Abstract
It has been reported that oncostatin M (OSM) could initiate cardiomyocyte dedifferentiation both in vivo and in vitro. OSM-induced cardiomyocyte dedifferentiation might be a new target for the treatment of diabetic cardiomyopathy (DCM). This study was designed to determine the role of OSM in cardiomyocyte dedifferentiation and the progression of DCM. A mouse DCM model was established to evaluate the effects of OSM in vivo. Echocardiography was applied to determine cardiac function. Sirius red staining was used to detect fibrosis area. Transmission electron microscopy was used to evaluate mitochondria impairment. Real-time polymerase chain reaction and western blot analysis were performed to detect relative mRNA expressions and cardiomyocyte dedifferentiation-related protein expressions, respectively. OSM treatment induced similar impaired cardiac function and cardiac ultrastructure impairment to those detected in DCM mice. The expressions of dedifferentiation markers of cardiomyocyte (Runx1, and α-SM-actin) were up-regulated in the OSM-treated mice compared with those in the control group. To further demonstrate the important role of OSM, OSM receptor knockout (Oβko) mice were used. In Oβko mice, cardiomyocytes dedifferentiation markers of c-kit, Runx1, and atrial natriuretic peptide were down-regulated, with attenuated DCM injury and abrogated OSM/B-Raf/Mek/Erk signaling pathway. In conclusion, OSM-induced cardiomyocyte dedifferentiation plays a crucial role in the progression of DCM. The mechanism of OSM-induced cardiomyocyte dedifferentiation is associated with B-Raf/Mek/Erk signaling pathway through the OSM receptor Oβ.
Keywords: oncostatin M, cardiomyocyte dedifferentiation, diabetic cardiomyopathy
Introduction
Diabetic cardiomyopathy (DCM) was first reported in 1972 when Rubler et al. [1] described four diabetic patients with congestive heart failure. Since then, DCM has been defined as ventricular dysfunction that occurs independently of coronary artery disease and hypertension [2]. There are three stages in the progression of DCM. The early stage is the phase when cellular and metabolic changes occur without obvious systolic dysfunction; the middle stage is characterized by increased apoptosis, left ventricular size, and diastolic dysfunction with normal ejection fraction (EF); and the late stage is characteristic of alteration in microvasculature compliance, increased left ventricular size, and decreased cardiac performance which leads to heart failure [3]. As the diabetic population increased rapidly in recent years, DCM has gained much attention. However, the mechanisms of DCM are still not well understood [4,5].
Cardiomyocyte dedifferentiation is an adaptive process which reacts to the outside stimulation [6]. The main presentations of cardiomyocyte dedifferentiation focus on the cardiomyocyte structure changes, the expressions of the related markers including Runx1, c-kit, α-SM-actin, and atrial natriuretic peptide (ANP) [7–9]. Kubin et al. [10] revealed that partial dedifferentiation of cardiomyocyte initially protected the damaged myocardium, but promoted heart failure in the chronic phase characterized by prolonged induction of dedifferentiation, indicating that cardiomyocytes dedifferentiation might be an important target for the treatment of DCM.
Oncostatin M (OSM), an inflammatory cytokine of the interleukin 6 (IL-6) family members, could exert multiple physiological functions. There are two kinds of OSM receptors, namely, Type I receptor formed by LIFR (leukemia inhibitory factor receptor)-gp130 and Type II receptor composed of OSM receptor Oβ and gp130. Murine OSM binds exclusively to Type II receptor, while human OSM has the exceptional capability to recruit both two receptors [11]. Previous studies have demonstrated that OSM initiates dedifferentiation of adipocytes [12]. Elevated expression levels of OSM have been found in patients with dilative cardiomyopathy. Furthermore, OSM initiates dedifferentiation of cardiomyocyte in dilative cardiomyopathy and acute myocardial infarction [10].
However, whether OSM takes part in the dedifferentiation of cardiomyocyte in DCM remains unclear. In this study, we aimed to determine whether OSM-related dedifferentiation is associated with DCM progression, and to explore the underlying mechanisms.
Materials and Methods
Animals
129-Osmrtm1.1Nat/J mice possessing loxP sites on both sides of the second exon (first coding exon) of the OSM receptor (Oβ) gene were purchased from Jackson Laboratory (Bar Harbor, USA). 129-Osmrtm1.1Nat/J mice were crossed with C-Tg (CMV-cre) 1 Cgn/J mice (Jackson Laboratory) to knockout OSM receptor Oβ. Polymerase chain reaction (PCR) was used to screen Oβ−/− mice, and Oβ+/+ mice (body weight 23–25 g) were used as controls.
Mice were randomly allocated into the following groups, with 20 mice in each group: (i) control group, (ii) DCM group, (iii) OSM group, (iv) DCM-Oβ−/− group, (v) DCM-Oβ−/−+OSM group, (vi) DCM+OSM group, (vii) DCM-Oβ−/−+OSM group, and (viii) DCM+UO126+OSM group. Diabetes mellitus was induced in 8–12 weeks Oβ−/− mice and Oβ+/+ mice (body weight 23–25 g) by intraperitoneal injections (i.p.) of streptozotocin (STZ) (Sigma, St Louis, USA). STZ was dissolved in 0.1 M citrate buffer, pH 4.5 and injected (50 mg/kg) for five consecutive days as previously described [13]. Control mice were administered an equivalent volume of citrate buffer. Random blood glucose concentration was determined using a reflectance meter (Accu-Chek; Roche Diagnostics GmbH, Mannheim, Germany) at 3 days, 7 days, and 3 months post STZ injection. The glucose value of ≥16.7 mM was considered as a cutoff point for diabetes.
OSM-treated mice were then received OSM injection for 2 months after STZ or citrate buffer injection. OSM (R&D Systems, Minneapolis, USA) was dissolved in sterile phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin and injected intraperitoneally twice per day with 60 ng/g of body weight for 14 days. The control group received the same volume of sterile PBS containing 0.1% bovine serum albumin for 14 days. The chemical 1,4-diamino-2,3-dicyano-1,4-bis (o-aminophenylmercapto)butadiene (UO126) was purchased from Calbiochem (La Jolla, USA) and dissolved in sterile saline. Mice in the UO126-treatment group received UO126 injection at a dose of 1 μg/g body weight, together with OSM through the tail vein twice per day for 14 days.
The experiments were approved by the Ethic Committee on Animal Care of the Fourth Military Medical University and performed according to the instructions of the National Institutes of Health Guidelines on the Use of Laboratory Animals.
Echocardiography for the determination of cardiac function and hemodynamic evaluation
Echocardiography was conducted 12 weeks after STZ injection. Mice were sedated using 3% isoflurane, and transthoracic echocardiography was performed by Vevo 2100 ultrasound system (Visual-Sonics, Toronto, Canada) with a 30-MHz linear transducer. Left ventricular end-systolic volume (LVESV), left ventricular end-diastolic volume (LVEDV), left ventricular EF, and left ventricular fractional shortening (FS) were calculated by the use of computer algorithms. All parameters were calculated based on the statistics of four consecutive cardiac cycles, and all of these measurements were performed in a blinded manner. FS and EF were calculated as follows: [14], and [15]. The first derivative of the left ventricular pressure (+dp/dt max and -dp/dt max) was measured in mice anesthetized with 3% isoflurane as previously described [16].
Transmission electron microscopy
After echocardiography assessment, mice were anesthetized with 3% sodium pentobarbital. Hearts were rapidly removed and washed with PBS solution. At a low temperature, a specimen of the left ventricular myocardium was removed with ophthalmic scissors and cut into ultra-thin sections with the thickness of 60–64 nm. Images were taken after fixation, soaking, stepwise alcohol dehydration, displacement, embedding, polymerization, sectioning, and staining, and then observed with an electron microscope (JEM-2000EX TEM, JEOL Ltd., Tokyo, Japan). Random sections were taken and analyzed by two technicians blinded to the treatments [17].
Sirius red staining
After catheterization, cardiac puncture was performed as previously described [18]. The heart was then fixed with 10% buffered formalin and assigned a numerical code to conceal identification of the treatment group. Tissues were subsequently processed through graded alcohols, embedded in paraffin, sliced to 4 μm thickness, placed onto microscope slides, and stained using a picro sirius red-fast green (Sigma) staining technique. Sirius red binds to collagen and fast green binds to noncollagenous protein. Images were captured under a light microscope equipped with a DFC490 digital camera (Leica Microsystems, Wetzlar, Germany).
Quantitative real-time PCR
RNA was isolated using a NucleoSpin RNA II kit (Macherey-Nagel GmbH, Mannheim, Germany), and cDNA was synthesized with a Reverse Transcription System kit (Promega, Madison, USA). Quantitative real-time PCR was performed using predesigned Taqman Gene Expression Assays and AmpliTaq Gold DNA polymerase following the manufacturer's instructions (Applied Biosystems, Foster City, USA). PCR was performed in a GeneAmp PCR system 2400 Thermal Cycler (Perkin-Elmer, Norwalk, USA) and the conditions were 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C (30 cycles). Primers used are listed in Table 1. The ratio of the mRNA levels for each sample was calculated by normalizing the comparative quantitation values to those of GAPDH mRNA.
Table 1.
Sequence of primers used in Quantitative real-time PCR
| Gene | Primer sequence |
|---|---|
| OSM | 5′-TGCCCGGCACAATATCCTC-3′ (forward) |
| 5′-GTGTGGGCTCAGGTATCTCCAG-3′ (reverse) | |
| Oβ | 5′-GATTCGCATCAC AGCCAACAA-3′(forward) |
| 5′-CCAGATACGGGCTCCCAAGA-3′ (reverse) | |
| Runx1 | 5′-AACCAGGTAGCGAGATTCAACGAC-3′ (forward) |
| 5′-CAACTTGTGGCGGATTTGTAAAGA-3′ (reverse) | |
| c-kit | 5′- CGGGCTAGCCAGAGA CATCA-3′ (forward) |
| 5′-TCTCTGGTGCCATCCACTTCA-3′ (reverse) | |
| ANP | 5′- TGACAGGATTGGAGCCCAGA-3′ (forward) |
| 5′-GACACACCACAAGGGCTTAGG A-3′ (reverse) | |
| GAPDH | 5′-GGCACAGTCAAGGCTGAGAATG-3′ (forward) |
| 5′-ATG GTGGTGAAGACGCCAGTA-3′ (reverse) |
Western blot analysis
Protein samples were prepared from homogenized heart tissue according to standard protocols. Total proteins were loaded onto the sodium dodecyl sulfate polyacrylamide gel electrophoresis gel and electrophoretically transferred to nitro-cellulose membranes (Millipore, Billerica, USA). After being blocked with 5% skim milk, the membranes were incubated with the appropriate primary antibody of the recommended dilution at 4°C overnight. The membranes were washed and further incubated with horseradish peroxidase-conjugated secondary antibody at 37°C for 60 min. The blots were developed with an enhanced chemiluminescence reagent kit (Millipore) and visualized with UVP Bio-Imaging Systems (UVP, LLC, Upland, USA). Vision Works LS Acquisition and Analysis Software were used to analyze blot densities [18].
Primary antibodies against the following proteins were used in this study: β-actin (1:2000; Cell Signaling, Beverly, USA), GADPH (1:2000; Cell Signaling), OSM (1:2000; R&D systems), Oβ (1:500, R&D system), c-kit (1:1000; Abcam, Cambridge, USA), Sca-1 (1:2000; Abcam), Runx1 (1:500; Abcam,), ANP (1:200; Abcam), α-SM-actin (1:2000; Abcam), p-gp130 (ser782, 1:100; Abcam), B-raf (1:500; Becton Dickinson, Franklin Lakes, USA), Mek (1:1000; Becton Dickinson), Erk (1:1000; Abcam), and p-Erk (Thr202/Tyr204, 1:2000; Cell Signaling). Secondary antibodies were horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5000; Santa Cruz Biotechnology, Santa Cruz, USA), and rabbit anti-goat IgG (1:5000; Santa Cruz Biotechnology).
Immunofluorescence staining
Hearts were prepared as previously described and then fixed by immersion in cold 4% formaldehyde for 24 h and then processed for paraffin embedding. Microtome sections (4 μm) were used for immunofluorescence analysis. Paraffin sections were treated with xylene and ethanol, washed with PBS and then subject to antigen retrieval. Slices were blocked with 3% bovine serum albumin and incubated with primary antibodies against Runx1 (Abcam) and ANP (Abcam) at the dilution of 1:100 at 4°C overnight, washed with PBS for three times (5 min each), and then incubated with tetraethyl rhodamine isothiocyanate conjugated goat anti-rabbit IgG polyclonal secondary antibody at a 1:400 dilution for 60 min at 37°C. F-actin was co-stained with CytoPainter F-actin Staining kit (Abcam) for 30 min at 37°C. Then the slices were washed again with PBS for three times (5 min each). Nuclei were counterstained with 4',6-diamidino-2-phenylindole and then washed with PBS. After the final wash, the slices were covered with cover-slips and examined with a confocal microscope (Olympus FV 1000, Tokyo, Japan).
Statistical analysis
Continuous variables that approximated the normal distribution were expressed as means ± standard deviation. Comparisons between groups were done with analysis of variance followed by Bonferroni correction for the post hoc t-test. Data expressed as proportions were assessed with a χ2-test. Two-sided tests were used throughout, and P < 0.05 was considered statistically significant. SPSS software package version 14.0 (SPSS, Chicago, USA) was used for data analysis.
Results
DCM mice exhibited impaired cardiac function and increased expressions of OSM and Oβ receptor in heart
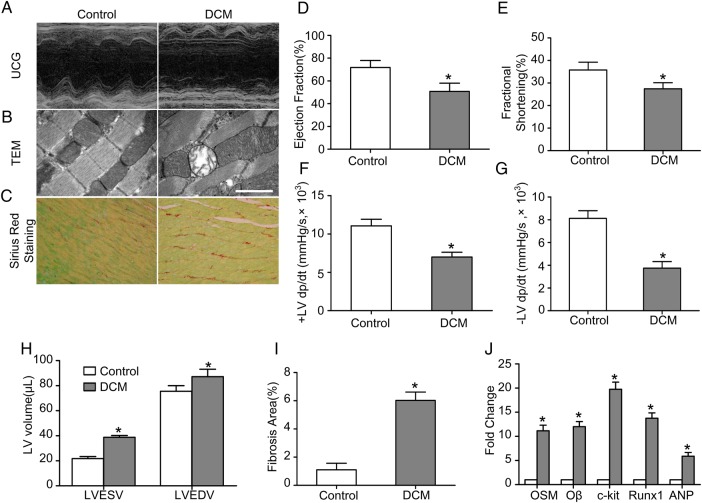
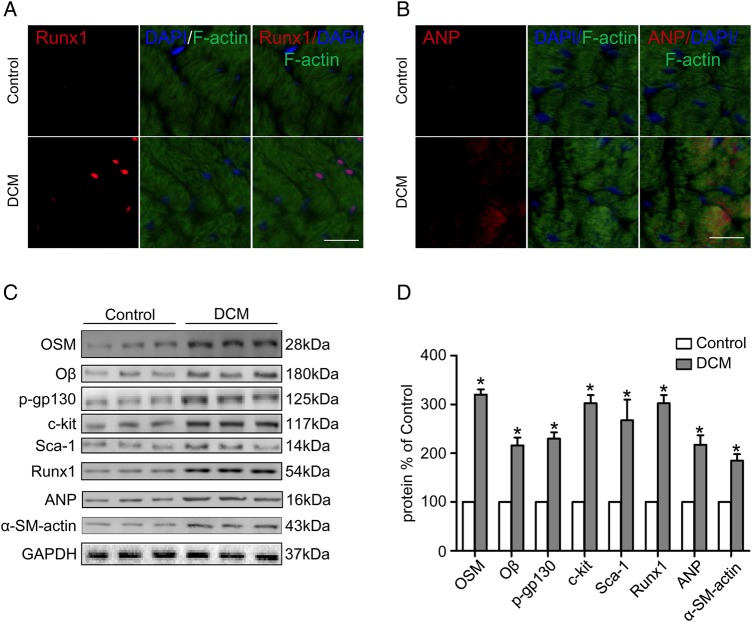
Cardiac function of mice in the control group and the DCM group was evaluated by ultrasound cardiography (UCG) (Fig. 1A). The LVEF (50.75% ± 7.30% vs. 71.78% ± 6.18%, P < 0.05) and the FS (27.45% ± 2.71% vs. 35.77% ± 3.46%, P < 0.05) were decreased while the LVEDV (87.25 ± 11.79 µl vs. 75.50 ± 9.04 µl, P < 0.05) and LVESV (38.75 ± 2.99 µl vs. 21.75 ± 3.30 µl, P < 0.05) were increased in the DCM group compared with the control group (Fig. 1D,E,H). The +LV dp/dt max (6999.2 ± 1244.1 mmHg/s vs. 11061.5 ± 1716.2 mmHg/s, P < 0.05) and the −LV dp/dt max (3748.6 ± 1152.3 mmHg/s vs. 8125.6 ± 1343.5 mmHg/s, P < 0.05) were decreased in the DCM group compared with the control group (Fig. 1F,G). Sirius red staining (Fig. 1C) showed that the fibrosis area was increased in the DCM group (6.03% ± 0.59% vs. 1.10% ± 0.47%, P < 0.05) compared with the control group (Fig. 1I). Moreover, transmission electron microscopy (TEM) revealed mitochondria impairment in the DCM mice (Fig. 1B). Immunofluorescence staining of heart specimens from DCM mice revealed significant up-regulation of dedifferentiation markers including Runx1 and ANP (Fig. 2A,B). The expressions of OSM, Oβ, p-gp130 (ser782), dedifferentiation markers and typical fetal gene proteins including c-kit, scal-1, Runx1, ANP, and α-SM-actin were detected by western blot analysis (Fig. 2C). The expressions of OSM, Oβ, c-kit, Runx1, and ANP were also detected by real-time PCR. Results showed that the expressions of OSM, Oβ, p-gp130 (ser782), c-kit, scal-1, Runx1, ANP, and α-SM-actin in the DCM group were much higher than those in the control group (Figs. 1J and 2D; P < 0.05).
Figure 1.
DCM mice exhibited impaired cardiac function (A) Echocardiography showed normal cardiac function in the control group and impaired cardiac function in the DCM group compared with the control group. (B) TEM showed mitochondria impairment in the DCM mice (scale bar = 2 μm). (C) Sirius red staining showed increased fibrosis area in the DCM group compared with the control group (magnification, ×400). (D–H) Statistic analysis of EF, FS, ±LV dp/dt max, LVEDV, and LVESV. (I) Statistic analysis of fibrosis area. (J) RT-PCR analysis of OSM, Oβ, c-kit, Runx1, and ANP. *P < 0.05 vs. control group.
Figure 2.
DCM mice showed increased expressions of OSM and its receptor Oβ in heart (A, B) Fluorescent immunostaining of Runx1 and ANP expression (red) of cardiomyocytes (green) (scale bar = 30 μm). (C) Western blot analysis showed increased expression of OSM, Oβ, p-gp130, c-kit, scal-1, Runx1, ANP, and α-SM-actin in the DCM group. (D) Statistical analysis of western blot results. *P < 0.05 vs. control group.
OSM treatment induced cardiac function impairment and up-regulation of dedifferentiation-related markers
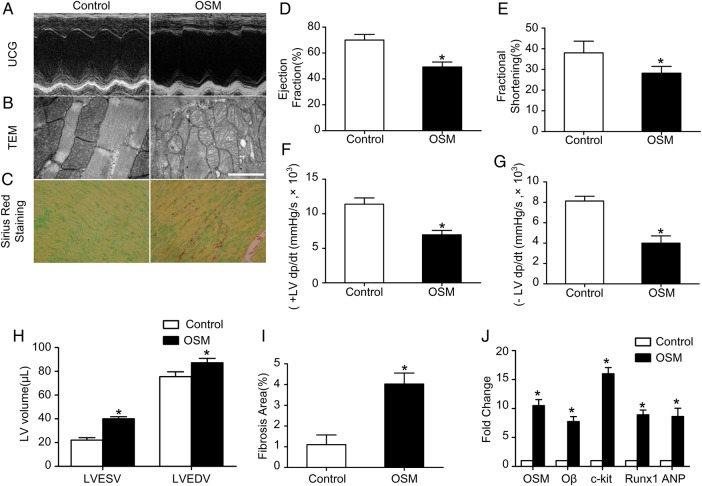
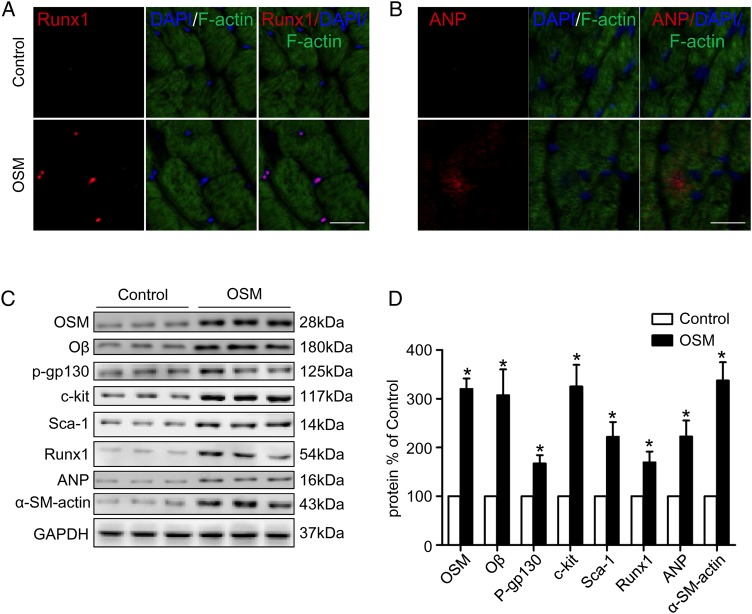
To investigate whether OSM treatment is sufficient to induce similar changes of DCM in vivo, 6-month-old mice were treated with 60 ng OSM per gram of body weight twice per day. After 14 days of treatment, UCG (Fig. 3A) revealed that cardiac function was impaired as evidenced by decreased LVEF (49.25% ± 3.75% vs. 70.07% ± 4.37%, P < 0.05) and FS (28.20% ± 3.30% vs. 38.02% ± 5.66%, P < 0.05) (Fig. 3D,E), while the LVEDV (87.18 ± 7.47 µl vs. 75.50 ± 8.23 µl, P < 0.05) and LVESV (40.09 ± 2.99 µl vs. 22.15 ± 4.32 µl, P < 0.05) were increased (Fig. 3H) in the OSM group in comparison with the control group. The ±LV dp/dt max were also decreased significantly (+LV dp/dt max: 6949.2 ± 1257.6 mmHg/s vs. 11386.5 ± 1794.6 mmHg/s, P < 0.05; −LV dp/dt max: 3998.0 ± 1411.3 vs. 8125.6 ± 932.3 mmHg/s, P < 0.05) in the OSM group compared with the control group (Fig. 3F,G). Similar to the DCM mice, increased fibrosis area (4.03% ± 0.53% vs. 1.11% ± 0.44%, P < 0.05) (Fig. 3C,I) and mitochondria impairment (Fig. 3B) were also observed in the OSM-treated mice. Moreover, immunofluorescence revealed increased expressions of Runx1 and ANP in the OSM-treated mice (Fig. 4A,B). The expressions of OSM, Oβ, c-kit, Runx1, and ANP were also detected by real-time PCR and western blot analysis. Results demonstrated the increased expressions of OSM, Oβ, and the increased phosphorylation level of gp130 at serine 782 in the OSM group. The expressions of the progenitor cell markers including c-kit, scal-1, Runx1, ANP, and α-SM-actin were also increased significantly in the OSM group compared with the control group (Figs. 3J and 4C,D; P < 0.05).
Figure 3.
OSM treatment induced cardiac function impairment (A) Echocardiography showed normal cardiac function in the control group and impaired cardiac function in the OSM group. (B) TEM showed mitochondria impairment in the OSM mice (scale bar = 2 μm). (C) Sirius red staining showed increased fibrosis area in the OSM group compared with the control group (magnification, ×400). (D–H) Statistic analysis of EF, FS, ±LV dp/dt max, LVEDV, and LVESV. (I) Statistic analysis of fibrosis area. (J) RT-PCR analysis of OSM, Oβ, c-kit, Runx1, and ANP in the DCM group relative to the control group. *P < 0.05 vs. control group.
Figure 4.
OSM treatment induced up-regulation of dedifferentiation-related markers (A,B) Fluorescent immunostaining of Runx1 and ANP expression (red) of cardiomyocytes (green) (scale bar = 30 μm). (C) Western blot analysis showed increased expression of OSM, Oβ, p-gp130, c-kit, scal-1, Runx1, ANP, and α-SM-actin in the OSM group in comparison with the control group. (D) Statistical analysis of western blot results. *P < 0.05 vs. control group.
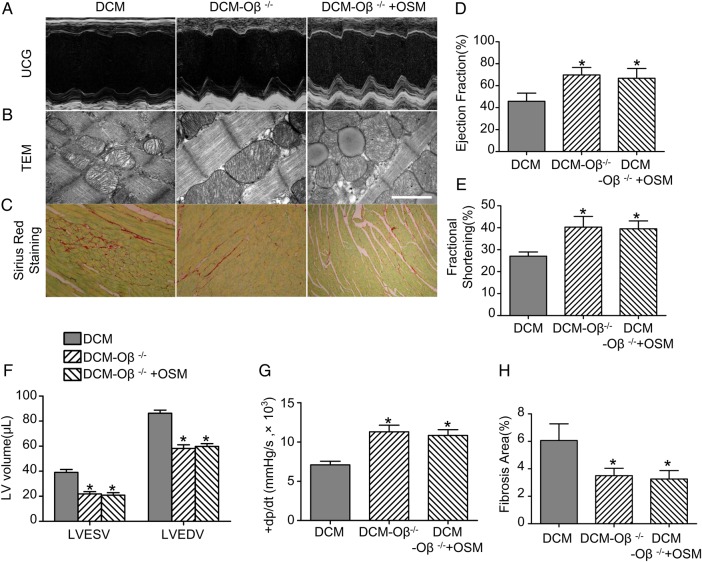
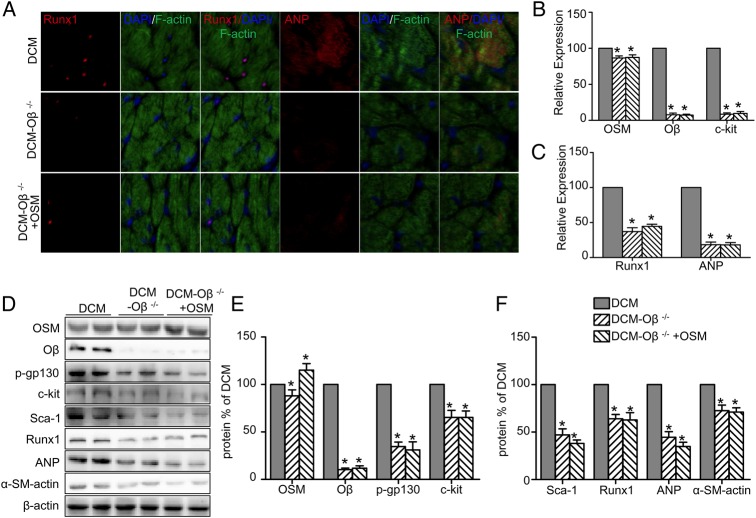
OSM receptor Oβ-knockout attenuated DCM injury and abrogated OSM signaling pathway
The DCM model was constructed by Oβ+/+ mice (DCM group) and Oβ-knockout (Oβ-KO) mice (DCM-Oβ−/− group). Oβ-KO mice in the DCM-Oβ−/−+OSM group received additional OSM injection as described above. Interestingly, the deletion of Oβ significantly attenuated DCM injury. UCG (Fig. 5A) showed that the LVEF (69.82% ± 6.82% vs. 45.75% ± 7.45%, P < 0.05) and the FS (43.50% ± 5.22% vs. 31.07% ± 5.86%, P < 0.05) were increased (Fig. 5D,E), while the LVEDV (58.17 ± 7.20 µl vs. 86.32 ± 6.01 µl, P < 0.05) and LVESV (21.83 ± 4.49 µl vs. 39.07 ± 5.87 µl, P < 0.05) were decreased (Fig. 5F) in the Oβ-KO group in comparison with the DCM group. The ±LV dp/dt max (+LV dp/dt max: 11291.2 ± 1675.3 mmHg/s vs. 7082.4 ± 924.8 mmHg/s, P < 0.05) were also increased significantly (Fig. 5G) in the DCM-Oβ−/− group compared with the DCM group. The mitochondria impairment was relieved (Fig. 5B) and the fibrosis area was decreased (6.066% ± 1.214% vs. 3.504% ± 0.532%, P < 0.05) (Fig. 5C,H) in the DCM-Oβ−/− group compared with the DCM group. No expression of Runx1 and ANP was found in the DCM-Oβ−/− group (Fig. 6A). Similarly, western blot analysis showed that the deletion of OSM receptor Oβ significantly down-regulated the expressions of Oβ, p-gp130, and the progenitor cell markers including c-kit, scal-1, Runx1, ANP, and α-SM-actin (Fig. 6D–F; P < 0.05), and real-time PCR results also showed the down-regulation of OSM, Oβ, c-kit, Runx1, and ANP (Fig. 6B,C; P < 0.05). Furthermore, Oβ-KO abolished the dedifferentiation effects of OSM in diabetic mice as described above, indicating that OSM-mediated cardiomyocyte dedifferentiation was involved in the progression DCM through OSM receptor Oβ.
Figure 5.
OSM receptor Oβ knockout (Oβ-KO) attenuated DCM injury (A) Representative images as evaluated by echocardiography. (B) TEM showed relieved mitochondria impairment in the DCM-Oβ−/− group and OSM could no longer lead to mitochondria impairment in DCM-Oβ−/− group mice (scale bar = 2 μm). (C) Sirius red staining showed decreased fibrosis area in the DCM-Oβ−/− group compared with the DCM group, and no increased fibrosis area was detected in the DCM-Oβ−/− +OSM group (magnification, ×400). (D–G) Statistical analysis of EF, FS, ±LV dp/dt max, LVEDV and LVESV. (H) Statistical analysis of fibrosis area. *P < 0.05 vs. DCM group.
Figure 6.
OSM receptor Oβ-KO abrogated OSM signaling pathway (A) Fluorescent immunostaining of Runx1 and ANP expression (red) of cardiomyocytes (green) (scale bar = 30 μm). (B,C) RT-PCR analysis of mRNA expressions of OSM, Oβ, c-kit, Runx1, and ANP. (D) Western blot analysis shows decreased expression of OSM, Oβ, p-gp130, c-kit, scal-1, Runx1, ANP, and α-SM-actin in the DCM-Oβ−/− group and the DCM-Oβ−/− +OSM group. (E,F) Statistical analysis of western blot results. *P < 0.05 vs. DCM group.
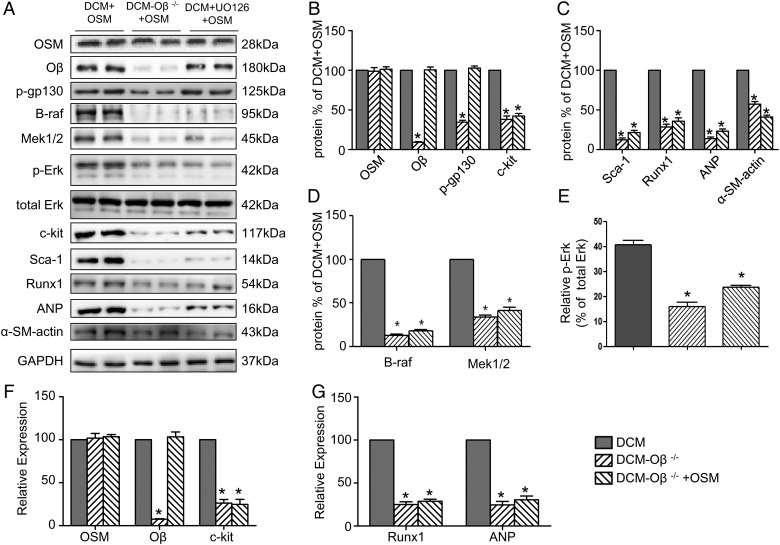
OSM induced cardiomyocyte dedifferentiation in DCM by stimulation of the B-Raf/Mek/Erk cascade via its receptor Oβ
OSM was injected into DCM mice and Oβ-KO DCM mice. Western blot analysis (Fig. 7A–D; P < 0.05) demonstrated a marked decrease of Erk1/2 phosphorylation together with strong down-regulation of the B-Raf, Mek1/2, and the progenitor cell markers c-kit, scal-1, and dedifferentiation markers Runx1, ANP, and α-SM-actin in Oβ-KO DCM mice. Real-time PCR showed the down-regulation of OSM, Oβ, c-kit, Runx1, and ANP (Fig. 7E–G; P < 0.05), indicating that the B-Raf/Mek/Erk cascade might play an important role in the OSM-mediated cardiomyocyte dedifferentiation. The Mek1/2 inhibitor UO126 pretreatment of Oβ+/+ mice abolished the effects of OSM on cardiomyocyte dedifferentiation in DCM, as evidenced by decreased expressions of B-Raf, Mek1/2, Erk1/2 phosphorylation and the progenitor cell markers c-kit, scal-1, Runx1, ANP, and α-SM-actin (Fig. 7A–D; P < 0.05).
Figure 7.
OSM induced cardiomyocyte dedifferentiation in DCM by stimulation of the B-Raf/Mek/Erk cascade via OSM receptor Oβ (A–D) Western blot analysis showed decreased expressions of OSM, Oβ, B-Raf, p-Erk and the progenitor cell markers c-kit, scal-1, Runx1, ANP, and α-SM-actin relative to the DCM+OSM group, and the UO126-treatment group showed decreased expression of B-Raf, p-Erk and the progenitor cell markers c-kit, scal-1, Runx1, ANP, and α-SM-actin relative to the DCM + OSM group. (E-G) RT-PCR analysis of mRNA expressions of OSM, Oβ, c-kit, Runx1, and ANP. *P < 0.05 vs. DCM + OSM group.
Discussion
DCM is defined as the structural and functional changes in the myocardium associated with diabetes in the absence of ischemic heart diseases or other cardiac pathologies [3]. Although several decades have passed since DCM was first reported, the pathogenesis and underlying mechanisms were not fully understood. Risk factors of DCM include a high fat diet/obesity, cardiovascular autonomic neuropathy, inflammation, and elevated levels of free fatty acid, advanced glycation end products with their receptors, and reactive oxygen species [3]. Recent studies have revealed that differential expression of miRNAs [19,20] and stem cell survival and differentiation [21] might play roles in the progression of DCM. In addition, dedifferentiation of cardiomyocyte was reported to be seen in ischemic hearts and probably participated in heart remodeling after injury [6,22,23]. Huang et al. [24] reported that myocardial remodeling in DCM was associated with cardiac mast cell activation. Jopling et al. [25] observed heart regeneration induced by cardiomyocyte dedifferentiation and proliferation in zebrafish. Tsyplenkova et al. [26] reported that cardiomyocyte dedifferentiation might be a new target for the development of DCM, indicating that cardiomyocyte dedifferentiation might participate in the progression of DCM. Modulating the extent of cardiomyocyte dedifferentiation might provide some clues to the treatment of DCM.
OSM, an inflammatory cytokine of the IL-6 family, exerts multiple functions such as induction of dedifferentiation of adipocytes [12] and maintenance of megakaryocyte and erythroid progenitor pools [27]. In the heart, members of this family have so far been mostly associated with cardiac hypertrophy and cardiomyocyte survival due to activation of the common co-receptor gp130 [28]. Hohensinner et al. [29] has reported that OSM might play a key role in cardiac regeneration via the induction of SDF-1. Additionally, OSM could initiate cardiomyocyte dedifferentiation both in vivo and in vitro [30]. Previous studies have shown that OSM-mediated cardiomyocyte dedifferentiation protected the heart after acute myocardial damage, but promoted functional deterioration upon chronic activation in dilative cardiomyopathy [10]. Our results revealed that mice with DCM exhibited deteriorated cardiac function as evidenced by echocardiography and hemodynamic evaluation. Increased cardiac fibrosis and cardiac ultrastructure impairment were also found in the DCM group. The expressions of Runx1 and ANP, two dedifferentiation markers of cardiomyocyte, were significantly increased in the DCM group. The expressions of typical fetal genes, including c-kit, scal-1, and α-SM-actin, were increased in the DCM group, indicating that cardiomyocyte dedifferentiation may involve in the development of DCM. Furthermore, DCM mice also exhibited increased expressions of OSM and Oβ.
We were also interested in whether OSM treatment can mimic the pathological changes of DCM. As expected, OSM treatment induced similar cardiac dysfunction and increased cardiac fibrosis and mitochondria impairment as DCM. The expressions of dedifferentiation markers (Runx1 and ANP) and typical fetal genes expressions were also increased in the OSM-treated group, indicating that OSM might play an important role in the progression of the DCM, possibly via initiating cardiomyocyte dedifferentiation, and that modulating OSM expression may attenuate cardiac function impairment, mitochondria injury and relieve fibrosis extent.
As murine OSM binds exclusively to Type II receptor, OSM receptor Oβ-KO mice were constructed to further test whether cardiomyocyte dedifferentiation in DCM is initiated by OSM through OSM receptor Oβ. Our results clearly showed that OSM receptor deletion alleviated DCM injury as evidenced by improved cardiac function, decreased cardiac fibrosis area, and relieved mitochondrial impairment, indicating that cardiomyocyte dedifferentiation critically depended on OSM signaling under DCM conditions. OSM administration in Oβ-KO mice did not induce increased expressions of dedifferentiation markers and typical fetal genes as noticed in the Oβ+/+ mice. Thus, the correlation of OSM/Oβ signaling, cardiomyocyte dedifferentiation, and development of DCM suggested a decisive role of OSM in these events.
However, the signaling pathways that might mediate OSM signaling in the progression of DCM have not been fully delineated. OSM downstream signaling events include the activation of p38, extracellular signal-regulated kinase 1/2, phosphatidylinositol 3-kinase, and janus kinase/signal transducer and activator of transcription [11]. As B-Raf/Mek/Erk cascade is closely related to cell proliferation, survival, differentiation, motility, and angiogenesis [31], we believe that it might play a key role in DCM. Interestingly, we found that OSM up-regulation was closely related to the activation of B-Raf/Mek/Erk cascade, and knock out of OSM receptor Oβ abrogated the effects of OSM on B-Raf/Mek/Erk signaling pathway. UO126, an inhibitor of Mek1/2, also abolished the effects of OSM through B-Raf/Mek/Erk signaling pathway, indicating that OSM-mediated cardiomyocyte differentiation and DCM development might be related to the activation of B-Raf/Mek/Erk signaling pathway.
In summary, our study demonstrated that OSM-induced cardiomyocyte dedifferentiation plays a crucial role in the progression of DCM. The mechanism of OSM-induced cardiomyocyte dedifferentiation is associated, at least in part, with B-Raf/Mek/Erk signaling pathway through Oβ receptor. The findings of this study may provide a new target for the further treatment of DCM.
Funding
This work was supported by the grants from the National Funds for Distinguished Young Scientists of China (No. 81325009), the National Natural Science Foundation of China (Nos. 81270168, 81530058, and 81570272), Beijing Natural Science Foundation (No. 7152131), the National Basic Research Program of China (No. 2012CB518101), and Shanxi Province Program (2014KCT-20).
References
- 1.Rubler S, Dlugash J, Yuceoglu YZ, Kumral T, Branwood AW, Grishman A. New type of cardiomyopathy associated with diabetic glomerulosclerosis. Am J Cardiol 1972, 30: 595–602. [DOI] [PubMed] [Google Scholar]
- 2.Bertoni AG, Tsai A, Kasper EK, Brancati FL. Diabetes and idiopathic cardiomyopathy: a nationwide case-control study. Diabetes Care 2003, 26: 2791–2795. [DOI] [PubMed] [Google Scholar]
- 3.Chavali V, Tyagi SC, Mishra PK. Predictors and prevention of diabetic cardiomyopathy. Diabetes Metab Syndr Obes 2013, 6: 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols GA, Hillier TA, Erbey JR, Brown JB. Congestive heart failure in type 2 diabetes: prevalence, incidence, and risk factors. Diabetes Care 2001, 24: 1614–1619. [DOI] [PubMed] [Google Scholar]
- 5.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation 2007, 115: 3213–3223. [DOI] [PubMed] [Google Scholar]
- 6.Taegtmeyer H, Sen S, Vela D. Return to the fetal gene program: a suggested metabolic link to gene expression in the heart. Ann N Y Acad Sci 2010, 1188: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H et al. . Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114: 763–776. [DOI] [PubMed] [Google Scholar]
- 8.Driesen RB, Verheyen FK, Debie W, Blaauw E, Babiker FA, Cornelussen RN, Ausma J et al. . Re-expression of alpha skeletal actin as a marker for dedifferentiation in cardiac pathologies. J Cell Mol Med 2009, 13: 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 2008, 9: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubin T, Poling J, Kostin S, Gajawada P, Hein S, Rees W, Wietelmann A et al. . Oncostatin M is a major mediator of cardiomyocyte dedifferentiation and remodeling. Cell Stem Cell 2011, 9: 420–432. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 2003, 374: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song HY, Kim MR, Lee MJ, Jeon ES, Bae YC, Jung JS, Kim JH. Oncostatin M decreases adiponectin expression and induces dedifferentiation of adipocytes by JAK3- and MEK-dependent pathways. Int J Biochem Cell Biol 2007, 39: 439–449. [DOI] [PubMed] [Google Scholar]
- 13.Rajesh M, Mukhopadhyay P, Batkai S, Patel V, Saito K, Matsumoto S, Kashiwaya Y. Cannabidiol attenuates cardiac dysfunction, oxidative stress, fibrosis, and inflammatory and cell death signaling pathways in diabetic cardiomyopathy. J Am Coll Cardiol 2010, 56: 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salloum FN, Chau VQ, Hoke NN, Abbate A, Varma A, Ockaili RA, Toldo S et al. . Phosphodiesterase-5 inhibitor, tadalafil, protects against myocardial ischemia/reperfusion through protein-kinase g-dependent generation of hydrogen sulfide. Circulation 2009, 120: S31–S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YC, Leu S, Sun CK, Yen CH, Kao YH, Chang LT, Tsai TH et al. . Early combined treatment with sildenafil and adipose-derived mesenchymal stem cells preserves heart function in rat dilated cardiomyopathy. J Transl Med 2010, 8: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun D, Huang J, Zhang Z, Gao H, Li J, Shen M, Cao F et al. . Luteolin limits infarct size and improves cardiac function after myocardium ischemia/reperfusion injury in diabetic rats. PLoS One 2012, 7: e33491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun D, Shen M, Li J, Li W, Zhang Y, Zhao L, Zhang Z et al. . Cardioprotective effects of tanshinone IIA pretreatment via kinin B2 receptor-Akt-GSK-3beta dependent pathway in experimental diabetic cardiomyopathy. Cardiovasc Diabetol 2011, 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Luo P, Wang Y, Li W, Wang C, Sun D, Zhang R et al. . Glucagon-like peptide-1 protects against cardiac microvascular injury in diabetes via a cAMP/PKA/Rho-dependent mechanism. Diabetes 2013, 62: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra PK, Tyagi N, Kumar M, Tyagi SC. MicroRNAs as a therapeutic target for cardiovascular diseases. J Cell Mol Med 2009, 13: 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyagi AC, Sen U, Mishra PK. Synergy of microRNA and stem cell: a novel therapeutic approach for diabetes mellitus and cardiovascular diseases. Curr Diabetes Rev 2011, 7: 367–376. [DOI] [PubMed] [Google Scholar]
- 21.Mishra PK, Chavali V, Metreveli N, Tyagi SC. Ablation of MMP9 induces survival and differentiation of cardiac stem cells into cardiomyocytes in the heart of diabetics: a role of extracellular matrix. Can J Physiol Pharmacol 2012, 90: 353–360. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Li TS, Lee ST, Wawrowsky KA, Cheng K, Galang G, Malliaras K et al. . Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One 2010, 5: e12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dispersyn GD, Mesotten L, Meuris B, Maes A, Mortelmans L, Flameng W, Ramaekers F et al. . Dissociation of cardiomyocyte apoptosis and dedifferentiation in infarct border zones. Eur Heart J 2002, 23: 849–857. [DOI] [PubMed] [Google Scholar]
- 24.Huang ZG, Jin Q, Fan M, Cong XL, Han SF, Gao H, Shan Y et al. . Myocardial remodeling in diabetic cardiomyopathy associated with cardiac mast cell activation. PLoS One 2013, 8: e60827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464: 606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsyplenkova VG. Cardiomyocytic dedifferentiation, “hibernation”, and apoptosis are possible factors of progressive diabetic cardiomyopathy. Arkh Patol 2009, 71: 30–33. [PubMed] [Google Scholar]
- 27.Tanaka M, Hirabayashi Y, Sekiguchi T, Inoue T, Katsuki M, Miyajima A. Targeted disruption of oncostatin M receptor results in altered hematopoiesis. Blood 2003, 102: 3154–3162. [DOI] [PubMed] [Google Scholar]
- 28.Hirota H, Chen J, Betz UA, Rajewsky K, Gu Y, Ross J Jr, Muller W et al. . Loss of a gp130 cardiac muscle cell survival pathway is a critical event in the onset of heart failure during biomechanical stress. Cell 1999, 97: 189–198. [DOI] [PubMed] [Google Scholar]
- 29.Hohensinner PJ, Kaun C, Rychli K, Niessner A, Pfaffenberger S, Rega G, Furnkranz A et al. . The inflammatory mediator oncostatin M induces stromal derived factor-1 in human adult cardiac cells. FASEB J 2009, 23: 774–782. [DOI] [PubMed] [Google Scholar]
- 30.Pöling J, Gajawada P, Lörchner H, Polyakowa V, Szibor M, Böttger T, Warnecke H et al. . The Janus face of OSM-mediated cardiomyocyte dedifferentiation during cardiac repair and disease. Cell Cycle 2012, 11: 439–445. [DOI] [PubMed] [Google Scholar]
- 31.Akinleye A, Furqan M, Mukhi N, Ravella P, Liu D. MEK and the inhibitors: from bench to bedside. J Hematol Oncol 2013, 6: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]