Abstract
Integration of hepatitis B virus (HBV) DNA into the human liver cell genome is believed to promote HBV-related carcinogenesis. This study aimed to quantify the integration of HBV DNA into the leukocyte genome in hepatocellular carcinoma (HCC) patients in order to identify potential biomarkers for HBV-related diseases. Whole-genome comparative genomic hybridization (CGH) chip array analyses were performed to screen gene copy number variations (CNV) in the leukocyte genome, and the results were confirmed by quantitative polymerase chain reaction (qPCR). The commonly detected regions included chromosome arms 19p, 5q, 1q and 15p, where 200 copy number gain events and 270 copy number loss events were noted. In particular, gains were observed in 5q35.3 (OR4F3) and 19p13.3 (OR4F17) in 90% of the samples. Successful homologous recombination of OR4F3 and the HBV P gene was demonstrated, and the amplification at 5q35.3 is potentially associated with the integration of HBV P gene into natural killer cells isolated from peripheral blood mononuclear cells (PBMCs). Receiver operating characteristic (ROC) curve analysis indicated that the combination of OR4F3 and OR4F17 a novel potential biomarker of HBV-related diseases.
Keywords: HBV-HCC, CGH, integration, biomarker
INTRODUCTION
Hepatocellular carcinoma (HCC), the third leading cause of cancer-related mortality worldwide [1], is associated with various etiological factors, including hepatitis B virus (HBV) or hepatitis C virus (HCV) infections, alcohol consumption, and dietary aflatoxin [2]. Among those, HBV is a particularly important factor in China [3]. Previous studies showed that HBV carriers have a 100-fold greater risk of HCC than non-carriers [4]. Moreover, HBV-HCC exhibits a propensity to metastasize shortly after diagnosis [5]. Although surveillance and surgical interventions have improved prognoses, a large proportion of HBV-positive HCC patients experience metastasis or postsurgical recurrence. Consequently, the five-year survival rate for HCC is only 30% to 40% [6].
The integration of HBV DNA into the human genome is of paramount importance for altering the function of endogenous genes or inducing chromosomal instability in HBV-HCC [7]. Studies have shown that HBV DNA integration into the hepatocyte genome occurs in 85% to 90% of HBV-HCC cases [8] and leads to multiple genomic aberrations [9]. Viral DNA integration disrupts the function of several genes important for normal cell growth and differentiation. In addition to point mutations, structural variations, and DNA methylation, the virus-associated aberrations include gene disruption or activation, formation of chimeric viral-human fusion genes, and DNA copy number variations (CNV) in some genes (e.g., cyclin A2), which cumulatively lead to hepatocyte carcinogenesis [10]. HBV DNA may also integrate into kidney and pancreas cells, indicating HBV integration is not liver cell-specific [11, 12].
The interaction between HBV and the immune system appears to be a crucial event during HBV-HCC development [13]. Numbers of immunocytes, including monocytes, are sharply decreased in HBV infected patients [14] and genes regulating immune function, such as HLA, IL-28B, STAT4 and MICA, are mutated by HBV [15]. These mutational events may disrupt immune clearance of HBV. Following clinically resolved infections, covalently closed circular (ccc) HBV DNA emerges not only in hepatocytes but also in leukocytes [16], which indicates that HBV infection may be responsible for leukocyte dysfunction in the peripheral blood [17]. Whether HBV can integrate into the leukocyte genome remains unknown, however.
In recent years, comparative genomic hybridization (CGH) array chips have become a useful tool in detecting and mapping gene CNV [18]. To provide an overview of CNV in the leukocyte genome of HBV-HCC patients, we employed CGH array chips to analyze peripheral blood mononuclear cells (PBMCs) from 10 HCC patients who were HBV carriers. Our study identified common gene gains and losses in the leukocyte genome of the HBV-HCC patients. We identified 14 immune-related genes. Of those, two genes coding for olfactory receptors, OR4F3 and OR4F17, exhibited increased copy numbers in approximately 90% of the samples. Thus HBV DNA does appear to integrate into the leukocyte genome, and the CNV correlated with HBV-related disease diagnoses and evaluations.
RESULTS
Genomic DNA copy number variations (CNV) in the leukocyte genome of HBV-HCC patients
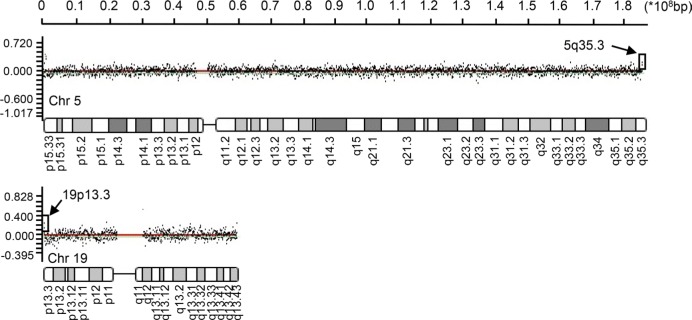
Whole genome CGH array analyses of the peripheral blood leukocytes from 10 HBV-HCC patients was performed to study the CNV at a 70-kb average resolution (Supplementary Figure 1). The commonly detected regions included chromosome arms 19p, 5q, 1q, and 15p. The most frequent losses were observed at 1p34.1 (MSAT2), 8p21.3 (SEC22B), 9p12 (ZNF658B) 1q21.1 (PPLAL4E), 19p21.3 (HIST2H4A), 15q11.2 (BCL8), 8p23.1 (FAM66E), 16p11.2 (NF1P1) and 16p13 (MIR1977). Gains were observed at 1p11.2 (FAM72B), 1q21.2 (NBPF16), 15q11.2 (OR4M2) (GOLGA8C), 1p36.33 (LOC133331), 15q26.3 (POTEB), 16p11.2 (TP5STG3B), 19p13.3 (OR4F17), 8p27.2(SPAGE11A), 9p24.3 (FAM138C), 1p36 (SMN1) and 5q35.3 (OR4F3). 200 copy number gain events and 270 copy number loss events were noted. We analyzed the 82 most frequently mutated genes (Table 1), which included 14 immune-related genes with high CNV (Table 2) using the process presented in Supplementary Figure 2. Although multiple chromosome regions were affected, the most frequent gains (log2 ratio > 0.75) were observed in the 5q35.3 and 19p13.3 regions in the olfactory receptor genes in 90% of the samples (Figure 1, indicated by arrows). The observed olfactory receptor (OR) genes included two members: OR4F3 (chromosome5, 181367287-181368225) and OR4F17 (chromosome 19, 107152-111690), presented in Figure 1.
Table 1. Genomic gains and losses of genes in the leukocyte genome.
| Gene | Chromosome | Ref.seq | Start | End | Gene size(kb) | Train start | Train end | Frequency (%) | Gene Location | Gain or loss |
|---|---|---|---|---|---|---|---|---|---|---|
| FAM72B | 1 | NM_001100910 | 120279658 | 143991624 | 23711966 | 120640527 | 120657204 | 60 | 1p11.2 | Gain |
| MAST2 | 1 | NM_015112 | 46051093 | 46114083 | 62990 | 46041871 | 46274383 | 60 | 1p34.1 | Loss |
| NBPF15 | 1 | NM_173638 | 146330841 | 148150199 | 1819358 | 146827613 | 146862782 | 60 | 1q21.2 | Gain |
| NBPF16 | 1 | NM_001102663 | 146330841 | 148150199 | 1819358 | 146843980 | 146862891 | 60 | 1q21.2 | Gain |
| NBPF20 | 1 | NM_001037675 | 120279658 | 143991624 | 23711966 | 143326315 | 143540167 | 60 | 1q21.2 | Gain |
| POTEB | 15 | NM_207355 | 18304999 | 20124999 | 1820000 | 19305252 | 19336667 | 50 | 15q26.3 | Gain |
| PPIAL4A | 1 | NM_178230 | 146330841 | 148150199 | 1819358 | 147819626 | 147820286 | 60 | 1q21.1 | Gain |
| PPIAL4E | 1 | NM_001144032 | 146330841 | 148150199 | 1819358 | 146910634 | 146911417 | 60 | 1q21.2 | Gain |
| BCL8 | 15 | NR_027992 | 18304660 | 20080292 | 1775632 | 19134810 | 19221496 | 40 | 15q11.2 | Gain |
| FAM138A | 1 | NR_026818 | 20214 | 239271 | 219057 | 27220 | 28690 | 50 | 1p36.33 | Gain |
| FAM138C | 9 | NR_026822 | 20214 | 239271 | 219057 | 27220 | 28690 | 50 | 9p24.3 | Gain |
| FAM138F | 19 | NR_026820 | 20214 | 239271 | 219057 | 27220 | 28690 | 50 | 19p13.3 | Gain |
| FAM66A | 8 | NR_026789 | 11874577 | 12618127 | 743550 | 12263898 | 12312881 | 40 | 8p23.1 | Gain |
| FAM66E | 8 | NR_027424 | 7169866 | 7903147 | 733281 | 7849944 | 7903687 | 40 | 8p23.1 | Gain |
| FLJ45445 | 7 | NR_028324 | 20214 | 239271 | 219057 | 148015 | 153209 | 50 | 7q32.1 | Loss |
| LOC 100132602 | 14 | NR_028325 | 30634 | 765731 | 735097 | 313754 | 318443 | 60 | 14q11.1 | Gain |
| LOC 100133331 | 1 | NR_028327 | 30634 | 717252 | 686618 | 651002 | 655594 | 60 | 1p36.33 | Gain |
| LOC 727924 | 15 | NR_015416 | 18304660 | 20080292 | 1775632 | 19779395 | 19872450 | 40 | 15q11.2 | Loss |
| *MIR1977 | 1 | NR_031741 | 30634 | 717252 | 686618 | 556050 | 556128 | 60 | MT | Loss |
| CXADRP2 | 15 | NR_024387 | 18304999 | 20124999 | 1820000 | 19267798 | 19270255 | 50 | 15q11.2 | Gain |
| GOLGA8C | 15 | NR_027411 | 18304999 | 20124999 | 1820000 | 19027687 | 19041040 | 50 | 15q11.2 | Loss |
| LOC 646214 | 15 | NR_027053 | 18304999 | 20124999 | 1820000 | 19185890 | 19194116 | 50 | 15q11.2 | Gain |
| NF1P1 | 15 | NR_028506 | 18304999 | 20124999 | 1820000 | 19386652 | 19399284 | 50 | 15q11.2 | Gain |
| OR4N3P | 15 | NR_028067 | 18304999 | 20124999 | 1820000 | 19914825 | 19915757 | 50 | 15q11.2 | Gain |
| ZNF658B | 9 | NR_027861 | 38751600 | 70210086 | 31458486 | 39433819 | 39437188 | 40 | 9p12 | Gain |
| GOLGA6L6 | 15 | NM_001145004 | 18304999 | 20124999 | 1820000 | 18997107 | 19007128 | 50 | 15q11.2 | Loss |
| FAM90A7 | 8 | NM_001136572 | 7244999 | 7804999 | 560000 | 7401070 | 7404644 | 40 | 8p23.1 | Gain |
| DEFB103A | 8 | NM_018661 | 7244999 | 7804999 | 560000 | 7273825 | 7275092 | 40 | 8p23.1 | Gain |
| DEFB103B | 8 | NM_001081551 | 7244999 | 7804999 | 560000 | 7273900 | 7275280 | 40 | 8p23.1 | Gain |
| DEFB104A | 8 | NM_080389 | 7244999 | 7804999 | 560000 | 7315239 | 7320014 | 40 | 8p23.1 | Gain |
| DEFB104B | 8 | NM_001040702 | 7244999 | 7804999 | 560000 | 7315239 | 7320014 | 40 | 8p23.1 | Gain |
| DEFB105A | 8 | NM_152250 | 7244999 | 7804999 | 560000 | 7716939 | 7718770 | 40 | 8p23.1 | Gain |
| DEFB105B | 8 | NM_001040703 | 7244999 | 7804999 | 560000 | 7716939 | 7718770 | 40 | 8p23.1 | Gain |
| DEFB106A | 8 | NM_152251 | 7244999 | 7804999 | 560000 | 7327435 | 7331319 | 40 | 8p23.1 | Gain |
| DEFB106B | 8 | NM_001040704 | 7244999 | 7804999 | 560000 | 7327435 | 7331319 | 40 | 8p23.1 | Gain |
| DEFB107A | 8 | NM_001037668 | 7244999 | 7804999 | 560000 | 7706651 | 7710648 | 40 | 8p23.1 | Gain |
| DEFB107B | 8 | NM_001040705 | 7244999 | 7804999 | 560000 | 7706651 | 7710648 | 40 | 8p23.1 | Gain |
| SPAG11B | 8 | NM_058202 | 7244999 | 7804999 | 560000 | 7292685 | 7308602 | 40 | 8p23.1 | Loss |
| OR4F16 | 1 | NM_001005484 | 34999 | 524999 | 490000 | 58953 | 59871 | 40 | 1p36.33 | Gain |
| OR4F17 | 19 | NM_001005240 | 34999 | 174999 | 140000 | 61678 | 62596 | 90 | 19p13.3 | Gain |
| OR4F29 | 1 | NM_001005221 | 34999 | 524999 | 490000 | 357521 | 358460 | 40 | 1p36.33 | Gain |
| OR4F3 | 5 | NM_001005224 | 34999 | 524999 | 490000 | 357521 | 358460 | 90 | 5q35.3 | Gain |
| OR4F5 | 1 | NM_001005277 | 34999 | 524999 | 490000 | 357521 | 358460 | 40 | 1p36.33 | Gain |
| OR4N4 | 15 | NM_001005241 | 18304999 | 20124999 | 1820000 | 19883836 | 19884787 | 50 | 15q11.2 | Gain |
| OR4M2 | 15 | NM_001004719 | 18304999 | 20124999 | 1820000 | 19869939 | 19870881 | 50 | 15q11.2 | Gain |
| SEC22B | 1 | NM_004892 | 120279658 | 143991624 | 23711966 | 143807763 | 143828279 | 60 | 1q21.1 | Loss |
| BOLA1 | 1 | NM_016074 | 146330841 | 148150199 | 1819358 | 148137778 | 148138971 | 60 | 1q21 | Gain |
| FAM75A5 | 9 | NM_001113541 | 38751600 | 70210086 | 31458486 | 41311106 | 41317364 | 40 | 9p12 | Loss |
| FAM75A7 | 9 | NM_015667 | 38751600 | 70210086 | 31458486 | 41311109 | 41317364 | 40 | 9q12 | Loss |
| HIST2H2AA3 | 1 | NM_003516 | 146330841 | 148150199 | 1819358 | 148089251 | 148089785 | 60 | 1q21.2 | Gain |
| HIST2H2AA4 | 1 | NM_001040874 | 146330841 | 148150199 | 1819358 | 148089251 | 148089785 | 60 | 1q21.2 | Gain |
| HIST2H2AC | 1 | NM_003517 | 146330841 | 148150199 | 1819358 | 148125148 | 148125585 | 60 | 1q21.2 | Gain |
| HIST2H3A | 1 | NM_001005464 | 146330841 | 148150199 | 1819358 | 148090804 | 148091311 | 60 | 1q21.2 | Gain |
| HIST2H3C | 1 | NM_021059 | 146330841 | 148150199 | 1819358 | 148090804 | 148091311 | 60 | 1q21.2 | Gain |
| HIST2H4A | 1 | NM_003548 | 146330841 | 148150199 | 1819358 | 148070844 | 148071240 | 60 | 1q21.2 | Gain |
| HIST2H4B | 1 | NM_001034077 | 146330841 | 148150199 | 1819358 | 148070844 | 148071240 | 60 | 1q21.2 | Gain |
| FAM66D | 8 | NR_027425 | 11874577 | 12618127 | 743550 | 12010699 | 12046107 | 40 | 8p23.1 | Loss |
| GTF2H2D | 5 | NM_001042490 | 68914999 | 70664999 | 1750000 | 68891829 | 68924108 | 40 | 5q13.2 | Loss |
| GTF2H2C | 5 | NM_001098728 | 68914999 | 70664999 | 1750000 | 68891806 | 68924485 | 40 | 5q13.2 | Loss |
| GTF2H2B | 5 | NM_001098729 | 68914999 | 70664999 | 1750000 | 68891824 | 68924485 | 40 | 5q13.2 | Loss |
| TP53TG3B | 16 | NM_001099687 | 32444999 | 32724999 | 280000 | 32592341 | 32596379 | 40 | 16p11.2 | Loss |
| TP53TG3 | 16 | NM_016212 | 32444999 | 32724999 | 280000 | 32592349 | 32595554 | 40 | 16p13 | Loss |
| MSTP2 | 1 | NR_027504 | 16834999 | 16974999 | 140000 | 16844655 | 16849501 | 40 | 1p36.2 | Gain |
| LOC 100132287 | 1 | NR_028322 | 34999 | 524999 | 490000 | 313754 | 318443 | 40 | 1p36.33 | Gain |
| LCE3C | 1 | NM_178434 | 150822555 | 150852635 | 30080 | 150839761 | 150840186 | 60 | 1q21.3 | Loss |
| NAIP | 5 | NM_022892 | 68888924 | 70682363 | 56632 | 70300065 | 70356697 | 50 | 5q13.2 | Gain |
| SMN1 | 5 | NM_022874 | 69386142 | 70704534 | 1318392 | 70256523 | 70284592 | 50 | 5q13.2 | Gain |
| SMN2 | 5 | NM_022877 | 69386142 | 70704534 | 1318392 | 69381105 | 69409177 | 40 | 5q13.2 | Gain |
| DEFB109P1B | 8 | NR_003668 | 7169866 | 7903147 | 733281 | 7885347 | 7892453 | 30 | 8p23.1 | Gain |
| LOC 653391 | 5 | NR_029426 | 68888924 | 70682363 | 1793439 | 68713442 | 69917303 | 40 | 5q13.2 | Gain |
| LOC 653501 | 9 | NR_003528 | 38751600 | 70210086 | 31458486 | 39433813 | 39454526 | 30 | 9p13.1 | Gain |
| BTNL8 | 5 | NM_197975 | 180355922 | 45982 | 18034890 | 180348506 | 180366333 | 20 | 5q35.3 | Loss |
| SPAG11A | 8 | NM_001081552 | 7153912 | 7982049 | 828137 | 7742811 | 7758729 | 60 | 8p27.2 | Gain |
| ZNF705G | 8 | NM_001164457 | 7153912 | 7982049 | 828137 | 7202908 | 7207900 | 50 | 8p23.1 | Gain |
| FAM86B2 | 8 | NM_001137610 | 11874577 | 12618127 | 743550 | 12327496 | 12338223 | 50 | 8p23.1 | Loss |
| FAM90A10 | 8 | NM_001164447 | 7169866 | 7903147 | 733281 | 7663908 | 7666919 | 60 | 8p23.1 | Loss |
| FAM90A13 | 8 | NM_001164456 | 7169866 | 7903147 | 733281 | 7610374 | 7613384 | 60 | 8p23.1 | Loss |
| FAM90A14 | 8 | NM_001164452 | 7169866 | 7903147 | 733281 | 7101835 | 7104845 | 50 | 8p23.1 | Loss |
| FAM90A18 | 8 | NM_001164451 | 6891623 | 7903147 | 1011524 | 7618022 | 7621032 | 40 | 8p23.1 | Loss |
| FAM90A19 | 8 | NM_001164449 | 7153912 | 7982049 | 828137 | 7648613 | 7651623 | 40 | 8p23.1 | Loss |
| FAM90A20 | 8 | NM_001164453 | 6891623 | 7903147 | 1011524 | 7139945 | 7142955 | 40 | 8p23.1 | Loss |
| FAM90A5 | 8 | NM_001164455 | 6891623 | 7903147 | 1011524 | 7132323 | 7135333 | 30 | 8p23.1 | Loss |
Ref.seq: The Reference Sequence (RefSeq) collection provides a comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins. Frequency: Mutated frequency for genes in the 10 PBMC samples of CGH chips for HBV-HCC patients. *MIR1977 belongs to the MT (mitochondrial) genome.
Table 2. Copy number variations of immune-related genes in the leukocyte genome.
| Gene | Chromosome | Ref.seq | Start | End | Gene size (kb) | Train start | Train end | Frequency (%) | Gene Location | Gain or loss |
|---|---|---|---|---|---|---|---|---|---|---|
| DEFB103A | 8 | NM_018661 | 7244999 | 7804999 | 560000 | 7273825 | 7275092 | 40 | 8p23.1 | Gain |
| DEFB103B | 8 | NM_001081551 | 7244999 | 7804999 | 560000 | 7273900 | 7275280 | 40 | 8p23.1 | Gain |
| DEFB104A | 8 | NM_080389 | 7244999 | 7804999 | 560000 | 7315239 | 7320014 | 40 | 8p23.1 | Gain |
| DEFB104B | 8 | NM_001040702 | 7244999 | 7804999 | 560000 | 7315239 | 7320014 | 40 | 8p23.1 | Gain |
| DEFB105A | 8 | NM_152250 | 7244999 | 7804999 | 560000 | 7716939 | 7718770 | 40 | 8p23.1 | Gain |
| DEFB105B | 8 | NM_001040703 | 7244999 | 7804999 | 560000 | 7716939 | 7718770 | 40 | 8p23.1 | Gain |
| DEFB106A | 8 | NM_152251 | 7244999 | 7804999 | 560000 | 7327435 | 7331319 | 40 | 8p23.1 | Gain |
| DEFB106B | 8 | NM_001040704 | 7244999 | 7804999 | 560000 | 7327435 | 7331319 | 40 | 8p23.1 | Gain |
| DEFB107A | 8 | NM_001037668 | 7244999 | 7804999 | 560000 | 7706651 | 7710648 | 40 | 8p23.1 | Gain |
| DEFB107B | 8 | NM_001040705 | 7244999 | 7804999 | 560000 | 7706651 | 7710648 | 40 | 8p23.1 | Gain |
| DEFB109P1B | 8 | NR_003668 | 7169866 | 7903147 | 733281 | 7885347 | 7892453 | 30 | 8p23.1 | Gain |
| BTNL8 | 5 | NM_197975 | 180355922 | 45982 | 18034890 | 180348506 | 180366333 | 20 | 5q35.3 | Loss |
| SPAG11A | 8 | NM_001081552 | 7153912 | 7982049 | 828137 | 7742811 | 7758729 | 60 | 8p27.2 | Gain |
| LCE3C | 1 | NM_178434 | 150822555 | 150852635 | 150839761 | 150840186 | 353144 | 30 | 1p21.3 | Loss |
*Ref.seq: The Reference Sequence (RefSeq) collection provides a comprehensive, integrated, non-redundant, well-annotated set of sequences, including genomic DNA, transcripts, and proteins. Frequency: Mutated frequency for genes in the 10 PBMC samples of CGH chips for HBV-HCC patients.
Figure 1. OR4F3 and OR4F17 amplifications.
A chromosome view of the CGH array results showing the terminal copy number gains on 5q35.3 and 19p13.3 including OR4F3 and OR4F17. Arrowheads indicate targets with upward-shifted log2 ratios. Chromosome (Chr).
We assessed OR4F3 and OR4F17 copy number levels by absolute qPCR in healthy controls (Con, n = 5), chronic hepatitis B patients (CHB, n = 10), hepatitis B virus-liver cirrhosis patients (HBV-LC, n = 10), and hepatitis B virus- HCC patients (HBV-HCC, n = 10). The results were consistent with the CGH array results. Compared with healthy controls, we observed up-regulation of OR4F3 and OR4F17 in HBV-related patients (p<0.05) (Supplementary Figure 3). The absolute amount of OR4F3 and OR4F17 DNA in the peripheral blood was highest in CHB patients, followed by HBV-LC patients, and lowest for HBV-HCC patients. All qPCR reaction efficiency values were between 1.95 and 1.99. The inter-assay variation in the qPCR efficiency was < 1%, confirming that the qPCR products were amplified at approximately the same rate.
The HBV DNA integration into the leukocyte genome
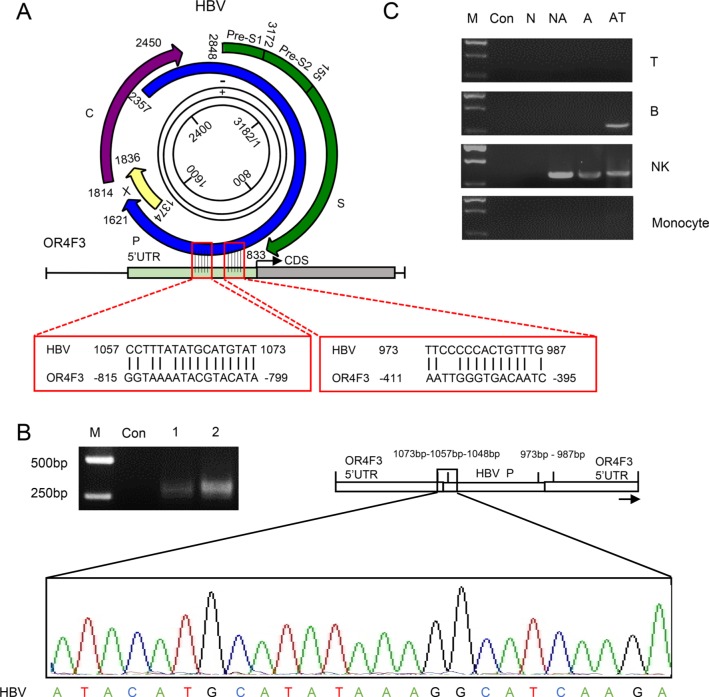
Wing-Kin Sung's study found that the HBV DNA integration breakpoints appear to be associated with increased CNV [19]. To determine whether HBV DNA integrates into the leukocyte genome, a pairwise sequence alignment was performed between HBV and OR4F3 as a proof-of-principle study. Two complementary sites between these two sequences were identified (Figure 2A). In the human leukocyte genome, two sites at the OR4F3 5′ untranslated region (UTR) position (−395bp to −411bp and −799bp to −815bp) with an interval of 421 bp were complementary to the HBV P gene position sites (987bp to 973bp and 1073bp to 1057bp) with an interval of 101 bp. To test whether the 101 bp interval of the HBV P gene may interchange with the 421 bp interval that was located in the OR4F3 5′ UTR, PCR amplification was performed with a pair of exclusive primers, 5′–CTTGATGCCTTTATATGCATGTAT (HBV), 3′—CAACATGTGAGCTGCAAC CT (OR4F3, Supplementary Figure 4). As shown in Figure 2B, the specific recombination fragments, which were approximately 310 bp for HBV-HCC sample 1 and HBV-HCC sample 2, were derived from HBV in the OR4F3 5′ UTR. Additionally, the chimeric fusion genes (accession number: KT715476) were confirmed by sequencing the region between nucleotides 1048 and 1073 of the HBV P gene (Figure 2B, indicated by the black frames). This finding indicated the successful homologous recombination of the HBV P gene and the OR4F3 5′ UTR, thus confirming the integration of HBV DNA directly into the leukocyte genome.
Figure 2. The homologous recombination of the OR4F3 5′UTR and HBV P gene.
A. A schematic alignment of OR4F3 and HBV is presented. The complementary sites between the OR4F3 gene and the HBV genome P gene are indicated by red frames. B. Compared with the control (Con), the agarose gel electrophoresis results show the OR4F3 and HBV chimeric fusion genes in the HBV-HCC samples. Sequencing results show one complementary site and a short HBV P gene sequence. The M lanes contain the DNA size markers (DL2000). The Con lane is healthy control, and lanes 1 and 2 are the HBV-HCC samples. UTR (untranslated region). C. The OR4F3 and HBV P gene chimeric fusion genes in the leukocyte genome are shown. Agarose gel electrophoresis showing the PCR amplification products for the healthy control (Con), non-HBV-HCC (N), non-active HBV-HCC (NA), active HBV-HCC (A), and antiviral therapy (AT) samples as well as marker (M). The chimeric fusion DNA fragments of P gene and OR4F3 were observed in the sorted NK cells of HBV- samples and B cells of samples from patient receiving antiviral therapy. T-lymphocyte cells (T cells); B-lymphocyte cells (B cells); Natural killer cells (NK cells).
Subsequently, CD3+ T (34.03%), CD19+ B (2.95%), CD3−CD56+ natural killer (NK) cells (26.89%) and CD14+ monocytes (1.89%) from PBMCs were sorted to determine the specific cell-lineages into which the HBV P gene integrated. The peripheral blood samples were obtained from healthy controls (Con, n = 3), non-HBV HCC patients (N, n = 3), non-active-HBV HCC patients (NA, n = 3), active-HBV HCC patients (A, n = 3), and from patients receiving antiviral therapy (AT, n = 3). As illustrated in Supplementary Figure 5, CD3+ T cells, CD19+ B cells, CD3−CD56+ natural killer (NK) cells and CD14+ monocytes were successfully isolated from PBMCs. Then, PCR analyses with the exclusive primers were used to identify the homologous recombination fragments of the P gene and OR4F3. As shown in Figure 2C, no specific P gene integration band was displayed in the four cell-lineages that were sorted from the healthy controls and the HBV-negative HCC samples. However, in the non-active HBV HCC sample lanes, active HBV HCC lanes, and samples from patient receiving antiviral therapy, bands that represented the chimeric fusion DNA fragments of P gene and OR4F3 were observed in the sorted NK cells. Moreover, bands that represented the chimeric fusion DNA fragments of P gene and OR4F3 were also observed in the sorted B cells of samples from patients receiving antiviral therapy. As shown in Figure 2C, the observed integration of the HBV P gene into the OR4F3 5 'UTR was cell-lineage specific.
OR4F3 and OR4F17 are candidate biomarkers for HBV-related disease diagnoses
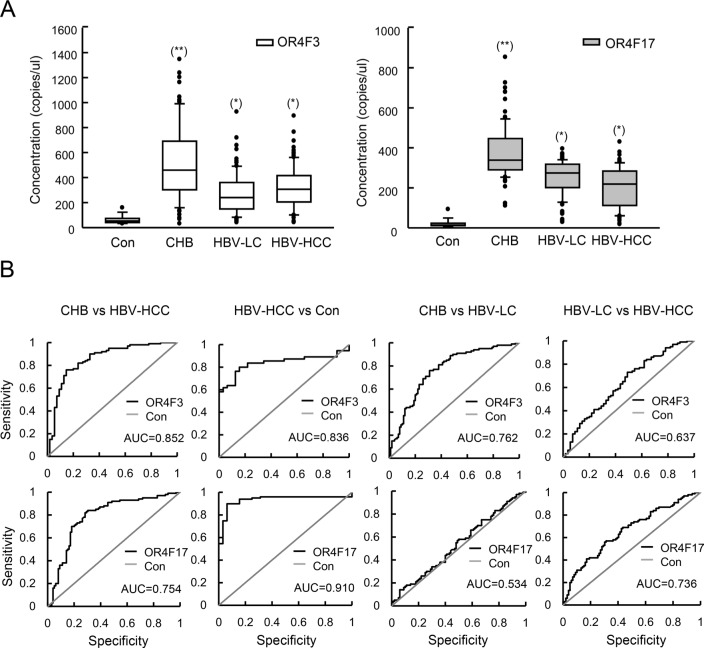
We collected 320 samples of DNA from PBMCs from chronic hepatitis B (CHB), hepatitis B virus-liver cirrhosis (HBV-LC), HBV-HCC patients, and normal subjects (Supplementary Figure 6). As illustrated in Figure 3A, the absolute amounts of OR4F3 and OR4F17 approximately 9-fold and 16-fold higher in the CHB samples (P < 0.005), respectively, and 5-fold and 13-fold higher in the HBV-LC and HBV-HCC samples (P < 0.05), respectively, relative to healthy controls.
Figure 3. OR4F3 and OR4F17 as biomarkers for HBV-related diseases.
A. Quantification of OR4F3 and OR4F17 levels in healthy controls (n = 20), CHB patients (n = 100), HBV-LC patients (n = 100), and HBV-HCC patients (n = 100). The copy number levels were determined using absolute qPCR. Results are presented as copies per microliter. The unpaired t-test was performed to assess significance of differences between HBV-related groups and control groups. P-values of less than 0.05 were deemed to be significant. (*p < 0.05; **p < 0.005). Control (Con); chronic hepatitis B (CHB); hepatitis B virus-liver cirrhosis (HBV-LC); hepatitis B virus- hepatocellular carcinoma (HBV-HCC); OR4F3, olfactory receptor, family 4, subfamily F, member 3; OR4F17, olfactory receptor, family 4, subfamily F, member 17. B. ROC curves for the OR4F3 and OR4F17 genes between the CHB and HBV-HCC as well as HBV-HCC and normal are presented. OR4F3 yielded AUCs (areas under the ROC curve) of 0.852 for discriminating HBV-HCC from CHB and 0.836 for discriminating HBV-HCC from normal. OR4F17 yielded AUCs of 0.754 for discriminating HBV-HCC from CHB and 0.910 for discriminating HBV-HCC from normal. The ROC curves between CHB and HBV-LC as well as HBV-HCC and HBV-LC are presented. OR4F3 yielded AUCs of 0.762 for discriminating HBV-LC from CHB and 0.637 for discriminating HBV-HCC from HBV-LC. OR4F17 yielded AUCs of 0.534 for discriminating HBV-LC from CHB and 0.736 for discriminating HBV-HCC from HBV-LC. Control (Con).
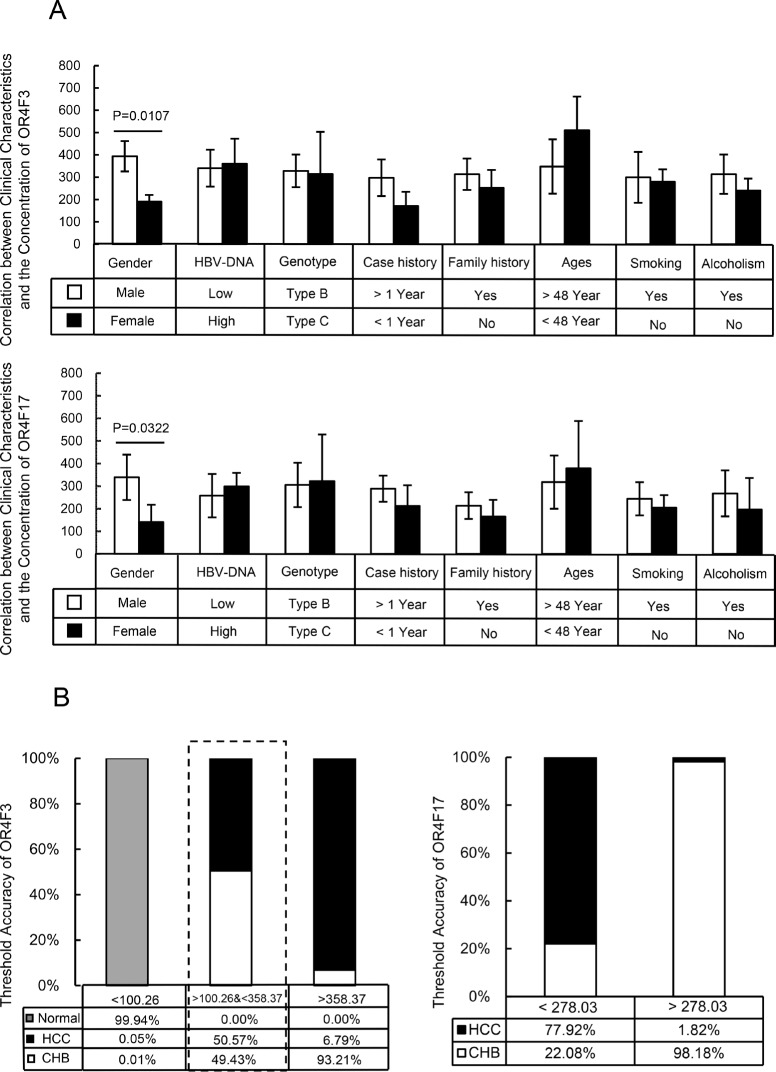
Subsequently, we evaluated whether OR4F3 and OR4F17 copy number could be used as diagnostic biomarkers for HBV-related diseases. ROC curve analyses were performed in pooled data sets. As shown in Figure 3B, we observed that OR4F3 copy number variation could distinguish CHB from HBV-HCC with an area under curves (AUC) of 0.852 (95% confidence interval (CI), 0.829-0.895) as well as HBV-HCC from healthy controls (Con) with an AUC of 0.836 (95% CI, 0.824-0.872). OR4F17 was also a potential biomarker for discriminating CHB from HBV-HCC with an AUC of 0.754 (95% CI, 0.715-0.803) as well as HBV-HCC from healthy controls (Con) with an AUC of 0.910 (95% CI, 0.894-0.936). However, OR4F3 could not discriminate CHB from HBV-LC with AUC of 0.762 or HBV-HCC from HBV-LC with AUC of 0.637, (95% CI not shown). OR4F17 was also not a potential biomarker for discriminating CHB from HBV-LC with AUC of 0.534 or for HBV-HCC from healthy controls with AUC of 0.736 (95% CI not shown). Furthermore, as shown in Figure 4A, the concentration of the OR genes did not correlate with the HBV-DNA level, genotype, case history, family history, age (group defined on the median of ages, Table 3), smoking, or alcoholism (P > 0.05). However, the concentrations of OR4F3 and OR4F17 were significantly different between males and females (P < 0.05).
Figure 4. Clinical analysis of the samples and the threshold accuracy of OR4F3 and OR4F17.
A. Correlation between clinical characteristics and the concentration of OR4F3 and OR4F17. According to the imaging and histological examination of liver, the patients were grouped into 100 CHB patients, 100 HBV-LC patients and 100 HBV-HCC patients. The concentration of OR gene did not correlate with the HBV-DNA level, genotype, case history, family history, ages, smoking, or alcoholism (P>0.05); however, the concentrations of OR4F3 and OR4F17 were varied between males and females (P = 0.0107 and P = 0.0322, respectively). Units: copies/ul. B. Threshold accuracy of OR4F3 and OR4F17. The threshold accuracy for OR4F3 and OR4F17. The cut-off value unit was copies/ul.
Table 3. Observed clinical characteristics of CHB, HBV-LC and HBV-HCC patients.
| Copy number(/ml) | OR3<15.66 | 15.66<OR3<56.08 | OR3>56.08 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | CHB | HBV-LC | HBV-HCC | CHB | HBV-LC | HBV-HCC | CHB | HBV-LC | HBV-HCC |
| Number | 1 | 6 | 4 | 24 | 62 | 46 | 75 | 32 | 50 |
| Age (yrs) | 40 | 48±6.9 | 44±4 | 41±11.8 | 51±8.6 | 47±10.6 | 43±13.4 | 50±7.9 | 55±9.4 |
| Gender | |||||||||
| male | 1 | 6 | 3 | 16 | 48 | 26 | 48 | 26 | 28 |
| female | 0 | 0 | 1 | 8 | 14 | 20 | 27 | 5 | 22 |
| Biochemistry(IU/L) | |||||||||
| ALT | 14 | 40.0-44.0 | 9.0-12.0 | 25.5-137.0 | 37.8-139.8 | 18.5-40.9 | 33.3-147.8 | 37-158 | 17.5-38.6 |
| AST | 21 | 35.0-59.0 | 9.0-12.0 | 26.3-58.0 | 44.3-123.8 | 22.0-44.5 | 41.5-89.8 | 56-212 | 18.0-38.5 |
| AFP | 1.3 | 1.1-6.7 | 6.8-1629.7 | 8.0-127.9 | 1.1-7.1 | 2.9-32380.8 | 3.1-14.3 | 1.0-7.3 | 3.9-41283.8 |
| Tumor Sizes(cm) | \ | \ | 1.9-8.6* | \ | \ | 1.2-11.5* | \ | \ | 1.1-15.5* |
Data are presented as mean +/− standard deviation.
P <0.05.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; AFP, alpha-fetoprotein; yrs, years; HBV, hepatitis B virus; CHB, chronic hepatitis B; HBV-liver cirrhosis (HBV-LC); HBV-hepatocellular carcinoma (HBV-HCC).
DISCUSSION
In this study, we identified multiple copy number alterations to the leukocyte genome in HBV-HCC patients. Of these, gains in two genes, OR4F3 and OR4F17, with a CNV frequency greater than 90% were noted at chromosome loci 5q35.3 and 19p13.3, respectively. Moreover, in combination OR4F3 and OR4F17 may provide a new biomarker for HBV-related disease diagnoses.
A specific fragment of the HBV genome, the P gene, was commonly integrated into the leukocyte genome upstream of OR4F3. Although the HBV cccDNA was observed in the cytoplasm of leukocytes, the fact that its integration is widespread in NK and B cells from patients receiving antiviral therapy has not been reported previously [20]. HBV DNA integration into the hepatocyte genome is a major mechanism underlying the etiologic role of HBV in HBV-HCC development [21]. A number of reports have indicated that HBx is one of the viral ORFs most commonly integrated into the hepatocyte genome, and its sequence variants play a crucial role in the progression of HBV-HCC [22]. HBx acts via the HBx-P3K-Akt-Bad pathway to inhibit caspase 3 function, thereby preventing p53-induced apoptosis and mediating induction of H-ras oncogene via phosphatidylinositol-3 kinase and Akt pathways [23]. Additionally, integration of the P region of HBV into the hepatocyte genome promotes genetic instability through mechanisms that include p53-mediated repression of gene transcription [24]. HBV integration also leads to elevated expression of several other cancer-related genes, including TERT, MLL4 and CCNE1 [25]. The dysregulation of numerous signaling pathways, including the MAPK, P53, sex steroid, Wnt/β-catenin, transforming growth factor β (TGFβ), PI3K/AKT, cytokine, IKK/NF-kB, and Hedgehog (Hh) pathways, has been observed in patients, and these changes are closely related with HBV-HCC development [26].
Our study revealed that genes exhibiting multiple copy number alterations, as detected using CGH array chips, may be associated with HBV DNA integration into the leukocyte genome. Alterations in chromosomal stability and changes in copy numbers are characteristic of HBV integrated events in HCC. The variation in copy numbers located within or near the integration sites suggests HBV DNA integration induces chromosomal instability. Jiang et al. previously showed that HBV DNA frequently integrates into the human genome, causing diverse changes, including DNA copy number variations, chimeric viral-human transcript fusions, and transcriptional activation [7]. Additional studies exploring possible tumorigenic roles of these genes with copy number changes would be of interest. Moreover, the underlying mechanisms of OR genes in leukocyte dysfunction, including weakened immune surveillance and HBV- HCC development, and the novel functions of HBV DNA integrated into the leukocyte genome provide interesting areas for future study.
Surprisingly, OR4F3 and OR4F17 copy numbers in the leukocyte genome serve as noninvasive biomarkers for diagnosing HBV-related diseases. We found that when the OR4F3 copy number is lower than 100.26 copies/μL, the probability that the samples were healthy controls was 99.94%, 0.01% for CHB, and 0.05% for HBV-HCC. When the OR4F3 copy number was higher than 358.37 copies/μL, the probability that the samples were from CHB patients, HBV-HCC patients or healthy controls was 93.21%, 6.79% and 0%, respectively. However, when the copy numbers were between 100.26 and 358.37 copies/μL, the probability that the samples were from CHB patients or HBV-HCC patients was 49.43% and 50.57%, respectively. Within the range of 100.26 to 358.37 copies/μL, these samples could be distinguished based on OR4F17. When the OR4F17 copy number was less than 278.03 copies/μL, the probability that the samples were from CHB or HBV-HCC patients was 22.08% and 77.92%, respectively. When the OR4F17copy number was greater than 278.03 copies/μL, the probability that the samples were from CHB or HBV-HCC patients was 98.18% and 1.82%, respectively (Figure 4B). However, these biomarkers could not distinguish HBV-LC from HBV-HCC. Given that liver cirrhosis development is the closest step to HBV-HCC, we hypothesize that the genomic aberrations in the HBV-LC and HBV-HCC patients were approximately the same. However, this hypothesis needs to be tested.
In summary, our results demonstrate that both integration of the HBV P gene into the 5′ UTR of OR4F3 genes and the copy numbers of OR genes were significantly increased within the leukocyte genome of HBV-related patients. Thanks to their reasonable sensitivity and specificity values for HBV-related diseases, it appears that OR4F3 and OR4F17 could potentially serve as diagnostic biomarkers. Several studies have also demonstrated that olfactory receptors possess oncogenic potential and can induce tumor formation when overexpressed in cells [27]. However, studies on the oncogenic potential of OR genes in the development of HBV-HCC have not been performed. Based on our present results, we are currently investigating the primary functions of OR genes duing the development and progression of HBV-HCC.
MATERIALS AND METHODS
Clinical samples
We performed clinical CGH array testing on 10 PBMCs samples from HBV-HCC patients from Eastern Hepatobiliary Surgery Hospital, Shanghai. HBV-HCC was diagnosed by the elevation of alpha-fetoprotein (AFP) (>400ng/mL) combined with at least one positive iconography examination result, including computed tomography (CT) and magnetic resonance imaging (MRI). The pathologic data of these 10 patients are summarized in Supplementary Table 1. They had pathological stage III (n = 7), pathological stage IV (n = 2), and pathological stage II (n = 1) according to the Edmondson-Steiner grading system. Four patients (40%) had tumor size less than 5 cm while one patient (10%) had tumor size of 11 cm. The rest (50%) had tumor sizes between 5 and 10 cm. These patients' ages ranged from 35-68 years with a median age of 43. They all had Child-Pugh class A liver function and were positive for HBV antigen (HBV-Ag). Their AFP levels ranged from 12-12430 ug/L. The ethics committee of the Eastern Hepatobiliary Surgery Hospital, Second Military Medical University (Shanghai, China), approved this study. Written informed consent was obtained from the patients according to the regulations of the committee.
CGH array chips
For the CGH array experiments, 1 μg of purified patient DNA and control DNA were digested with 10U Alui and 10U RsaI and differentially labeled with cyanine-5 (Cy5) and cyanine-3 (Cy3) fluorescent dyes (PerkinElmer, Waltham, Massachusetts, USA) by random priming with the Bioprime CGH Array genomic labeling module (Invitrogen, Carlsbad, California, USA). Hybridization was performed for at least 20 h at 65°C in a rotating microarray hybridization chamber and then samples were washed according to the manufacturer's protocols (Agilent Technologies, Santa Clara, California, USA). The slide hybridization results were scanned into image files using a GenePix 4000B microarray scanner (Molecular Devices, Sunnyvale, California, USA). Then, Agilent's Feature Extraction v9.1 was used to quantify the array features, and the CGH analytics software was used to analyze the text file outputs for relative copy number changes. Common copy number variations were assessed by comparing these sample results with CNV databases and parental CGH patterns, when available.
Microarray data analysis
Microarray data were analyzed with BRB-array tools. The Agilent Genomic Workbench was used to calculate the log2 ratios for every probe and to identify any genomic aberrations. The mean log2 ratios of all the probes in the chromosome regions between 0.25 and 0.75 were classified as genomic gains, > 0.75 was classified as high-level DNA amplification, < 0.25 was classified as homozygous losses, and < −0.75 was classified as homozygous deletions.
Sample collection
Peripheral blood samples collected from 320 subjects were obtained from Eastern Hepatobiliary Hospital, Shanghai. According to imaging and histological examination of liver, patients were grouped into 100 CHB patients, 100 HBV-LC patients and 100 HBV-HCC patients. Twenty healthy control samples were obtained from the Physical Examination Center of Eastern Hepatobiliary Hospital. Five milliliters (mL) of peripheral blood was collected in K3-EDTA containing tubes, and PBMCs were isolated with 75% Ficoll Hypaque. PBMCs were distributed into aliquots and stored at −80°C until use.
The clinical characteristics of patients and controls, including age, gender etc., were investigated at the time of blood collection. The following laboratory parameters were obtained for each participant: aspartate aminotransferase (AST), alanine aminotransferase (ALT), and AFP. The main clinical characteristics of patients are summarized in Table 3.
Patients were divided into three main groups: (1) Group I-the CHB group (65 males and 35 females). CHB was diagnosed based on HBV-Ag seropositivity and continuously elevated ALT. Their ages ranged from 40-57 years with a median age of 48. The degree of inflammation of the CHB patients was between G1 and G2 and the fibrosis staging was between S0 and S2. (2) Group II-the HBV-LC group (81 males and 19 females). HBV-LC was diagnosed on the basis of pathology biopsy. Their ages ranged from 42-59 years with a median age of 51. The fibrosis staging of the HBV-LC patients was between S3 and S4. (3) Group III-the HBV-HCC group (57 males and 43 females). HBV-HCC was diagnosed by the elevation of AFP (>400ng/mL) combined with at least one positive iconography examination results, including CT and MRI. Their ages ranged from 37-65 years with a median age of 50. Twenty-three patients (23%) had tumor size less than 3 cm while sixteen patients (16%) had tumor size greater than 10 cm. The rest (61%) had tumor size between 3 and 10 cm. The fibrosis staging of the HBV-LC and HBV-HCC patients was between S3 and S4. All had Child-Pugh class A liver function. In addition, the patients with CHB, HBV-LC, and HBV-HCC were HBV-Ag positive in the serum and had not undergone anti-viral treatment.
DNA purification
The genomic DNA of the PBMCs was extracted with a D70K PureGene DNA isolation kit (Qiagen/Gentra, Dusseldorf, Germany) according to the manufacturer's protocols. The genomic DNA was eluted in a final volume of 50 μL sterile water and was stored at −20°C until use.
Sequences homology analysis
We performed pairwise sequence alignment analyses between the HBV and OR4F3 sequences using the basic local alignment search tool (BLAST, http://www.ncbi.nlm.nih.gov/).
Construction of standard curves for qPCR
The partial OR4F3 and OR4F17 gene sequence were amplified by PCR. Amplicon sizes were verified by agarose gel electrophoresis and compared to a 2000-bp DNA ladder (Invitrogen, Burlington, Ontario, Canada). PCR fragments were cloned using the pMDTM 18-T Vector Cloning kit, according to the manufacturer's manual (Takara). Plasmids were extracted using the DirectPrep 96 miniprep plasmid extraction kit (Qiagen, Mississauga, Ontario, Canada), and cloned fragments were verified by PCR and agarose gel electrophoresis. The plasmid copy number was quantified by spectrophotometry using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies), and calculation of the gene copy numbers was performed according to the molar mass derived from the plasmid and amplicon sequences. A ten-fold dilution series over five points was prepared from 106 copies/ul. The standard curves were constructed by plotting Cp values against the logarithmic concentration of the starting OR4F3 and OR4F17 with different concentrations. The amount of an unknown sample was quantified by interpolating the Cp values in the standard curves.
Quantitative PCR
We used quantitative PCR (qPCR) to detect the copy number gains and losses in the genomic material to validate the representative CGH array results. qPCR was conducted in duplicate on a LightCycler 480II (Roche, Basel, Switzerland). Each 20 μL reaction consisted of 10 μL SYBR Premix DimerEraserTM (Takara, Dalian, China), 250 nmol of each primer, 5 ng DNA sample, and 9 μL RNase-free water (Takara, Dalian, China). qPCR was conducted at 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 72°C for 10 s. Melting curve analysis was performed to confirm the specificity of the PCR products.
T cell, B cell, NK cell and monocyte purification with flow cytometry sorting
PBMCs were stained with CD3-APC, CD19-APC, CD56-PE, and CD14-APC antibodies (eBioscience, State of California, USA) at 4°C for 30 minutes [28]. Isotype controls with irrelevant specificities were included as negative controls. Human TruStain FcX™ (Fc Receptor Blocking Solution, Biolegend, San Diego, California, USA) was used to block Fc-receptors. Cells were washed three times with sterilized phosphate-buffered saline (PBS). CD3+ T cells, CD19+ B cells, CD3−CD56+ NK cells, and CD14+ monocytes were gated and isolated with a MoFlo XDP high-speed flow cytometry sorter (Beckman, Brea, California, USA). The purified cells were collected for CNV analysis of the OR gene.
Statistical analysis
The distribution of the normalized OR4F3 and OR4F17 genes was characterized by their mean values and standard deviations. One-way analysis of variance (ANOVA) was used to compare the log2-transformed Cp values of the candidate reference genes among the groups. Student t-tests were used for the qPCR data to assess differences in CNV of the OR genes and the HBV-related samples. P-values < 0.05 were considered significant. Using the Statistical Package for Social Sciences (SPSS), the data were summarized in ROC curves to visualize the efficacy of OR4F3 and OR4F17 as biomarkers for HBV-related diseases. In these curves, the sensitivity (true positives) was plotted on the Y-axis, and the specificity (false positives) on the X-axis. All of the observed values were considered as arbitrary cutoff values. The AUC values were calculated as a measure for the discriminative efficacy of the tested biomarkers.
SUPPLEMENTARY DATA FIGURES AND TABLE
Acknowledgments
We would like to thank Anfei Liu for critical reading and Zixin Liu for collecting samples.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by the China National Funds for Distinguished Young Scientists (No: 81425019, 81125018); Chang Jiang Scholars Program (2012) of the China Ministry of Education; The grants of the Science Fund for Creative Research Groups (No: 81221061); The National Key Basic Research Program “973 project” (No: 2015CB554000); The China Natural Science Foundation Program (No: 81301876, 81302116); The New Excellent Young Talents of Shanghai Municipal Health Bureau (No: XQY 2013112); The Excellent Young Scholar of SMMU; Shanghai Science and Technology Committee Program (No. 12ZR1439600, 13ZR1409200); The New Excellent Talents Program of Shanghai Municipal Health Bureau (No: XBR2011025); Shanghai Science and Technology Committee (No: 134119a0200); SMMU Innovation Alliance for Liver Cancer Diagnosis and Treatment (2012).
REFERENCES
- 1.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. Journal of clinical oncology. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z. Genomic landscape of liver cancer. Nature genetics. 2012;44:1075–1077. doi: 10.1038/ng.2412. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. The New England journal of medicine. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 5.Ryu SH, Jang MK, Kim WJ, Lee D, Chung YH. Metastatic tumor antigen in hepatocellular carcinoma: golden roads toward personalized medicine. Cancer metastasis reviews. 2014;33:965–980. doi: 10.1007/s10555-014-9522-4. [DOI] [PubMed] [Google Scholar]
- 6.Tse EY, Ching YP. The role of p21-activated kinases in hepatocellular carcinoma metastasis. Journal of molecular signaling. 2014;9:7. doi: 10.1186/1750-2187-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S, Diao J, Yeung S, Jubb A, Ye W, Wu TD, Kapadia SB, et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome research. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Williams V, Filippova M, Filippov V, Duerksen-Hughes P. Viral carcinogenesis: factors inducing DNA damage and virus integration. Cancers. 2014;6:2155–2186. doi: 10.3390/cancers6042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamori A, Yamanishi Y, Kawashima S, Kanehisa M, Enomoto M, Tanaka H, Kubo S, Shiomi S, Nishiguchi S. Alteration of gene expression in human hepatocellular carcinoma with integrated hepatitis B virus DNA. Clinical cancer research. 2005;11:5821–5826. doi: 10.1158/1078-0432.CCR-04-2055. [DOI] [PubMed] [Google Scholar]
- 10.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nature genetics. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 11.Sanz G, Leray I, Dewaele A, Sobilo J, Lerondel S, Bouet S, Grebert D, Monnerie R, Pajot-Augy E, Mir LM. Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PloS one. 2014;9:e85110. doi: 10.1371/journal.pone.0085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiberg IL, Pallett LJ, Winther TN, Hogh B, Maini MK, Peppa D. Defective Natural Killer cell antiviral capacity in paediatric HBV infection. Clin Exp Immunol. 2015;179:466–476. doi: 10.1111/cei.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader J, Iredale JP. The inflammatory microenvironment of HCC - the plot becomes complex. Journal of hepatology. 2011;54:853–855. doi: 10.1016/j.jhep.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 14.Ayub A, Ashfaq UA, Haque A. HBV induced HCC: major risk factors from genetic to molecular level. BioMed research international. 2013;2013:810461. doi: 10.1155/2013/810461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hai H, Tamori A, Kawada N. Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World journal of gastroenterology. 2014;20:6236–6243. doi: 10.3748/wjg.v20.i20.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durmus O, Tekin L, Carli AB, Cakar E, Acar A, Ulcay A, Dincer U, Kiralp MZ. Hepatitis B virus reactivation in a juvenile rheumatoid arthritis patient under treatment and its successful management: a complicated case. Rheumatology international. 2013;33:1345–1349. doi: 10.1007/s00296-011-2244-9. [DOI] [PubMed] [Google Scholar]
- 17.Bourne EJ, Dienstag JL, Lopez VA, Sander TJ, Longlet JM, Hall JG, Kwiatkowski RW, Wright T, Lai CL, Condreay LD. Quantitative analysis of HBV cccDNA from clinical specimens: correlation with clinical and virological response during antiviral therapy. Journal of viral hepatitis. 2007;14:55–63. doi: 10.1111/j.1365-2893.2006.00775.x. [DOI] [PubMed] [Google Scholar]
- 18.Bollig-Fischer A, Michelhaugh SK, Wijesinghe P, Dyson G, Kruger A, Palanisamy N, Choi L, Alosh B, Ali-Fehmi R, Mittal S. Cytogenomic profiling of breast cancer brain metastases reveals potential for repurposing targeted therapeutics. Oncotarget. 2015;6:14614–14624. doi: 10.18632/oncotarget.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, Gao H, Hao K, Willard MD, Xu J, Hauptschein R, Rejto PA, Fernandez J, Wang G, Zhang Q, Wang B, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome research. 2013;23:1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao T, Li X, Yin W, Cai X, Zhang W, Chen F, Lai G, Huang A. Establishment of an HBV chronic hepatitis B infection mouse model by vivo transduction of HBV cccDNA [Article in Chinese] Zhonghua Gan Zang Bing Za Zhi. 2014;22:260–265. doi: 10.3760/cma.j.issn.1007-3418.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Lupberger J, Hildt E. Hepatitis B virus-induced oncogenesis. World journal of gastroenterology. 2007;13:74–81. doi: 10.3748/wjg.v13.i1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu JW, Yang WY, Tsai SM, Lin YM, Chang PH, Chen JR, Wang HD, Wu JL, Jin SL, Yuh CH. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PloS one. 2013;8:e76951. doi: 10.1371/journal.pone.0076951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SG, Rho HM. Transcriptional repression of the human p53 gene by hepatitis B viral X protein. Oncogene. 2000;19:468–471. doi: 10.1038/sj.onc.1203312. [DOI] [PubMed] [Google Scholar]
- 25.Tang S, Yuan Y, He Y, Pan D, Zhang Y, Liu Y, Liu Q, Zhang Z, Liu Z. Genetic polymorphism of interleukin-6 influences susceptibility to HBV-related hepatocellular carcinoma in a male Chinese Han population. Human immunology. 2014;75:297–301. doi: 10.1016/j.humimm.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Tarocchi M, Polvani S, Marroncini G, Galli A. Molecular mechanism of hepatitis B virus-induced hepatocarcinogenesis. World journal of gastroenterology. 2014;20:11630–11640. doi: 10.3748/wjg.v20.i33.11630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weng J, Wang J, Hu X, Wang F, Ittmann M, Liu M. PSGR2, a novel G-protein coupled receptor, is overexpressed in human prostate cancer. International journal of cancer. 2006;118:1471–1480. doi: 10.1002/ijc.21527. [DOI] [PubMed] [Google Scholar]
- 28.Levi I, Amsalem H, Nissan A, Darash-Yahana M, Peretz T, Mandelboim O, Rachmilewitz J. Characterization of tumor infiltrating natural killer cell subset. Oncotarget. 2015;6:13835–13843. doi: 10.18632/oncotarget.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.