Abstract
Mutation of oncogene KRAS is common in non-small cell lung cancer (NSCLC), however, its clinical significance is still controversial. Independent studies evaluating its prognostic and predictive value usually drew inconsistent conclusions. Hence, We performed a meta-analysis with 41 relative publications, retrieved from multi-databases, to reconcile these controversial results and to give an overall impression of KRAS mutation in NSCLC. According to our findings, KRAS mutation was significantly associated with worse overall survival (OS) and disease-free survival (DFS) in early stage resected NSCLC (hazard ratio or HR=1.56 and 1.57, 95% CI 1.39-1.76 and 1.17-2.09 respectively), and with inferior outcomes of epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) treatment and chemotherapy (relative risk or RR=0.21 and 0.66 for objective response rate or ORR, 95% CI 0.12-0.39 and 0.54-0.81 respectively; HR=1.46 and 1.30 for progression-free survival or PFS, 95%CI 1.23-1.74 and 1.14-1.50 respectively) in advanced NSCLC. When EGFR mutant patients were excluded, KRAS mutation was still significantly associated with worse OS and PFS of EGFR-TKIs (HR=1.40 and 1.35, 95 % CI 1.21-1.61 and 1.11-1.64). Although KRAS mutant patients presented worse DFS and PFS of chemotherapy (HR=1.33 and 1.11, 95% CI 0.97-1.84 and 0.95-1.30), and lower response rate to EGFR-TKIs or chemotherapy (RR=0.55 and 0.88, 95 % CI 0.27-1.11 and 0.76-1.02), statistical differences were not met. In conclusion, KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes in NSCLC. There's a need for developing target therapies for KRAS mutant lung cancer and other tumors.
Keywords: non-small cell lung cancer, KRAS mutation, prognosis, EGFR-TKI, meta-analysis
INTRODUCTION
Lung cancer, with NSCLC accounts for 85% of all cases, is the most common human malignant disease and the leading cause of cancer-related mortality worldwide [1, 2]. Early as the beginning of this century, novel molecular targeted agents like EGFR-TKIs represented by gefitinib or erlotinib, which interfere with EGFR signaling, have been proved dramatically effective for selected advanced NSCLC patients with sensitive EGFR mutations [3]. Since then, molecular target therapies provided promising treatment alternatives to surgery, radiation therapy and chemotherapy. Personalized, genotype-directed therapy for NSCLC couldn't be more popular. Besides EGFR, KRAS is the most frequently mutated oncogene in NSCLC (15-20%) with most cases affect exon 2 and 3 (G12, G13 and Q61). It seemed that KRAS mutation occurs more frequently in lung adenocarcinomas (approximately 30%), in the Caucasian population, and in the population with smoking history [4–6].
KRAS mutation was described as a negative prognostic marker for OS and DFS in lung adenocarcinoma more early in 1990 [7]. Not until the last ten years, clinical significance of KRAS mutation in NSCLC has been attracted more and more attention. Although a lot of published studies reported that KRAS mutation is associated with poor prognosis and outcomes of EGFR-TKIs treatment [8–11] and chemotherapy [10, 12–15], more than a few independent studies argued that it predicts neither worse prognosis [8, 10–12, 14, 16–28] nor inferior outcomes of EGFR-TKIs treatment or chemotherapy [14, 18, 19, 29–32]. Therefore, we carried out a comprehensively search and review of relevant publications in multi-database. Useful data was extracted and then aggregated by using a meta-analysis methodology to give an overall impression of KRAS mutation in NSCLC.
Moreover, it is accepted that sensitive EGFR mutation predicts benefit from EGFR-TKIs treatment and even from chemotherapy in NSCLC [8, 9, 18, 19, 26, 32-34]. Mutations of KRAS and EGFR are common and mutually exclusive in NSCLC [35–37]. Thus EGFR mutation predominantly coexists with wild type KRAS, which made us overestimate the prognostic and predictive value of KRAS mutation. Therefore, analyses were re-performed in EGFR wild-type NSCLC to obtain objective and unassertive conclusions.
RESULTS
Study characteristics and quality assessment
Based on our search criteria, a total of 41 studies, which enrolled 13,103 KRAS assessable patients with 18 percent (2,374) KRAS mutant positive cases, were eligible for inclusion in the present analyses. The process of selecting publications was presented in Figure 1 and the clinical characteristics of the included studies were listed in Table 1. All of the studies were published from 2005-2015, consisting of 40 cohort studies [8–31, 34, 35, 38-51] and one randomized controlled trial (RCT) [32]. Thirty studies [8, 9, 11–19, 21–28, 31, 35, 38, 40–43, 46–48, 51] conducted in Europe and North America, ten studies [10, 20, 29, 30, 32, 34, 39, 44, 45, 50] in Asia, and one study [49] in Latin America. All of the studies focused on NSCLC or lung adenocarcinoma only except one [46] on lung squamous cell carcinoma. Ten studies [16, 20, 24, 25, 29, 30, 39, 43, 47, 48] dealt with stage I-IIIa resected tumors, twenty-nine studies [8, 9, 11–15, 17–19, 21–23, 26–28, 31, 34, 35, 38, 40–42, 44–46, 49–51] with stage IIIb-IV unresectable tumors, and two studies [10, 32] with all stage tumors. Thirteen studies [10, 12, 13, 18, 29, 30, 34, 35, 41, 44, 46, 47, 49] used a polymerase chain reaction (PCR) or modified PCR method to test gene mutation, while the others used a direct sequencing method. Four studies [12, 13, 38, 45] assessed KRAS mutation in plasma DNA and the others in tumor specimens. In consistent with large-scaled demographic results [6], the majority of KRAS mutation occurs in codon 12, with G12C the most, occasionally in codon 13, and rarely in codon 61. All the studies selected patients randomly without concerning gender or smoking status and most results were adjusted for gender, age, stage and Karnofsky performance score.
Figure 1. Flow Chart of publication search and selection.
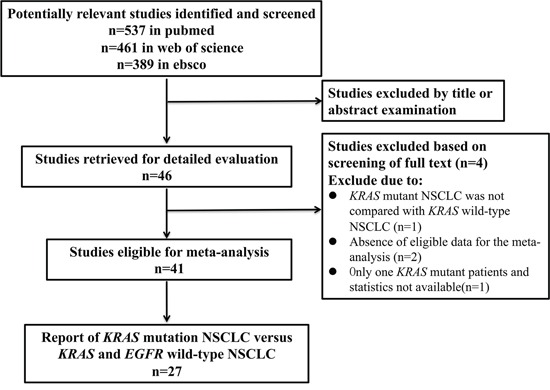
Table 1. Clinical characteristics of included studies.
| First Author | Year | Race | Patients Number | KRAS MUT Number(%) | Gene Testing Method | Specimens Assessed | Pathology | Stage | Treatments | Outcome | KRAS MUT Versus | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| William [35] | 2005 | Caucasian | 60 | 9 (15.0) | PCR | Tumor | ADC | IIIB-IV | TKI | ORR | WT/WT | 6 |
| David [8] | 2005 | Caucasian | 274 | 55 (20.0) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI / CT | OS / ORR / PFS | WT/WT | 8 |
| Erminia [9] | 2007 | Caucasian | 70 | 16 (22.9) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | ORR / PFS | WT/WT | 8 |
| Oliver [38] | 2007 | Caucasian | 175 | 16 (9.1) | Sequencing | Plasma | NSCLC | I-IV | Various | OS | WT | 6 |
| Young [29] | 2008 | Asian | 71 | 5 (7.0) | PCR | Tumor | ADC | I-III | R | OS / DFS | WT | 6 |
| Jenifer [16] | 2008 | Caucasian | 296 | 50 (16.9) | Sequencing | Tumor | ADC | I-III | R | OS | WT/WT | 9 |
| Chang-qi [17] | 2008 | Caucasian | 206 | 30 (14.6) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / ORR | NA | 9 |
| Takayuki [39] | 2009 | Asian | 254 | 32 (12.6) | Sequencing | Tumor | ADC | I-III | R | OS | WT/WT | 9 |
| MILOS [18] | 2009 | Caucasian | 208 | 32 (15.4) | PCR | Tumor | NSCLC | IIIB-IV | TKI | OS / PFS | WT/WT | 7 |
| David [19] | 2009 | Caucasian | 175 | 41 (23.4) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / ORR / PFS | WT/WT | 8 |
| Tetsukan [30] | 2009 | Asian | 168 | 24 (14.3) | PCR | Tumor | ADC | I | R | DFS | WT | 9 |
| Antonio [40] | 2009 | Caucasian | 83 | 16 (19.3) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / ORR / PFS | WT/WT | 7 |
| Hui-ping [20] | 2010 | Asian | 156 | 7 (4.5) | Sequencing | Tumor | NSCLC | I-III | R | OS | WT/WT | 7 |
| Laura [41] | 2010 | Caucasian | 62 | 12 (19.4) | PCR | Tumor | ADC | IIIB-IV | TKI | OS / PFS | WT | 8 |
| Vienna [21] | 2011 | Caucasian | 161 | 11 (6.8) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / ORR / PFS | WT/WT | 8 |
| Carlos [12] | 2011 | Caucasian | 308 | 27 (8.8) | PCR | Plasma | NSCLC | IIIB-IV | CT | OS / PFS | WT | 8 |
| Hye [10] | 2011 | Asian | 229 | 19 (8.3) | PCR | Tumor | NSCLC | I-IV | R / CT / TKI | OS / DFS / ORR / PFS | WT/WT | 6 |
| Wolfram [22] | 2011 | Caucasian | 493 | 90 (18.3) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / PFS | WT/WT | 9 |
| Vienna [42] | 2012 | Caucasian | 162 | 11 (6.8) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / ORR / PFS | WT/WT | 9 |
| Chiara [43] | 2012 | Caucasian | 249 | 46 | Sequencing | Tumor | NSCLC | I-III | R | DFS | WT/WT | 9 |
| Melissa [23] | 2012 | Caucasian | 1036 | 241 (23.3) | Sequencing | Tumor | ADC | IV | CT / TKI | OS | WT/WT | 9 |
| Jie [44] | 2012 | Asian | 104 | 9 (8.7) | PCR | Tumor | NSCLC | IIIB-IV | Various | OS | WT | 6 |
| Giulio [11] | 2012 | Caucasian | 67 | 18 (26.9) | Sequencing | Tumor | ADC | IIIB-IV | TKI | OS / ORR / PFS | WT/WT | 7 |
| Jacques [31] | 2012 | Caucasian | 307 | 42 (13.7) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS / PFS | WT/WT | 7 |
| Seung [45] | 2013 | Asian | 57 | 14 (24.6) | Sequencing | Plasma | NSCLC | IIIB-IV | TKI / BSC | OS / ORR | WT | 8 |
| Anneli [13] | 2013 | Caucasian | 246 | 43 (17.5) | PCR | Plasma | NSCLC | IIIB-IV | CT | OS / ORR / PFS | WT | 8 |
| Ondrej [46] | 2013 | Caucasian | 215 | 16 (7.4) | PCR | Tumor | SCC | IIIB-IV | TKI | OS / PFS | WT | 7 |
| Ji-lin [32] | 2013 | Asian | 1935 | 98 (5.1) | Sequencing | Tumor | ADC | I-IV | R / TKI / CT | OS / DFS / ORR / PFS | WT/WT | 3* |
| Jong-Mu [34] | 2013 | Asian | 484 | 39 (8.1) | PCR | Tumor | ADC | IIIB-IV | TKI / CT | OS / ORR / PFS | WT/WT | 9 |
| Frances [25] | 2013 | Caucasian | 1543 | 300 (19.4) | Sequencing | Tumor | NSCLC | I-III | R | OS / DFS | WT | 9 |
| Gerald [26] | 2013 | Caucasian | 368 | 110 (29.9) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI | OS | WT/WT | 6 |
| Wouter [14] | 2013 | Caucasian | 161 | 60 (37.3) | Sequencing | Tumor | ADC | IIIB-IV | CT | OS / ORR / PFS | WT | 8 |
| Giulio [15] | 2014 | Caucasian | 204 | 77 (37.7) | Sequencing | Tumor | ADC | IIIB-IV | CT | OS / ORR / PFS | WT/WT | 9 |
| Marianna [27] | 2014 | Caucasian | 108 | 39 (36.1) | Sequencing | Tumor | NSCLC | IIIB-IV | CT | OS / ORR / PFS | WT/WT | 9 |
| Mihaly [28] | 2014 | Caucasian | 1125 | 361 (32.1) | Sequencing | Tumor | ADC | IIIB-IV | CT | OS / ORR / PFS | WT/WT | 9 |
| Mark [24] | 2014 | Caucasian | 230 | 39 (17.0) | Sequencing | Tumor | ADC | I-III | R | OS / DFS | WT/WT | 8 |
| Benjamin [47] | 2014 | Caucasian | 312 | 127 (40.7) | PCR | Tumor | ADC | I | R | OS / DFS | WT/WT | 8 |
| Ernest [48] | 2015 | Cacasian | 179 | 85 (47.5) | Sequencing | Tumor | ADC | I-III | R | OS / DFS | WT | 8 |
| Alma [49] | 2015 | Other | 225 | 40 (17.8) | PCR | Tumor | NSCLC | IIIB-IV | TKI / CT | OS / ORR / PFS | WT | 8 |
| Shigehiro [50] | 2015 | Asian | 119 | 16 (13.4) | Sequencing | Tumor | ADC | IIIB-IV | CT | OS / ORR / PFS | WT/WT | 8 |
| Eliana [51] | 2015 | Caucasian | 218 | 51 (23.4) | Sequencing | Tumor | NSCLC | IIIB-IV | TKI / CT | OS / ORR / PFS | WT/WT | 8 |
MUT, mutation; NSCLC, non-small cell lung cancer; ADC lung adenocarcinoma; SCC, lung squamous cell carcinoma; CT, chemotherapy; R, surgical resection; WT, KRAS wild-type; WT/WT, KRAS and EGFR wild-type.
randomized controlled trial was evaluated based on Jadad Scale.
The quality of cohort study was assessed using the Newcastle-Ottawa Scale (NOS) on three perspectives: patient selection, comparability of groups, and assessment of outcome. Full score is nine stars, and a study with more stars was considered to be of higher quality. Quality scores of 40 cohort studies ranged from six to nine with a median score of eight. The quality of RCT was assessed using the Jadad Scale on three perspectives: randomization, double blinding, withdraws and dropouts. Full score is five points, and a study with score no less than three points is defined the high-quality study. The only included RCT gained a score of three points. No “poor quality” study was found and all of the studies were considered acceptable for inclusion in the present meta-analysis. The study specific scores were summarized in Table S1.
KRAS mutation and clinical features
Data of clinical features stratified by KRAS mutational status was reported in 25 studies [9, 10, 12–17, 21, 23, 24, 26–28, 30, 32, 34, 39, 41, 44–48, 50]. Data was extracted from individual studies and then aggregated. The result indicated that KRAS mutation occurs more frequently in lung adenocarcinoma (RR=1.16 p=0.016), and in former or current smokers (RR=1.13 p=0.017), but not in male gender (RR=1.07 p=0.142) (Table S2). Reported gene mutation rate ranged from 4.4% to 24.5% in the Asians and from 6.7% to 47.4% in the Caucasians. Additionally, an increased incidence of presence of stage IV disease and distant metastasis in KRAS mutant patients was reported in several studies [27, 50].
Prognostic and predictive value of KRAS mutation in unselected NSCLC
Thirty-seven studies [8, 10–29, 31, 32, 34, 38–42, 44–51] provided HRs for OS comparing KRAS mutant NSCLC with KRAS wild-type NSCLC. Pooled HR was 1.56 for OS (95%CI 1.39-1.76, p=0.00) (Figure 2A), indicating a significantly worse survival for KRAS mutant patients. Significant heterogeneity among studies (I2 =54.6%, p=0.00) and publication bias (Begg's test p=0.053, Egger's test p=0.014) (Figure 2B) was detected. Meta-regression analysis showed that only race (adjusted R2=77.12%, p=0.00) might contribute to the heterogeneity, but not other factors such as disease stage (p=0.885), pathology (p=0.454), gene mutation testing method (p=0.029) and specimens (plasma/tumor foci) for mutation assessment (p=0.560). As shown in Figure 2A, subgroup analysis according to race showed that KRAS mutation is a more powerful negative prognostic factor for OS in the Asians (HR=2.39 with 95% CI 1.97-2.90 and p=0.00, I2=0.0% and p=0.648 for heterogeneity) than in the Caucasians (HR=1.37 with 95%CI 1.24-1.51 and p=0.00, I2=30.5 and p=0.066 for heterogeneity).
Figure 2.

Forrest plot A, D. with influence analysis C, F. of hazard ratio for overall survival and disease-free-survival comparing KRAS mutant patients with KRAS wild-type patients. Begg's funnel plot of enrolled studies for estimating the hazard ratio for overall survival B. and disease-free-survival E.
Nine studies [10, 24, 25, 29, 30, 32, 43, 47, 48] dealt with stage I-IIIa resected NSCLC and provided HRs for DFS comparing KRAS mutant tumors with KRAS wild-type tumors. All cases received R0 resection and lobectomies were performed mostly. Pooled HR was 1.57 for DFS (95% CI 1.17-2.09, p=0.002) (Figure 2D), indicating an increased hazard for disease recurrence after tumor resection for KRAS mutant patients. Neither significant heterogeneity (I2 =47.6%, p=0.054) nor publication bias (Begg's test p=0.754, Egger's test p=0.062) (Figure 2E) was detected. However, meta-regression analysis showed that significant heterogeneity did exist between two races (adjusted R2=−85.65%, p=0.042). Similarly, subgroup analysis according to race showed that KRAS mutation is a more powerful negative prognostic factor for DFS in the Asians (HR=2.59, 95% CI 1.55-4.30 and p=0.00, I2=0.0% and p=0.847 for heterogeneity) than in the Caucasians (HR=1.31, 95% CI 0.99-1.73 and p=0.057, I2=42.7% and p=0.137 for heterogeneity) (Figure 2D).
Eighteen studies [9–11, 17–19, 21, 22, 31, 32, 34, 35, 40–42, 46, 49, 51] investigated outcomes (response rate or PFS) of EGFR-TKIs treatment in stage IIIb-IV unresectable NSCLC comparing KRAS mutant tumors with KRAS wild-type tumors. Either gefitinib or erlotinib was administered in standard dosage as first to three-line treatment. The total ORR (complete response or CR + partial response or PR) was 2.5% (6/237) in KRAS mutant patients and 34.0% (499/1469) in KRAS wild-type patients. Pooled RR was 0.21 for ORR (95% CI 0.12-0.39, p=0.00) (Figure 3A) while pooled HR was 1.46 for PFS (95% CI 1.23-1.74, p=0.0) (Figure 3D), indicating a significant lower response rate and shorter remission period of EGFR-TKIs treatment for KRAS mutant patients. Neither significant heterogeneity (I2=0.0%, p=0.876 and I2 =44.3%, p=0.033 respectively) nor publication bias (Begg's test p=0.502, Egger's test p=0.086 and Begg's test p=0.06, Egger's test p=0.053 respectively) (Figure 3B and 3E) was detected. Meta-regression analysis showed that neither race (p=0.440) nor gene mutation testing method (p=0.807) contributes significantly to the heterogeneity.
Figure 3.

Forrest plot of relative ratio for objective response rate A. and hazard ratio for progression-free-survival D. with influence analysis C, F. comparing KRAS mutant patients with KRAS wild-type patients treated with EGFR TKIs. Begg's funnel plot of enrolled studies for estimating the relative ration for overall response B. and hazard ratio for progression-free-survival E.
Thirteen studies [8, 10, 12–15, 27, 32, 34, 41, 49–51] investigated outcomes of chemotherapy in stage IIIb-IV unresectable NSCLC comparing KRAS mutant tumors with KRAS wild-type tumors. Platinum-based doublet was used for first to second-line treatment. The total ORR was 21.1% (82/389) in KRAS mutant patients and 32.9% (486/1477) in KRAS wild-type patients. Pooled RR was 0.66 for ORR (95% CI 0.54-0.81, p=0.00) (Figure 4A) while pooled HR was 1.30 for PFS (95% CI 1.14-1.50, p=0.0) (Figure 4D), indicating a significant lower response and shorter remission period of chemotherapy for KRAS mutant patients. Neither significant heterogeneity (I2=0.0%, p=0.949 and I2 =23.8%, p=0.203 respectively) nor publication bias (Begg's test p=0.755, Egger's test p=0.506 and Begg's test p=0.583, Egger's test p=0.419 respectively) (Figure 4B and 4E) was detected. Meta-regression analysis showed that neither race (p=0.736) nor gene mutation testing method (p=0.389) contributes significantly to the heterogeneity.
Figure 4.
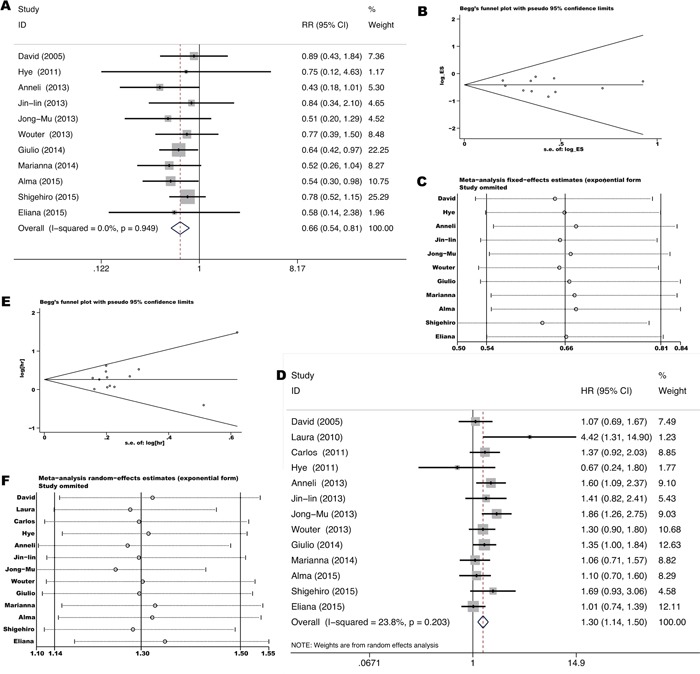
Forrest plot of relative ratio for objective response rate A. and hazard ratio for progression-free-survival D. with influence analysis C, F. comparing KRAS mutant patients with KRAS wild-type patients treated with chemotherapy. Begg's funnel plot of enrolled studies for estimating the relative ration for overall response B. and hazard ratio for progression-free-survival E.
Prognostic and predictive value of KRAS mutation in EGFR wild-type NSCLC
Additionally, twenty-seven studies [8–11, 15, 16, 18–24, 26–28, 31, 32, 34, 35, 39, 40, 42, 43, 47, 50, 51] including 9,383 both KRAS and EGFR assessable patients investigated the prognostic and predictive value of KRAS mutation in EGFR wild-type NSCLC. Although mutations of KRAS and EGFR were mutually exclusive in most cases [35–37], presence of both gene mutations could be seen occasionally [18, 34, 47].
As shown in Figure 5A, pooled HR was 1.40 for OS (95% CI 1.21-1.61, p=0.0) based on 21 studies [10, 11, 15, 16, 18–20, 22–24, 26–28, 31, 32, 34, 39, 42, 47, 50, 51] comparing KRAS mutant NSCLC with KRAS and EGFR wild-type NSCLC, indicating a significant worse survival for KRAS mutant patients. Significant heterogeneity among studies (I2 =57.3%, p=0.0) but not publication bias (Begg's test p=0.866, Egger's test p=0.486) (Figure S1A) was detected. Similarly, meta-regression analysis showed that only races (adjusted R2=95.14%, p=0.0) might contribute to the heterogeneity. Subgroup analysis according to races showed that KRAS mutation impairs survival more seriously in the Asians (HR=2.30 with 95% CI 1.84-2.88 and p=0.0, I2=6.1% and p=0.381 for heterogeneity) than in the Caucasians (HR=1.22 with 95% CI 1.11-1.33 and p=0.00, I2=0.0% and p=0.653 for heterogeneity) (Figure 5A).
Figure 5.

Forrest plot of hazard ratio for overall survival A. and disease-free-survival B. comparing KRAS mutant patients with KRAS and EGFR wild-type patients.
As shown in Figure 5B, pooled HR was 1.33 for DFS (95% CI 0.97-1.84, p=0.076) based on six studies [10, 24, 25, 32, 43, 47] conducted in stage I-IIIa resected NSCLC comparing KRAS mutant tumors with KRAS and EGFR wild-type tumors, exhibiting an insignificant trend towards increased hazard for disease recurrence after tumor resection for KRAS mutant patients. Neither significant heterogeneity (I2=36.0%, p=0.167) nor publication bias (Begg's test p=1.00, Egger's test p=0.334) (Figure S1B) was detected. Meta-regression analysis showed that neither race (p=0.242) nor gene mutation testing method (p=0.189) contributes significantly to the heterogeneity.
The total ORR to EGFR-TKIs was 2.3% (4/175) in KRAS mutant patients and 13.6% (101/740) in KRAS and EGFR wild-type patients based on 14 studies [9-11, 18, 19, 21, 22, 31, 32, 34, 35, 40, 42, 51] conducted in stage IIIb-IV unresectable NSCLC. As shown in Figure 6A and 6B, pooled RR was 0.55 for ORR (95% CI 0.27-1.11, p=0.095) while pooled HR was 1.35 for PFS (95% CI 1.11-1.64, p=0.002), exhibiting an insignificant trend towards lower response but significant shorter remission period of EGFR-TKIs treatment for KRAS mutant patients. Neither significant heterogeneity (I2=0.0%, p=0.996 and I2 =42.0%, p=0.069 respectively) nor publication bias (Begg's test p=1.00, Egger's test p=0.109 and Begg's test p=0.436, Egger's test p=0.256 respectively) (Figure S1C and S1D) was detected. Meta-regression analysis showed that neither race (p=0.159) nor gene mutation testing method (p=0.801) contributes significantly to the heterogeneity.
Figure 6.

Forrest plot of relative ratio for objective response rate A, C. and hazard ratio of progression-free-survival B, D. comparing KRAS mutant patients with KRAS and EGFR wild-type patients treated with EGFR TKIs and chemotherapy respectively.
The total ORR to chemotherapy was 35.8% (138/385) in KRAS mutant patients and 45.1% (381/845) in KRAS and EGFR wild-type patients based on eight studies [8, 10, 15, 27, 28, 32, 50, 51] conducted in stage IIIb-IV unresectable NSCLC. As shown in Figure 6C and 6D, pooled RR was 0.88 for ORR (95% CI 0.76-1.02, p=0.083) while pooled HR was 1.11 for PFS (95% CI 0.95-1.30, p=0.186), exhibiting an insignificant trend towards lower response and shorter remission period of chemotherapy for KRAS mutant patients. Neither significant heterogeneity (I2=1.6%, p=0.340 and I2 =18.0%, p=0.286 respectively) nor publication bias (Begg's test p=0.902, Egger's test p=0.3 and Begg's test p=0.764, Egger's test p=0.493 respectively) (Figure S1E and S1F) was detected. Meta-regression analysis showed that race (p=0.509) doesn't contribute significantly to the heterogeneity.
Sensitivity analyses
In general, no individual publication was found to be significantly biasing the results (Figure 2C, 2F, 3C, 3F, 4C, 4F and Figure S2A-D), but the associations between KRAS mutation with lower response rate and shorter remission period of chemotherapy in EGFR wild-type NSCLC were affected after the data set of Mihaly [28] was removed (Figure S2E, S2F). The associations shifted from statistically insignificant to significant with Mihaly et al.'s study excluded. However, this study enrolled the most patients assessed for outcomes of chemotherapy and gained a high quality score of nine stars, therefor it's unreasonable to role out this study for analyses. The sensitivity analyses showed that the cumulative results are stable.
DISCUSSION
The KRAS oncogene together with HRAS and NRAS encode a family of membrane-bound 21kd guanosine triphosphate binding proteins (GTPs) that regulate cell growth, differentiation, and apoptosis by interacting with multiple signaling including mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription (STAT), and phosphoinositide 3-kinase (PI3K) signaling cascades [52]. KRAS gene has been found frequently mutated in human tumors such as large intestine, lung and pancreas. Almost all KRAS-mutant cases affect exon 2 and 3 (G12, G13 and Q61), which impair the deactivation circuit of RAS proteins, thereby causing sustained activation of RAS signaling [53]. Meanwhile RAS is the most important downstream effector of EGFR, therefore sensitive mutation of KRAS gene might attenuate, even abolish the treatment efficacy of anti-EGFR agents such as EGFR-TKIs and EGFR monoclonal antibody (EGFR mAb). It is true that the benefit of cetuximab or panitumumab, two well-known EGFR mAb approved by FDA, is restricted to patients with KRAS wild-type colorectal cancer, and only this subset of patients should receive these agents [54]. Although KRAS is the most common mutated oncogene in NSCLC, its clinical significance is yet under debate. Should we test for it, and does it matter? Is KRAS testing necessary before EGFR-TKIs treatment?
The present meta-analysis with newest and largest quantity of relevant publications confirmed that KRAS mutation is significantly associated with worse OS (HR=1.56) and DFS (HR=1.57), and also with inferior ORR (RR=0.21 and 0.66 for TKI and chemotherapy respectively) and PFS (HR=1.46 and 1.30 respectively) of EGFR-TKIs treatment or chemotherapy, compared with KRAS wild-type NSCLC. While analyzing the association between KRAS mutation with OS, significant publication bias was detected by Egger's test (p=0.014). Thereby, a “trim and fill” method was applied. Elven hypothetical negative unpublished studies were imputed to produce a symmetrical funnel plot (Figure S3). The pooled analysis incorporating the hypothetical studies continued to show a statistically significant association between KRAS mutation and worse survival (HR=1.31, 95% CI 1.14-1.50 and p=0.00). Céline et al. [55] reported a significant worse survival (HR=1.35, 95% CI 1.16-1.56) of KRAS mutant NSCLC compared with KRAS wild-type NSCLC based on a meta-analysis of 28 studies early in 2004. The reported HR for OS was quit similar to ours, however, no significant survival hazard was observed in the subgroup analysis of nine studies using an immunohistochemistry (IHC) method to test RAS alternation (HR=1.08, 95% CI 0.86-1.34). Furthermore, none of the 28 studies used a direct sequencing method, which is a “gold standard” for gene testing and not spreading to clinical application until the last decade. On the contrary, none of the 41 studies included in the present meta-analysis used an IHC method. Instead, more than half of the included studies used a direct sequencing method. As more included studies, more enrolled cases and more developed gene testing method, our results are more reliable.
Resistance to EGFR-TKIs treatment for KRAS mutant NSCLC was also reported in other two meta-analysis conducted by Chen et al. [56] and Min et al. [57]. The reported pooled RR for ORR was 0.29 (95% CI 0.18-0.47) in Chen's study and 0.21 (95% CI 0.12-0.39) in ours while the reported pooled HR for PFS was 1.86 (95% CI 1.51-2.29) in Min's study and 1.46 (95% CI 1.23-1.74) in ours, showing highly consistent results among studies. Meanwhile, the present meta-analysis included more publications and presented more accurate confidence interval. Resistance to chemotherapy for KRAS mutant NSCLC was also reported by another meta-analysis [58]. The reported odds ratio (OR) was 0.67 (95% CI 0.50-0.88) for ORR with statistical significance and 0.75 (95% CI 0.54-1.04) for 6 month and 1-year PFS rate but without statistical significance. Only first-line chemotherapy was evaluated. We doubt that HR might be more suitable than OR for analyzing PFS, which displayed an abnormal distribution. Our results showed both significant inferior ORR (RR=0.67, 95% CI 0.50-0.88) and PFS (HR=1.30, 95% CI 1.14-1.50) for the KRAS mutant patients.
Additionally, we noticed that KRAS mutation impairs OS and DFS more obviously in the Asians (HR=2.39 and 2.59 respectively) than in the Caucasians (HR=1.37 and 1.31 respectively), which is not reported elsewhere. It is believed that KRAS mutation subtypes have diverse prognosis and respond differently to chemotherapy or EGFR-TKIs [15, 25, 28, 47, 48, 59]. The author speculated that different spectrum of KRAS mutation subtypes, especially increased proportion of G13, G12D and G12V in the Asians, might be partly responsible for the different hazard ratio between two races. Secondly, there were more KRAS wild-type cases than KRAS mutant cases enrolled in studies. This unbalanced situation was more obviously in studies conducted in Asia, which might exaggerate the HRs for OS and DFS in the Asians. More detailed mechanisms need to be exploited in future fundamental research focused on divergence of RAS signal transduction between two races.
Besides KRAS, oncogene EGFR is also frequently mutated in NSCLC, which predicts dramatic benefits from EGFR-TKIs treatment [3, 8, 17–20, 23, 31], and even from chemotherapy [32, 49]. Mutations of KRAS and EGFR are generally mutually exclusive in NSCLC, i.e. most EGFR mutations were existed in KRAS wild-type patients, which might bias the results toward an overestimation of the prognostic and predictive value of KAS mutation. Thus, we carried out further analyses in EGFR wild-type NSCLC to draw a more objective conclusion of clinical significance of KRAS mutation. While compared with KRAS and EGFR wild-type NSCLC, the prognostic and predictive value of KRAS mutation did decreased. Pooled HR decreased from 1.56 and 1.57 to 1.40 and 1.33 for OS and DFS respectively, yet statistically significant for OS (p=0.0) but not for DFS (p=0.076). Similarly, KRAS mutation impaired OS and DFS (without statistical significance, data not shown) more seriously in the Asians. Pooled RR for ORR increased from 0.21 and 0.66 to 0.55 and 0.88 for EGFR-TKIs treatment and chemotherapy respectively. No statistical significances were observed (p=0.095 and 0.813 respectively). Pooled HR for PFS decreased from 1.46 and 1.30 to 1.35 and 1.11 for EGFR-TKIs treatment and chemotherapy respectively. Statistical significance was observed in EGFR-TKIs treatment (p=0.002), but not in chemotherapy (p=0.186). Although associations of KRAS mutation with inferior treatment outcomes turned out to be statistically insignificant, the results seemed unstable. Sensitivity analyses showed that the associations of KRAS mutation with inferior chemotherapy outcomes were significantly affected after Mihaly et al.'s study was removed. It is noteworthy that there were fewer studies evaluating the associations of KRAS mutation with treatment outcomes in EGFR wild-type NSCLC. Besides, obvious trends towards inferior treatment outcomes and borderline confidence intervals were observed, the author speculated that KRAS mutation is still a valid predictor for poor treatment outcomes in EGFR wild-type NSCLC with more publications to be included. However, its prognostic and predictive value is not so remarkable as it was greatly affected by exclusion of EGFR mutant patients and the HRs for OS, DFS and PFS were no more than two fold. Actually only NSCLC patients with sensitive EGFR mutation are recommend to first line EGFR-TKIs treatment according to NCCN Guidelines. Based on the notion that mutations of EGFR and KRAS are generally mutually exclusive, a very few KRAS mutant patients are subjected to EGFR-TKIs treatment. Therefore KRAS testing is of limited value to optimize the use of EGFR-TKIs in clinic compared to EGFR testing.
Despite our efforts in performing a comprehensive and accurate analysis, yet several limitations should be taken into consideration when interpreting the findings. Firstly, fewer studies assed the predictive and prognostic value of KRAS mutation in EGFR wild-type NSCLC. Thus borderline significant associations of KRAS mutation with inferior treatment outcomes were observed. Secondly, the present study is a univariate analysis. Although several factors such as race, stage, gene testing method and EGFR mutational status were taken into consideration, other factors such as KRAS mutation subtypes, other gene mutational status as ALK rearrangement [10, 15] and PIK3CA mutation [21, 42, 46, 60], performance status and smoking status should not be neglected in the analysis with more available data provided in the future studies. Lastly, it is noteworthy that KRAS mutation, and even subtype-specific KRAS mutations, responds differently to different chemotherapeutics [34, 61]. Therefore, associations between subtype-specific KRAS mutations and responses to specific chemotherapeutics should be strictly exploited in future studies.
In conclusion, KRAS mutation is a weak, but valid predictor for poor prognosis and treatment outcomes for surgical resection, EGFR-TKIs treatment or chemotherapy. Its prognostic and predictive value is greatly impaired when EGFR mutant patients were excluded. One thing for sure is that it closely related to a worse survival irrespective of EGFR mutational status especially for the Asians. So far, no effective treatment method direct targeting mutant KRAS gene has been approved in clinic. Agents interrupting RAS signaling such as MEK inhibitor [62–64] or miR-126 [65] seemed selective effective for KRAS mutant tumors, which could be utilized for the development of target therapy for KRAS mutant tumors and might overcome the survival hazard induced by KRAS mutation.
MATERIALS AND METHODS
Publication search and selection
The identification of potential relevant studies was performed through a systemic search in PubMed, Embase and Web of Science databases using the following keywords “lung cancer”, “non-small cell lung cancer” or “NSCLC” and “KRAS”. The latest search was updated on September 2015. Bibliographies of eligible studies, review articles and other relevant publications were also reviewed to identify all potential studies.
A study had to fulfill the following criteria: (1) to deal with non-small cell lung cancer (any stage); (2) to stratified by KRAS mutational status; (3) to assess the correlation between KRAS mutation and survival or treatment outcome (surgery, EGFR-TKIs, platinum-based chemotherapy); (4) to have been published as a full paper in the English language and in the last ten years (2005-2015). The studies were excluded from the analysis if any of the cases occurred: (a) EGFR-TKIs and platinum-based chemotherapy were used as neo-adjuvant treatment; (b) critical information was missing or could not be obtained by our repeated quests.
Data extraction
Two investigators (Wei Pan and Yan Yang) independently screened the studies and extracted the data from included studies by using standard data-abstraction forms. Disagreements were resolved through discussion with another investigator (Hongcheng Zhu). For each study, the following characteristics and information were collected: first author, year of publication, number of patients assessed for KRAS gene and number of patients bearing KRAS mutation gene, gene mutation detection method, ethnicity, pathology, clinical stage and data linking KRAS mutation to treatment outcomes (i.e., CR+PR, SD, PD, and PFS). If a direct report of HR and 95% CI was not available, the total number of events, the number of patients at risk in each group and the log-rank statistic or its P-value was used to allow for an approximation of the HR estimate. If above parameters were yet unavailable, estimated value was derived indirectly from Kaplan-Meier curves using the methods described by Tierney et al. [66]. Survival rates on Kaplan-Meier curves were read by Engauge Digitizer version 4.1 (http://digitizer.sourceforge.net), and then the data read from the curves were entered in the calculation spreadsheet appended to Tierney's paper.
Statistical methods
We extracted relative risks (RRs) with its 95%CIs to show the strength of the association between KRAS mutation and objective response rate (CR + PR), and hazard ratios (HRs) with its 95%CIs to show the survival (OS, DFS or PFS) benefits of KRAS mutant tumors. The individual RRs and HRs were combined into pooled RR and HR, and the initial analyses were performed with a fixed effect model assuming homogeneity of the individual studies. Heterogeneity assumption was checked by Q-test and I2 test. A significant Q-test (p<0.05) or I2>50% indicate the heterogeneity among the studies, and the random-effect model was applied for meta-analysis.
Meta-regression analyses were generated to explore possible sources of heterogeneity (adjusted R2>50% and p<0.05 were consider significant).
Sensitivity analyses were conducted to identify whether results of the meta-analysis were signify affected by exclusion of any individual study and to testify the reliability of the conclusions.
Begg's and Egger's tests were used to evaluate the potential publication bias. The tests were considered statistically significant if p<0.05, and a non-parametric “trim-and-fill” method was applied. All p values were 2-sided and all analyses were performed using Stata SE 11.0 software.
SUPPLEMENTARY FIGURES AND TABLES
Acknowledgments
We thank Juejin Wang (Associate Professor, Department of Physiology, Nanjing Medical University) for the constructive comments on the revised manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflict of interests to disclose.
FUNDING
This study was supported by grants from Natural Science Foundation of China (No. 81272504, 81472809), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) (JX10231801) and Key Academic Discipline of Jiangsu Province “Medical Aspects of Specific Environments”.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2014;62:220–41. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard J-Y, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–46. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 4.Rodenhuis S, Slebos RJ. The ras oncogenes in human lung cancer. Am Rev Respir Dis. 1990;142:S27–30. doi: 10.1164/ajrccm/142.6_Pt_2.S27. [DOI] [PubMed] [Google Scholar]
- 5.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 6.Karachaliou N, Mayo C, Costa C, Magrí I. KRAS mutations in lung cancer. Clin Lung Cancer. 2013;14:205–14. doi: 10.1016/j.cllc.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Slebos RJ, Kibbelaar RE, Dalesio O, Kooistra A, Stam J, Meijer CJ, Wagenaar SS, Vanderschueren RG, van Zandwijk N, Mooi WJ. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–5. doi: 10.1056/NEJM199008303230902. [DOI] [PubMed] [Google Scholar]
- 8.Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–9. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 9.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS Mutation Is an Important Predictor of Resistance to Therapy with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. Clinical Cancer Research. 2007;13:2890–6. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 10.Kim HR, Shim HS, Chung J-H, Lee YJ, Hong YK, Rha SY, Kim SH, Ha S-J, Kim SK, Chung KY, Soo R, Kim JH, Cho BC. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2011;118:729–39. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]
- 11.Metro G, Chiari R, Duranti S, Siggillino A, Fischer MJ, Giannarelli D, Ludovini V, Bennati C, Marcomigni L, Baldi A, Giansanti M, Minotti V, Crinò L. Impact of specific mutant KRAS on clinical outcome of EGFR-TKI-treated advanced non-small cell lung cancer patients with an EGFR wild type genotype. Lung Cancer. 2012;78:81–6. doi: 10.1016/j.lungcan.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Camps C, Jantus-Lewintre E, Cabrera A, Blasco A, Sanmartín E, Gallach S, Caballero C, del Pozo N, Rosell R, Guijarro R, Sirera R. The identification of KRAS mutations at codon 12 in plasma DNA is not a prognostic factor in advanced non-small cell lung cancer patients. Lung Cancer. 2011;72:365–9. doi: 10.1016/j.lungcan.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Nygaard AD, Garm Spindler K-L, Pallisgaard N, Andersen RF, Jakobsen A. The prognostic value of KRAS mutated plasma DNA in advanced non-small cell lung cancer. Lung Cancer. 2013;79:312–7. doi: 10.1016/j.lungcan.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Mellema WW, Dingemans A-MC, Thunnissen E, Snijders PJF, Derks J, Heideman DAM, Van Suylen R, Smit EF. KRAS mutations in advanced nonsquamous non-small-cell lung cancer patients treated with first-line platinum-based chemotherapy have no predictive value. J Thorac Oncol. 2013;8:1190–5. doi: 10.1097/JTO.0b013e318298764e. [DOI] [PubMed] [Google Scholar]
- 15.Metro G, Chiari R, Bennati C, Cenci M, Ricciuti B, Puma F, Flacco A, Rebonato A, Giannarelli D, Ludovini V, Bellezza G, Ferolla P, Minotti V, et al. Clinical outcome with platinum-based chemotherapy in patients with advanced nonsquamous EGFR wild-type non-small-cell lung cancer segregated according to KRAS mutation status. Clin Lung Cancer. 2014;15:86–92. doi: 10.1016/j.cllc.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Marks JL, Broderick S, Zhou Q, Chitale D, Li AR, Zakowski MF, Kris MG, Rusch VW, Azzoli CG, Seshan VE, Ladanyi M, Pao W. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol. 2008;3:111–6. doi: 10.1097/JTO.0b013e318160c607. [DOI] [PubMed] [Google Scholar]
- 17.Zhu CQ, da Cunha Santos G, Ding K, Sakurada A, Cutz J-C, Liu N, Zhang T, Marrano P, Whitehead M, Squire JA, Kamel-Reid S, Seymour L, Shepherd FA, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol. 2008;26:4268–75. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 18.Pesek M, Benesova L, Belsanova B, Mukensnabl P, Bruha F, Minarik M. Dominance of EGFR and insignificant KRAS mutations in prediction of tyrosine-kinase therapy for NSCLC patients stratified by tumor subtype and smoking status. Anticancer Res. 2009;29:2767–73. [PubMed] [Google Scholar]
- 19.Jackman DM, Miller VA, Cioffredi L-A, Yeap BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV, Johnson BE. Impact of Epidermal Growth Factor Receptor and KRAS Mutations on Clinical Outcomes in Previously Untreated Non–Small Cell Lung Cancer Patients: Results of an Online Tumor Registry of Clinical Trials. Clin Cancer Res. 2009;15:5267–73. doi: 10.1158/1078-0432.CCR-09-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H-P, Isaac Wu H-D, Chang JW-C, Wu Y-C, Yang H-Y, Chen Y-T, Hsieh W-Y, Chen Y-T, Chen Y-R, Huang S-F. Prognostic implications of epidermal growth factor receptor and KRAS gene mutations and epidermal growth factor receptor gene copy numbers in patients with surgically resectable non-small cell lung cancer in Taiwan. J Thorac Oncol. 2010;5:1175–84. doi: 10.1097/JTO.0b013e3181e2f4d6. [DOI] [PubMed] [Google Scholar]
- 21.Ludovini V, Bianconi F, Pistola L, Chiari R, Minotti V, Colella R, Giuffrida D, Tofanetti FR, Siggillino A, Flacco A, Baldelli E, Iacono D, Mameli MG, et al. Phosphoinositide-3-kinase catalytic alpha and KRAS mutations are important predictors of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2011;6:707–15. doi: 10.1097/JTO.0b013e31820a3a6b. [DOI] [PubMed] [Google Scholar]
- 22.Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R, Ward C, Mayne K, Trunzer K, Cappuzzo F. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:4113–20. doi: 10.1200/JCO.2010.31.8162. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, Ladanyi M, Riely GJ. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer. 2012;119:356–62. doi: 10.1002/cncr.27730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ragusa M, Vannucci J, Ludovini V, Bianconi F, Treggiari S, Tofanetti FR, Flacco A, Colella R, Sidoni A, Crinò L, Puma F. Impact of epidermal growth factor receptor and KRAS mutations on clinical outcome in resected non-small cell lung cancer patients. American Journal of Clinical Oncology. 2014;37:343–9. doi: 10.1097/COC.0b013e31827a7e7a. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd FA, Domerg C, Hainaut P, Jänne PA, Pignon J-P, Graziano S, Douillard J-Y, Brambilla E, Le Chevalier T, Seymour L, Bourredjem A, Le Teuff G, Pirker R, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol. 2013;31:2173–81. doi: 10.1200/JCO.2012.48.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerner GSMA, Schuuring E, Sietsma J, Hiltermann TJN, Pieterman RM, de Leede GPJ, van Putten JWG, Liesker J, Renkema TEJ, van Hengel P, Platteel I, Timens W, Groen HJM. Common and Rare EGFR and KRAS Mutations in a Dutch Non-Small-Cell Lung Cancer Population and Their Clinical Outcome. PLoS ONE. 2013;8:e70346. doi: 10.1371/journal.pone.0070346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macerelli M, Caramella C, Faivre L, Besse B, Planchard D, Polo V, Ngo Camus M, Celebic A, Koubi-Pick V, Lacroix L, Pignon JP, Soria JC. Does KRAS mutational status predict chemoresistance in advanced non-small cell lung cancer (NSCLC)? Lung Cancer. 2014;83:383–8. doi: 10.1016/j.lungcan.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Cserepes M, Ostoros G, Lohinai Z, Raso E, Barbai T, Timar J, Rozsas A, Moldvay J, Kovalszky I, Fabian K, Gyulai M, Ghanim B, Laszlo V, et al. Subtype-specific KRAS mutations in advanced lung adenocarcinoma: a retrospective study of patients treated with platinum-based chemotherapy. Eur J Cancer. 2014;50:1819–28. doi: 10.1016/j.ejca.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Kim YT, Kim T-Y, Lee DS, Park SJ, Park J-Y, Seo S-J, Choi H-S, Kang HJ, Hahn S, Kang CH, Sung SW, Kim JH. Molecular changes of epidermal growth factor receptor (EGFR) and KRAS and their impact on the clinical outcomes in surgically resected adenocarcinoma of the lung. Lung Cancer. 2008;59:111–8. doi: 10.1016/j.lungcan.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Woo T, Okudela K, Yazawa T, Wada N, Ogawa N, Ishiwa N, Tajiri M, Rino Y, Kitamura H, Masuda M. Prognostic value of KRAS mutations and Ki-67 expression in stage I lung adenocarcinomas. Lung Cancer. 2009;65:355–62. doi: 10.1016/j.lungcan.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 31.Cadranel J, Mauguen A, Faller M, Zalcman G, Buisine M-P, Westeel V, Longchampt E, Wislez M, Coudert B, Daniel C, Chetaille B, Michiels S, Blons H, et al. Impact of systematic EGFR and KRAS mutation evaluation on progression-free survival and overall survival in patients with advanced non-small-cell lung cancer treated by erlotinib in a French prospective cohort (ERMETIC project—part 2) J Thorac Oncol. 2012;7:1490–502. doi: 10.1097/JTO.0b013e318265b2b5. [DOI] [PubMed] [Google Scholar]
- 32.Guan J-L, Zhong W-Z, An S-J, Yang J-J, Su J, Chen Z-H, Yan H-H, Chen Z-Y, Huang Z-M, Zhang X-C, Nie Q, Wu Y-L. KRAS mutation in patients with lung cancer: a predictor for poor prognosis but not for EGFR-TKIs or chemotherapy. Ann Surg Oncol. 2013;20:1381–8. doi: 10.1245/s10434-012-2754-z. [DOI] [PubMed] [Google Scholar]
- 33.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I. Gefitinib or chemotherapy for non–small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 34.Sun J-M, Hwang DW, Ahn JS, Ahn M-J, Park K. Prognostic and Predictive Value of KRAS Mutations in Advanced Non-Small Cell Lung Cancer. PLoS ONE. 2013;8:e64816. doi: 10.1371/journal.pone.0064816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS Mutations and Primary Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib. Plos Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dogan S, Shen R, Ang DC, Johnson ML, D'Angelo SP, Paik PK, Brzostowski EB, Riely GJ, Kris MG, Zakowski MF, Ladanyi M. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–77. doi: 10.4999/uhod.13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boch C, Kollmeier J, Roth A, Stephan-Falkenau S, Misch D, Gruning W, Bauer TT, Mairinger T. The frequency of EGFR and KRAS mutations in non-small cell lung cancer (NSCLC): routine screening data for central Europe from a cohort study. BMJ Open. 2013;3:e002560–0. doi: 10.1136/bmjopen-2013-002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gautschi O, Huegli B, Ziegler A, Gugger M, Heighway J, Ratschiller D, Mack PC, Gumerlock PH, Kung HJ, Stahel RA, Gandara DR, Betticher DC. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett. 2007;254:265–73. doi: 10.1016/j.canlet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–9. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- 40.Marchetti A, Milella M, Felicioni L, Cappuzzo F, Irtelli L, Del Grammastro M, Sciarrotta M, Malatesta S, Nuzzo C, Finocchiaro G, Perrucci B, Carlone D, Gelibter AJ, et al. Clinical implications of KRAS mutations in lung cancer patients treated with tyrosine kinase inhibitors: an important role for mutations in minor clones. Neoplasia. 2009;11:1084–92. doi: 10.1593/neo.09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonanno L, Schiavon M, Nardo G, Bertorelle R, Bonaldi L, Galligioni A, Indraccolo S, Pasello G, Rea F, Favaretto A. Prognostic and predictive implications of EGFR mutations, EGFR copy number and KRAS mutations in advanced stage lung adenocarcinoma. Anticancer Res. 2010;30:5121–8. [PubMed] [Google Scholar]
- 42.Ludovini V, Bianconi F, Pistola L, Pistola V, Chiari R, Colella R, Bellezza G, Tofanetti FR, Siggillino A, Baldelli E, Flacco A, Giuffrida D, Sidoni A, et al. Optimization of patient selection for EGFR-TKIs in advanced non-small cell lung cancer by combined analysis of KRAS, PIK3CA, MET, and non-sensitizing EGFR mutations. Cancer Chemother Pharmacol. 2012;69:1289–99. doi: 10.1007/s00280-012-1829-7. [DOI] [PubMed] [Google Scholar]
- 43.Scoccianti C, Vesin A, Martel G, Olivier M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK, Hainaut P. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: the EUELC cohort. European Respiratory Journal. 2012;40:177–84. doi: 10.1183/09031936.00097311. [DOI] [PubMed] [Google Scholar]
- 44.Gao J, Chen J-Q, Zhang L, Liang Z-Y. Relationship between EGFR and KRAS mutations and prognoisis in Chinese patients with non-small cell lung cancer: a mutation analysis with real-time polymerase chain reaction using scropion amplification refractory mutation system. Zhonghua Bing Li Xue Za Zhi. 2012;41:652–6. doi: 10.3760/cma.j.issn.0529-5807.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 45.Kim ST, Sung JS, Jo UH, Park KH, Shin SW, Kim YH. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol. 2013;30:328–10. doi: 10.1007/s12032-012-0328-3. [DOI] [PubMed] [Google Scholar]
- 46.Fiala O, Pesek M, Finek J, Benesova L, Bortlicek Z, Minarik M. Gene mutations in squamous cell NSCLC: insignificance of EGFR, KRAS and PIK3CA mutations in prediction of EGFR-TKI treatment efficacy. Anticancer Res. 2013;33:1705–11. [PubMed] [Google Scholar]
- 47.Izar B, Zhou H, Heist RS, Azzoli CG, Muzikansky A, Scribner EEF, Bernardo LA, Dias-Santagata D, Iafrate AJ, Lanuti M. The prognostic impact of KRAS, its codon and amino acid specific mutations, on survival in resected stage I lung adenocarcinoma. J Thorac Oncol. 2014;9:1363–9. doi: 10.1097/JTO.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 48.Nadal E, Beer DG, Ramnath N. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol. 2015;10:e9–10. doi: 10.1097/JTO.0000000000000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campos-Parra AD, Zuloaga C, Manríquez MEV, Avilés A, Borbolla-Escoboza J, Cardona A, Meneses A, Arrieta O. KRAS mutation as the biomarker of response to chemotherapy and EGFR-TKIs in patients with advanced non-small cell lung cancer: clues for its potential use in second-line therapy decision making. American Journal of Clinical Oncology. 2015;38:33–40. doi: 10.1097/COC.0b013e318287bb23. [DOI] [PubMed] [Google Scholar]
- 50.Yagishita S, Horinouchi H, Sunami KS, Kanda S, Fujiwara Y, Nokihara H, Yamamoto N, Sumi M, Shiraishi K, Kohno T, Furuta K, Tsuta K, Tamura T, et al. Impact of KRAS mutation on response and outcome of patients with stage III non-squamous non-small cell lung cancer. Cancer Sci. 2015;106:1402–07. doi: 10.1111/cas.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rulli E, Marabese M, Torri V, Farina G, Veronese S, Bettini A, Longo F, Moscetti L, Ganzinelli M, Lauricella C, Copreni E, Labianca R, Martelli O, et al. Value of KRAS as prognostic or predictive marker in NSCLC: results from the TAILOR trial. Ann Oncol. 2015;26:2079–84. doi: 10.1093/annonc/mdv318. [DOI] [PubMed] [Google Scholar]
- 52.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–5. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 53.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, Davies H, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 54.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–6. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 55.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert A-P, Noel S, Lafitte J-J, Sculier J-P. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2004;92:131–9. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mao C, Qiu L-X, Liao R-Y, Du F-B, Ding H, Yang W-C, Li J, Chen Q. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer. 2010;69:272–8. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 57.Ying M, Zhu X, Chen K, Sha Z, Chen L. Should KRAS mutation still be used as a routine predictor of response to EGFR-TKIs in advanced non-small-cell lung cancer? A revaluation based on meta-analysis. J Cancer Res Clin Oncol. 2015;141:1427–39. doi: 10.1007/s00432-015-1910-9. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Fang W, Yan Y, Wang M, Kang S, Sheng J, Zhan J, Chen N, Hong S, Yang Y, Ma Y, He D, Qin T, et al. The efficacy of first-line chemotherapy is associated with KRAS mutation status in patients with advanced non-small cell lung cancer: a meta-analysis. Med Oncol. 2015;32:61. doi: 10.1007/s12032-015-0489-y. [DOI] [PubMed] [Google Scholar]
- 59.Fiala O, Pesek M, Finek J, Benesova L, Belsanova B, Minarik M. The dominant role of G12C over other KRAS mutation types in the negative prediction of efficacy of epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Cancer Genet. 2013;206:26–31. doi: 10.1016/j.cancergen.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H, Ren-Heidenreich L, Shi B, Ren H, Chu X, Kang J, Wang W, Xu J, et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: a comprehensive mutation profiling from 5125 Chinese cohorts. Br J Cancer. 2014;110:2812–20. doi: 10.1038/bjc.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garassino MC, Marabese M, Rusconi P, Rulli E, Martelli O, Farina G, Scanni A, Broggini M. Different types of K-Ras mutations could affect drug sensitivity and tumour behaviour in non-small-cell lung cancer. Ann Oncol. 2011;22:235–7. doi: 10.1093/annonc/mdq680. [DOI] [PubMed] [Google Scholar]
- 62.Dingemans A-MC, Mellema WW, Groen HJM, van Wijk A, Burgers SA, Kunst PWA, Thunnissen E, Heideman DAM, Smit EF. A phase II study of sorafenib in patients with platinum-pretreated, advanced (Stage IIIb or IV) non-small cell lung cancer with a KRAS mutation. Clin Cancer Res. 2013;19:743–51. doi: 10.1158/1078-0432.CCR-12-1779. [DOI] [PubMed] [Google Scholar]
- 63.Metro G, Chiari R, Baldi A, De Angelis V, Minotti V, Crinò L. Selumetinib: a promising pharmacologic approach for KRAS-mutant advanced non-small-cell lung cancer. Future Oncol. 2013;9:167–77. doi: 10.2217/fon.12.198. [DOI] [PubMed] [Google Scholar]
- 64.Said R, Ye Y, Falchook GS, Janku F, Naing A, Zinner R, Blumenschein GR, Fu S, Hong DS, Piha-Paul SA, Wheler JJ, Kurzrock R, Palmer GA, et al. Outcomes of patients with advanced cancer and KRAS mutations in phase I clinical trials. Oncotarget. 2014;5:8937–46. doi: 10.18632/oncotarget.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hara T, Jones MF, Subramanian M, Li XL, Ou O, Zhu Y, Yang Y, Wakefield LM, Hussain SP, Gaedcke J, Ried T, Luo J, Caplen NJ, et al. Selective targeting of KRAS-mutant cells by miR-126 through repression of multiple genes essential for the survival of KRAS-mutant cells. Oncotarget. 2014;5:7635–50. doi: 10.18632/oncotarget.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


