [DOI] [PubMed] [Google Scholar]
- 15.Moon YS, Latasa M-J, Kim K-H, Wang D, Sul HS. Two 5′-Regions Are Required for Nutritional and Insulin Regulation of the Fatty-acid Synthase Promoter in Transgenic Mice. J Biol Chem. 2000;275:10121–10127. doi: 10.1074/jbc.275.14.10121. [DOI] [PubMed] [Google Scholar]
- 16.Shin DH, Paulauskis JD, Moustaïd N, Sul HS. Transcriptional regulation of p90 with sequence homology to Escherichia coli glycerol-3-phosphate acyltransferase. J Biol Chem. 1991;266:23834–9. [PubMed] [Google Scholar]
- 17.Moustaïd N, Beyer RS, Sul HS. Identification of an insulin response element in the fatty acid synthase promoter. J Biol Chem. 1994;269:5629–5634. [PubMed] [Google Scholar]
- 18.Sul HS, Wang D. NUTRITIONAL AND HORMONAL REGULATION OF ENZYMES IN FAT SYNTHESIS: Studies of Fatty Acid Synthase and Mitochondrial Glycerol-3-Phosphate Acyltransferase Gene Transcription. Annu Rev Nutr. 1998;18:331–351. doi: 10.1146/annurev.nutr.18.1.331. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Sul HS. Insulin stimulation of the fatty acid synthase promoter is mediated by the phosphatidylinositol 3-kinase pathway. Involvement of protein kinase B/Akt. J Biol Chem. 1998;273:25420–6. doi: 10.1074/jbc.273.39.25420. [DOI] [PubMed] [Google Scholar]
- 20.Paulauskis JD, Sul HS. Hormonal regulation of mouse fatty acid synthase gene transcription in liver. J Biol Chem. 1989;264:574–577. [PubMed] [Google Scholar]
- 21.Wong RH, et al. A role of DNA-PK for the metabolic gene regulation in response to insulin. Cell. 2009;136:1056–72. doi: 10.1016/j.cell.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong RHF, Sul HS. DNA-PK: Relaying the insulin signal to USF in lipogenesis. Cell Cycle. 2009;8:1973–1978. doi: 10.4161/cc.8.13.8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chanda D, et al. Hepatocyte growth factor family negatively regulates hepatic gluconeogenesis via induction of orphan nuclear receptor small heterodimer partner in primary hepatocytes. J Biol Chem. 2009;284:28510–21. doi: 10.1074/jbc.M109.022244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 25.Puc J, et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–80. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haince J-F, Rouleau M, Poirier GG. Gene Expression Needs a Break to Unwind Before Carrying On. Science. 2006;312:1752–1753. doi: 10.1126/science.1129808. [DOI] [PubMed] [Google Scholar]
- 27.Ryu KW, Kim D-S, Kraus WL. New Facets in the Regulation of Gene Expression by ADP-Ribosylation and Poly(ADP-ribose) Polymerases. Chemical Reviews. 2015;115:2453–2481. doi: 10.1021/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimano H, et al. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–54. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang T, et al. Diet-induced obesity in C57BL/6J mice causes increased renal lipid accumulation and glomerulosclerosis via a sterol regulatory element-binding protein-1c-dependent pathway. J Biol Chem. 2005;280:32317–25. doi: 10.1074/jbc.M500801200. [DOI] [PubMed] [Google Scholar]
- 30.Ponugoti B, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–70. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–24. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277:9520–8. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 33.Kim JB, et al. Nutritional and insulin regulation of fatty acid synthetase and leptin gene expression through ADD1/SREBP1. J Biol Chem. 1998;101:1–9. doi: 10.1172/JCI1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JB, et al. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–8. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latasa MJ, Griffin MJ, Moon YS, Kang C, Sul HS. Occupancy and function of the −150 sterol regulatory element and −65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol Cell Biol. 2003;23:5896–907. doi: 10.1128/MCB.23.16.5896-5907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin MJ, Wong RH, Pandya N, Sul HS. Direct interaction between USF and SREBP-1c mediates synergistic activation of the fatty-acid synthase promoter. J Biol Chem. 2007;282:5453–67. doi: 10.1074/jbc.M610566200. [DOI] [PubMed] [Google Scholar]
- 37.Latasa M-J, Moon YS, Kim K-H, Sul HS. Nutritional regulation of the fatty acid synthase promoter in vivo: Sterol regulatory element binding protein functions through an upstream region containing a sterol regulatory element. Proc Natl Acad Sci U S A. 2000;97:10619–10624. doi: 10.1073/pnas.180306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X, et al. Expression of the rat sterol regulatory element-binding protein-1c gene in response to insulin is mediated by increased transactivating capacity of specificity protein 1 (Sp1) J Biol Chem. 2007;282:17517–29. doi: 10.1074/jbc.M702228200. [DOI] [PubMed] [Google Scholar]
- 39.Yang L, et al. Ser1928 is a common site for Cav1.2 phosphorylation by protein kinase C isoforms. J Biol Chem. 2005;280:207–14. doi: 10.1074/jbc.M410509200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Shin DJ, Osborne TF. A simple promoter containing two Sp1 sites controls the expression of sterol-regulatory-element-binding protein 1a (SREBP-1a) Biochem J. 2005;386:161–8. doi: 10.1042/BJ20041514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Athanikar JN, Sanchez HB, Osborne TF. Promoter selective transcriptional synergy mediated by sterol regulatory element binding protein and Sp1: a critical role for the Btd domain of Sp1. Mol Cell Biol. 1997;17:5193–200. doi: 10.1128/mcb.17.9.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magana MM, Koo SH, Towle HC, Osborne TF. Different sterol regulatory element-binding protein-1 isoforms utilize distinct co-regulatory factors to activate the promoter for fatty acid synthase. J Biol Chem. 2000;275:4726–33. doi: 10.1074/jbc.275.7.4726. [DOI] [PubMed] [Google Scholar]
- 43.Lopez JM, Bennett MK, Sanchez HB, Rosenfeld JM, Osborne TF. Sterol regulation of acetyl coenzyme A carboxylase: a mechanism for coordinate control of cellular lipid. Proc Natl Acad Sci U S A. 1996;93:1049–53. doi: 10.1073/pnas.93.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jerkins AA, Liu WR, Lee S, Sul HS. Characterization of the Murine Mitochondrial Glycerol-3-phosphate Acyltransferase Promoter. J Biol Chem. 1995;270:1416–1421. doi: 10.1074/jbc.270.3.1416. [DOI] [PubMed] [Google Scholar]
- 45.Amemiya-Kudo M, et al. Promoter Analysis of the Mouse Sterol Regulatory Element-binding Protein-1c Gene. J Biol Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 46.Repa JJ, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yellaturu CR, et al. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 2009;284:7518–32. doi: 10.1074/jbc.M805746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Brown MS, Goldstein JL. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3441–6. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Owen JL, et al. Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc Natl Acad Sci U S A. 2012;109:16184–9. doi: 10.1073/pnas.1213343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakan I, Laplante M. Connecting mTORC1 signaling to SREBP-1 activation. Curr Opin Lipidol. 2012;23:226–34. doi: 10.1097/MOL.0b013e328352dd03. [DOI] [PubMed] [Google Scholar]
- 51.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci U S A. 2003;100:3155–60. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roth G, et al. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J Biol Chem. 2000;275:33302–7. doi: 10.1074/jbc.M005425200. [DOI] [PubMed] [Google Scholar]
- 54.Kotzka J, et al. Preventing phosphorylation of sterol regulatory element-binding protein 1a by MAP-kinases protects mice from fatty liver and visceral obesity. PLoS One. 2012;7:e32609. doi: 10.1371/journal.pone.0032609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu M, Shyy JY. Sterol regulatory element-binding protein 1 is negatively modulated by PKA phosphorylation. Am J Physiol Cell Physiol. 2006;290:C1477–86. doi: 10.1152/ajpcell.00374.2005. [DOI] [PubMed] [Google Scholar]
- 56.Kim KH, et al. Regulatory role of glycogen synthase kinase 3 for transcriptional activity of ADD1/SREBP1c. J Biol Chem. 2004;279:51999–2006. doi: 10.1074/jbc.M405522200. [DOI] [PubMed] [Google Scholar]
- 57.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–17. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee GY, et al. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol Cell Biol. 2014;34:926–38. doi: 10.1128/MCB.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W, Chen G, Head DL, Mangelsdorf DJ, Russell DW. Enzymatic reduction of oxysterols impairs LXR signaling in cultured cells and the livers of mice. Cell Metab. 2007;5:73–9. doi: 10.1016/j.cmet.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–31. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 61.Wagner BL, et al. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–9. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feldmann R, et al. Genome-wide analysis of LXRalpha activation reveals new transcriptional networks in human atherosclerotic foam cells. Nucleic Acids Res. 2013;41:3518–31. doi: 10.1093/nar/gkt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalaany NY, et al. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 2005;1:231–44. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 64.Beaven SW, et al. Reciprocal regulation of hepatic and adipose lipogenesis by liver X receptors in obesity and insulin resistance. Cell Metab. 2013;18:106–17. doi: 10.1016/j.cmet.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshikawa T, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci U S A. 2004;101:11245–50. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cha JY, Repa JJ. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem. 2007;282:743–51. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- 68.Mitro N, et al. The nuclear receptor LXR is a glucose sensor. Nature. 2007;445:219–23. doi: 10.1038/nature05449. [DOI] [PubMed] [Google Scholar]
- 69.Denechaud PD, et al. ChREBP, but not LXRs, is required for the induction of glucose-regulated genes in mouse liver. J Clin Invest. 2008;118:956–64. doi: 10.1172/JCI34314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joseph SB, et al. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem. 2002;277:11019–25. doi: 10.1074/jbc.M111041200. [DOI] [PubMed] [Google Scholar]
- 71.Schultz JR, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–8. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anthonisen EH, et al. Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J Biol Chem. 2010;285:1607–15. doi: 10.1074/jbc.M109.082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tobin KA, et al. Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J Biol Chem. 2002;277:10691–7. doi: 10.1074/jbc.M109771200. [DOI] [PubMed] [Google Scholar]
- 74.Bindesboll C, et al. Liver X receptor regulates hepatic nuclear O-GlcNAc signaling and carbohydrate responsive element-binding protein activity. J Lipid Res. 2015;56:771–85. doi: 10.1194/jlr.M049130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol. 2008;22:1312–9. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mouchiroud L, Eichner LJ, Shaw RJ, Auwerx J. Transcriptional coregulators: fine-tuning metabolism. Cell Metab. 2014;20:26–40. doi: 10.1016/j.cmet.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamamoto T, et al. Protein kinase A suppresses sterol regulatory element-binding protein-1C expression via phosphorylation of liver X receptor in the liver. J Biol Chem. 2007;282:11687–95. doi: 10.1074/jbc.M611911200. [DOI] [PubMed] [Google Scholar]
- 78.Hwahng SH, Ki SH, Bae EJ, Kim HE, Kim SG. Role of adenosine monophosphate-activated protein kinase-p70 ribosomal S6 kinase-1 pathway in repression of liver X receptor-alpha-dependent lipogenic gene induction and hepatic steatosis by a novel class of dithiolethiones. Hepatology. 2009;49:1913–25. doi: 10.1002/hep.22887. [DOI] [PubMed] [Google Scholar]
- 79.Ma L, Tsatsos NG, Towle HC. Direct role of ChREBP.Mlx in regulating hepatic glucose-responsive genes. J Biol Chem. 2005;280:12019–27. doi: 10.1074/jbc.M413063200. [DOI] [PubMed] [Google Scholar]
- 80.Stoeckman AK, Ma L, Towle HC. Mlx is the functional heteromeric partner of the carbohydrate response element-binding protein in glucose regulation of lipogenic enzyme genes. J Biol Chem. 2004;279:15662–9. doi: 10.1074/jbc.M311301200. [DOI] [PubMed] [Google Scholar]
- 81.Filhoulaud G, Guilmeau S, Dentin R, Girard J, Postic C. Novel insights into ChREBP regulation and function. Trends Endocrinol Metab. 2013;24:257–268. doi: 10.1016/j.tem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 82.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–30. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 83.Girard J, Ferre P, Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu Rev Nutr. 1997;17:325–52. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 84.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A. 2004;101:7281–6. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484:333–8. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Eissing L, et al. De novo lipogenesis in human fat and liver is linked to ChREBP-beta and metabolic health. Nat Commun. 2013;4:1528. doi: 10.1038/ncomms2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stiles B, et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected] Proc Natl Acad Sci U S A. 2004;101:2082–7. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dentin R, et al. Liver-specific inhibition of ChREBP improves hepatic steatosis and insulin resistance in ob/ob mice. Diabetes. 2006;55:2159–70. doi: 10.2337/db06-0200. [DOI] [PubMed] [Google Scholar]
- 89.Benhamed F, et al. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J Clin Invest. 2012;122:2176–94. doi: 10.1172/JCI41636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci U S A. 2003;100:5107–12. doi: 10.1073/pnas.0730817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci U S A. 2001;98:13710–5. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–35. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 93.Sakiyama H, et al. Regulation of nuclear import/export of carbohydrate response element-binding protein (ChREBP): interaction of an alpha-helix of ChREBP with the 14-3-3 proteins and regulation by phosphorylation. J Biol Chem. 2008;283:24899–908. doi: 10.1074/jbc.M804308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ge Q, et al. Structural characterization of a unique interface between carbohydrate response element-binding protein (ChREBP) and 14-3-3beta protein. J Biol Chem. 2012;287:41914–21. doi: 10.1074/jbc.M112.418855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arden C, et al. Fructose 2,6-bisphosphate is essential for glucose-regulated gene transcription of glucose-6-phosphatase and other ChREBP target genes in hepatocytes. Biochem J. 2012;443:111–23. doi: 10.1042/BJ20111280. [DOI] [PubMed] [Google Scholar]
- 96.Dentin R, et al. Glucose 6-phosphate, rather than xylulose 5-phosphate, is required for the activation of ChREBP in response to glucose in the liver. J Hepatol. 2012;56:199–209. doi: 10.1016/j.jhep.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 97.Bricambert J, et al. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120:4316–31. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55:1179–89. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 99.Li MV, Chen W, Poungvarin N, Imamura M, Chan L. Glucose-mediated transactivation of carbohydrate response element-binding protein requires cooperative actions from Mondo conserved regions and essential trans-acting factor 14-3-3. Mol Endocrinol. 2008;22:1658–72. doi: 10.1210/me.2007-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guinez C, et al. O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes. 2011;60:1399–413. doi: 10.2337/db10-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakiyama H, et al. The role of O-linked GlcNAc modification on the glucose response of ChREBP. Biochem Biophys Res Commun. 2010;402:784–9. doi: 10.1016/j.bbrc.2010.10.113. [DOI] [PubMed] [Google Scholar]
- 102.Wang Y, et al. Phosphorylation and recruitment of BAF60c in chromatin remodeling for lipogenesis in response to insulin. Mol Cell. 2013;49:283–97. doi: 10.1016/j.molcel.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abdulla A, et al. Regulation of lipogenic gene expression by lysine-specific histone demethylase-1 (LSD1) J Biol Chem. 2014;289:29937–47. doi: 10.1074/jbc.M114.573659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taniguchi CM, et al. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKClambda/zeta. Cell Metab. 2006;3:343–53. doi: 10.1016/j.cmet.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 105.Matsumoto M, et al. PKClambda in liver mediates insulin-induced SREBP-1c expression and determines both hepatic lipid content and overall insulin sensitivity. J Clin Invest. 2003;112:935–44. doi: 10.1172/JCI18816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li S, et al. Genome-wide coactivation analysis of PGC-1alpha identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 2008;8:105–17. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lamming Dudley W., Sabatini David M. A Central Role for mTOR in Lipid Homeostasis. Cell Metab. 18:465–469. doi: 10.1016/j.cmet.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peterson TR, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hagiwara A, et al. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 2012;15:725–38. doi: 10.1016/j.cmet.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 110.Yuan M, Pino E, Wu L, Kacergis M, Soukas AA. Identification of Akt-independent regulation of hepatic lipogenesis by mammalian target of rapamycin (mTOR) complex 2. J Biol Chem. 2012;287:29579–88. doi: 10.1074/jbc.M112.386854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Asher G, Sassone-Corsi P. Time for Food: The Intimate Interplay between Nutrition, Metabolism, and the Circadian Clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 112.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu S, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502:550–4. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bartok O, et al. The transcription factor Cabut coordinates energy metabolism and the circadian clock in response to sugar sensing. EMBO J. 2015 doi: 10.15252/embj.201591385. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giguere V, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–53. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 116.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bugge A, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–67. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cho H, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Delezie J, et al. The nuclear receptor REV-ERBalpha is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–35. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 120.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–9. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–9. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 122.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–82. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kang HS, et al. Transcriptional profiling reveals a role for RORalpha in regulating gene expression in obesity-associated inflammation and hepatic steatosis. Physiol Genomics. 2011;43:818–28. doi: 10.1152/physiolgenomics.00206.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang D, et al. Liver clock protein BMAL1 promotes de novo lipogenesis through insulin-mTORC2-AKT signaling. J Biol Chem. 2014;289:25925–35. doi: 10.1074/jbc.M114.567628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cretenet G, Le Clech M, Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 126.Basseri S, Austin RC. ER stress and lipogenesis: a slippery slope toward hepatic steatosis. Dev Cell. 2008;15:795–6. doi: 10.1016/j.devcel.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 127.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–27. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 128.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–90. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 129.Haeusler RA, et al. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang W, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–17. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 131.Otero YF, Stafford JM, McGuinness OP. Pathway-selective insulin resistance and metabolic disease: the importance of nutrient flux. J Biol Chem. 2014;289:20462–9. doi: 10.1074/jbc.R114.576355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gerhart-Hines Z, Lazar MA. Circadian Metabolism in the Light of Evolution. Endocr Rev. 2015 doi: 10.1210/er.2015-1007. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]