Abstract
Nosema bombycis is an obligate intracellular parasitic fungus that utilizes a distinctive mechanism to infect Bombyx mori. Germination, an indispensible process through which microsporidia infect the host cells, is regarded as a key developmental turning point for microsporidia from dormant state to reproduction state. Thus, elucidating the transcriptome changes before and after germination is crucial for parasite control. However, the molecular basis of germination of microsporidia remains unknown. To investigate this germination process, the transcriptome of N. bombycis ungerminated spores and germinated spores were sequenced and analyzed. More than 60 million high-quality transcript reads were generated from these two groups using RNA-Seq technology. After assembly, 2756 and 2690 unigenes were identified, respectively, and subsequently annotated based on known proteins. After analysis of differentially expressed genes, 66 genes were identified to be differentially expressed (P ≤ 0.05) between these two groups. A protein phosphatase-associated gene was first identified to be significantly up-regulated as determined by RNA-Seq and immunoblot analysis, indicating that dephosphorylation might potentially contribute to microsporidia germination. The DEGs that encode proteins involved in glycometabolism, spore wall proteins and ricin B lectin of N. bombycis were also analyzed. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes analyses revealed genes responsible for some specific biological functions and processes. The datasets generated in this study provide a basic characterization of the transcriptome changes in N. bombycis during germination. The analysis of transcriptome data and identification of certain functional genes which are robust candidate genes related to germination will help to provide a deep understanding of spore germination and invasion.
Keywords: microsporidia, Nosema bombycis, germination, transcriptome, dephosphorylation
Introduction
Microsporidia are obligate intracellular parasitic fungi with over 1300 species in ∼160 genera [1]. Most microsporidia are major pathogens of vertebrate and invertebrate. They can infect immunocompromised humans and are also important pathogens of silkworm [2]. Nosema bombycis, the first reported microsporidium, can transmit a highly fatal disease referred to as pébrine, which caused huge economic losses in the silk-producing industry in most nations around the world during the mid-nineteenth century [3] and still remains epidemic. N. bombycis infects silkworms through horizontal and vertical transmission. The infection of N. bombycis could result in continuous proliferation, chronically damaging the entire tissues and organs of the worm (including intestines, silk glands, muscles, and Malpighian tubules), ultimately leading to the death of the worm. The unusual lifecycle of microsporidia can be divided into three phases, namely the infective phase, the proliferative phase, and the sporogony phase. The unique mechanism through which microsporidia enter the host cells has been of particular interest. To survive outside of their hosts as environmentally resistant spores, microsporidia are protected by a thick two-layered wall [4]. These mature spores which are not germinated are considered to be dormant spores. The morphology and histology of dormant spores have already been investigated [4,5]. Once coming into contact with a host cell or triggered by appropriate stimuli, the spores rapidly evert the polar tube, penetrate the host cell membrane, and transfer the infective sporoplasm into the host cytoplasm where proliferation and the next round of spore production occur [6].
It is well accepted that germination is the first step in the infection of host cells by N. bombycis. Thus, spore germination may play a major role in host invasion, and the elucidation of spore germination will provide some valuable information for understanding the whole invasion process. Some genomes of important microsporidia have been reported and analyzed [7–9]. Based on the understanding of the microsporidia genome, transcriptome studies have been widely carried out [9–11], which generated information on the changes of total mRNA during a particular period. The genome of N. bombycis was published in 2013, and studies have shown that the genome of N. bombycis is expanded, rather than compact as some other parasitic microsporidian genomes are through several common molecular mechanisms [12]. Other studies on N. bombycis have mainly focused on its classification [13,14], detection and control [15,16], as well as its metabolism [17,18]. There are only a few findings regarding the genes that play a role in the germination process of microsporidia.
With the establishment of a method for the in vitro germination of microsporidia [19,20], researchers can efficiently observe how spores germinate and infect the host cells. It is universally accepted that an appropriate stimulus, such as pH, ionic species, osmotic pressure, temperature, and radiation [21–23], can make spores germinate, however the mechanism have not been fully elucidated. An increase in the intrasporal osmotic pressure can make spores germinate, and the change in the intracellular osmotic pressure has been proven to be associated with some proteins such as aquaporins (AQP) [24,25]. Many studies have shown that protease, such as metalloprotease and subtilisin-like protease, may play some roles in the germination and invasion processes [26–28]. Some spore wall proteins (SWPs) have recently been identified and proven to be related to germination and invasion. Spore phagocytosis assays indicated that NbSWP5 can protect spores from phagocytic uptake by cultured insect cells to modulate host cell invasion [29]. Another study showed that SWP5 is a spore wall protein that can interact with the polar tube during the germination process [30]. An IG-like protein denoted as BmTLP from Bombyx mori was proven to interact with SWP26 which is a spore wall protein involved in endospore formation, host cell adherence and infection in vitro [31,32]. It is possible that SWPs, as receptors, trigger spore germination under appropriate conditions after stimulation, but this needs to be further confirmed. Intrasporal sugars in some microsporidia change during germination [33,34]. However, the results of previous studies do not fully elucidate the pathway underlying the germination process.
Germination, which is regarded as the first step in host cell invasion from the dormant state, includes a series of changes at the biological level. An in-depth understanding of germination could provide some useful insights into the process of defense against microsporidia invasion. Furthermore, clarification of the unique invasion mode is also of interest. However, as a result of the lack of a system for genetic manipulation in microsporidia, it is extremely challenging to understand how spores germinate. Using high-throughput sequencing (HTS), we first investigated and compared the transcriptomes of N. bombycis ungerminated (UGS) and germinated spores (GS). Our results identified a number of DEGs after spore germination, and interestingly, protein dephosphorylation was found to be related to germination in microsporidia.
Materials and Methods
Purification and in vitro germination of microsporidia
N. bombycis spores were propagated and purified from laboratory-reared infected silkworm larvae as previously described [35]. Briefly, third instar molted silkworm larvae were fed with mulberry leaves that had been artificially contaminated with N. bombycis spores (∼3 × 104 spores per larvae). Heavily infected midguts and silk glands of fifth instar larvae at the fifth day after molt were collected and gently homogenized in physiological saline. Large silkworm debris was removed by filtration through four layers of cheesecloth. After centrifugation (3000 g, 5 min), the pellet was resuspended in 15 ml of a solution containing 0.05% Saponin and 0.05% (v/v) Triton X-100 in phosphate-buffered saline (PBS). The cell suspension was passed through a plastic syringe with an inserted wad of glass wool. A crude spore suspension was obtained by differential centrifugation after most of the contaminant was removed. The parasites were further purified through a continuous Percoll (pH 7.2; GE Healthcare, Milwaukee, USA) gradient centrifugation (46,000 g, 60 min). After washes with distilled water, the spore purity was examined by phase-contrast microscopy (Leica, Berlin, Germany). A total of 5 × 108 purified spores were divided into two equal parts and washed three times with PBS. The ungerminated control group (named UGS) was then treated with 200 µl of physiological saline. The group to be germinated (named GS) was treated with 200 µl of glycine-KOH-KCl germination buffer (0.05 M glycine, 0.05 M KOH, and 0.375 M KCl, pH 10.5), and both the UGS and GS groups were incubated at 30°C for 50 min. Germination was verified instantly by phase-contrast microscopy. The levels of germination in different biological replications were close to 70%. The in vitro germination experiments were performed in three independent biological replications.
RNA preparation for sequencing and sequence assembly
Total RNA was extracted from each sample using Trizol reagent (Invitrogen, Carlsbad, USA) and acid-washed glass beads (diameter: 425–600 µm; Sigma, St Louis, USA) in a Precellys-24 homogenizer (Bertin Technologies, Aix en Provence, France). The quantity and quality of all RNA samples were measured by gel electrophoresis and analyzed by 2100 Bioanalyser RNA Nanochip (Agilent, Palo Alto, USA). Samples with RNA integrity number (RIN) values of >8.0 were used in the subsequent analysis. Library construction and RNA-Seq were performed by the Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). Briefly, after DNase I treatment for 1 h at 37°C, PolyA RNA was isolated from 5 mg of total RNA, and the TruSeq libraries were prepared using the Illumina TruSeq™ RNA Sample Prep Kit (Illumina, San Diego, USA). Fifteen cycles of PCR were run to amplify the libraries, and the samples from the TruSeq libraries were loaded onto a flow cell and run on the HiSeq 2000 instrument using a paired-end 100-bp run. Three independent library constructions were performed for the GS and UGS groups, respectively. The raw Illumina reads obtained through RNA sequencing were filtered with SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) to remove the adapter sequences and low-quality reads (with a quality score <20 or a read length <20 bp). The filtered reads were first mapped with the N. bombycis genome database (http://silkpathdb.swu.edu.cn/silkpathd) to remove the non-target reads using the Tophat software (http://tophat.cbcb.umd.edu/) with a cut-off E-value of 1 × 10−5. The completely matched reads were then assembled using Cufflinks (http://cufflinks.cbcb.umd.edu/).
Functional annotation and analysis of UGS and GS transcriptome
To deduce their putative functions, the transcripts were subject to BLASTx analysis against the Non-Redundant (NR) Protein database (http://www.ncbi.nlm.nih.gov/) with a cut-off of 1 × 10−5. All of the transcripts were also annotated according to gene ontology (GO) terms using the Blast2Go program [36], which assigns GO terms describing biological process, molecular function, and cellular component to the query sequences. To gain an overview of the gene pathway networks, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was performed using the online KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/kegg/kaas/) [37]. The bidirectional best hit (BBH) method was used to obtain KEGG Orthology assignments for UGS and GS.
Differential gene expression analysis and clustering
To search for genes potentially involved in germination, the relative expression abundances for each group, which were calculated using the fragments per kilobase of per million mapped reads (FPKM), were compared between the two groups (UGS and GS). Differential expression analysis was conducted using the Tophat (http://tophat.cbcb.umd.edu/) and Cuffdiff (http://cufflinks.cbcb.umd.edu/) tools. Furthermore, the transcripts that were differentially expressed in comparison with the normalized expression values were visualized in a clustering heat map, which was constructed using the hcluster distance method associated with complete linkage.
Validation by qRT-PCR analysis
To validate the RNA-Seq data, 15 randomly selected candidate DEGs were subject to quantitative real-time PCR (qRT-PCR) analysis. Fifteen primers were designed using the Primer5 software (Premier Biosoft International) and are listed in Supplementary Table S1. Aliquots of the total RNA extracted for sequencing as described earlier were used for the qRT-PCR experiments according to the manufacturer's instructions (Roche, Basel, Switzerland), and the data were analyzed using the comparative CT method [38]. Each assay was performed in triplicate biological replicates and technical replicates. Each reaction was conducted in a volume of 20 µl using the Roche LightCyler 480 system (Roche). The β-tubulin gene was used as a reference in all of the qRT-PCR experiments. The expression level of each target gene was normalized to the level of β-tubulin. The relative expression values for the RNA-Seq data were then validated. The fold change of each gene was calculated by the 2−ΔΔCT method. The fold change of each gene was determined by three independent quantitative PCR amplifications of RNA extracted independently.
Protein isolation and western blot analysis
The total proteins from 1.2 × 109 UGS or GS were extracted with 500 µl of protein lysis buffer [100 mmol Tris–HCl, 4% sodium dodecyl sulfate (SDS) (w/v), and 0.05 mol dithiothreitol (DTT), pH 7.6] containing protease inhibitor cocktail (Sangon, Shanghai, China), and the protein concentration was determined with the Enhanced BCA Protein Assay Kit (Beyotime, Haimen, China). Then, 30 µg of protein from each group was subject to 10% SDS-PAGE and electroblotted onto a pure nitrocellulose blotting membrane (Pall Corporation, Port Washington, USA). The membrane was blocked for 60 min and the incubated overnight at 4°C with the first antibody (anti-phosphoserine antibody; Millipore, Billerica, USA). Then, the membrane was washed and incubated for 60 min with a horseradish peroxidase (HRP)-conjugated second antibody. The proteins were detected using enhanced chemiluminescence (ECL) kit (Thermo Scientific, Waltham, USA). Then the membranes were stripped using Restore™ Western Blot Stripping Buffer (Thermo Scientific), and the expression of the β-actin protein was determined. The band densities were measured using the software Quantify One (Bio-Rad, Hercules, USA).
Results
Comparison of UGS and GS and sequencing analysis of the N. bombycis transcriptome
The UGS and GS were observed with a phase-contrast microscope. The bright and smooth spores (Fig. 1A, arrow) were UGS, whereas the dark spores (Fig. 1B, arrow) were GS. In this study, whole genome mRNA sequencing was used to globally investigate the gene expression in the UGS and GS of N. bombycis using the HiSeq Illumina platform. After the sequencing, 32,284,160 and 32,755,830 reads (total numbers from three biological replicates) with an average length of 101 bp were obtained for the UGS and GS groups, respectively. After removing the adaptor sequences and filtering out the low-quality reads at high stringency using the SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) software, 31,969,334 and 32,457,679 high-quality reads were obtained for the UGS and GS groups. The sequence data generated in this study were deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) with the accession number SRP049542.
Figure 1.

Morphological analysis of N. bombycis using a phase-contrast microscope (A) UGS (arrow). (B) GS (arrow). Scale bar = 14 μm.
All of the high-quality reads were first mapped with the N. bombycis genome database (http://silkpathdb.swu.edu.cn/silkpathd) using the Tophat software (http://tophat.cbcb.umd.edu/) with a cut-off E-value of 1 × 10−5, and the mapped rate for UGS and GS reached 81.23% and 80.62%, respectively. Subsequently, the completely matched reads were assembled into unigenes using the Cufflinks software (http://cufflinks.cbcb.umd.edu/), which resulted in a total of 5446 unigenes with an average size of 1070 bp. The size distribution of the unigenes is shown in Fig. 2. All of the unmatched reads were excluded from further analysis. The summary of statistics for the N. bombycis transcriptome sequencing and assembly are shown in Table 1.
Figure 2.
Size distribution of unigenes
Table 1.
Summary of the results from the Illumina sequencing of the N. bombycis transcriptome
| UGS | GS | Total | |
|---|---|---|---|
| Number of raw reads 1a | 10,687,694 | 10,728,496 | 21,416,190 |
| Number of raw reads 2a | 10,877,295 | 10,915,671 | 21,792,966 |
| Number of raw reads 3a | 10,719,171 | 11,111,663 | 21,830,834 |
| Total length (bp) | 3,260,700,160 | 3,308,338,830 | 6,569,038,990 |
| Total number of high-quality reads after removing low-quality reads | 31,969,334 | 32,457,679 | 64,427,013 |
| Total length (bp) of high-quality reads | 3,173,730,123 | 3,223,483,816 | 6,397,213,939 |
| Total number of matched reads | 25,759,708 (81.23%) | 25,973,529 (80.62%) | 51,733,237 |
| Unigenes | 2756 | 2690 | 5446 |
aRaw reads 1, raw reads 2, and raw reads 3 represent three independent biological replications.
Gene annotation and differentially expressed gene analysis
The N. bombycis genome database has been submitted to the GenBank by the State Key Laboratory of Silkworm Genome Biology in Southwest University [12]. Therefore, all of the 2756 unigenes obtained after sequence assembly were annotated using the BLASTx program in the NR Protein database. Annotation using the BLASTx program was also conducted using some other protein databases, such as Nucleotide Sequence Database (NT), UniProt and String, to obtain supplementary gene function information. Approximately 2265 unigenes were successfully annotated in the NR Protein database with an E-value of 1 × 10−5. The sequences and annotation information of all of the unigenes are listed in Supplementary Dataset 01 and Supplementary Table S2. Supplementary Table S2 summarizes all the 2756 unigenes' identity (ID) and location in N. bombycis genome database. The count and FPKM for each unigene in Supplementary Table S2 were used to calculate the differential gene expression between UGS and GS. Supplementary Table S2 also lists the results of differentially expressed gene analysis and the detailed function annotation for all the 2756 unigenes. A total of 2265 (82.18%) and 2215 (82.34%) annotated unigenes were obtained for UGS and GS, respectively. In addition, 43 unigenes could not be matched in the NR Protein database, but were annotated with the NT, UniProt, or String Protein databases. The remaining unigenes were not annotated.
After annotation using the NR Protein database, the most possible biological functions of the unigenes were predicted. Analysis of the differences between UGS and GS in terms of gene expression and function will provide information of the biological changes that occur during germination. The DEGs were identified using the fragments per kilobase of exon model per million mapped reads (FPKM) to measure the gene expression levels. Transcriptome comparison revealed 66 differentially expressed genes (DEGs, P≤ 0.05, |logFC|≥1), of which 57 were down-regulated and nine were up-regulated in UGS compared with those in GS. Approximately 82% of these genes (49 down-regulated and 5 up-regulated genes) were annotated using the NR Protein database (Table 2). A number of un-annotated unigenes that are worthy of further investigation were also identified because their expression level changed significantly after spore germination. To confirm the DEGs obtained from the transcriptome analysis, qRT-PCR was performed to determine the differential expression levels of 15 randomly selected genes (Fig. 3). The expression patterns of all the selected genes measured by RNA-Seq and qRT-PCR were similar, and the results are consistent with the DEGs analysis. These genes encode proteins with functions associated with the polar tube, spore wall, glycometabolism, post-translational modifications (PTMs) and other functions.
Table 2.
Genes with differential transcriptional expression between UGS and GS
| ID | P-value | Annotation | FPKM in NGS | FPKM in GS |
|---|---|---|---|---|
| CUFF.321.1 | 1 × 10−7 | Hypothetical protein NBO_1272gi001, partial [Nosema bombycis CQ1] | 9.78 | 1.56 |
| CUFF.1046.1 | 5 × 10−7 | 60S ribosomal protein L6, partial [Nosema bombycis CQ1] | 13.3 | 2.73 |
| CUFF.303.1 | 8 × 10−7 | 60S ribosomal protein L6, partial [Nosema bombycis CQ1] | 29.3 | 5.28 |
| CUFF.17.1 | 1 × 10−5 | Polar tube protein 3 [Nosema bombycis CQ1] | 2.01 | 0.284 |
| CUFF.840.1 | 0.0001 | No annotation | 230 | 52.4 |
| CUFF.1727.1 | 0.0002 | Hypothetical protein NBO_48g0012 [Nosema bombycis CQ1] | 7.08 | 1.05 |
| CUFF.2690.1 | 0.0003 | Polar tube protein 1 [Nosema bombycis CQ1] | 4.56 | 0.516 |
| CUFF.2332.1 | 0.0004 | Polar tube protein 1 [Nosema bombycis CQ1] | 4.46 | 0.820 |
| CUFF.1729.1 | 0.0004 | Hypothetical protein NBO_48g0007 [Nosema bombycis CQ1] | 12.6 | 3.65 |
| CUFF.2396.1 | 0.0006 | Protein peanut [Nosema bombycis CQ1] | 5.81 | 0.360 |
| CUFF.21.1 | 0.0006 | MADS domain containing protein, partial [Nosema bombycis CQ1] | 2.27 | 0.223 |
| CUFF.1867.1 | 0.001 | DNA replication fork-blocking protein FOB1, partial [Nosema bombycis CQ1] | 2.65 | 0 |
| CUFF.1438.1 | 0.001 | Polar tube protein 3 [Nosema bombycis CQ1] | 2.46 | 0.696 |
| CUFF.1656.1 | 0.003 | Protein of unknown function GLTT, partial [Nosema bombycis CQ1] | 0.948 | 0 |
| CUFF.2325.1 | 0.004 | No annotation | 0.938 | 0 |
| CUFF.231.1 | 0.005 | Hypothetical protein NBO_72g0014 [Nosema bombycis CQ1] | 117 | 50.0 |
| CUFF.943.1 | 0.005 | Glutamate NMDA receptor-associated protein 1 [Nosema bombycis CQ1] | 4.14 | 0.904 |
| CUFF.1585.1 | 0.01 | Midasin [Nosema bombycis CQ1] | 13.8 | 6.12 |
| CUFF.673.1 | 0.01 | Pre-mRNA-splicing factor spp42, partial [Nosema bombycis CQ1] | 4.01 | 1.73 |
| CUFF.956.1 | 0.01 | Laminin subunit beta-4 [Nosema bombycis CQ1] | 30.8 | 13.8 |
| CUFF.1848.1 | 0.01 | glucose-6-phosphate isomerase [Nosema bombycis CQ1] | 4.62 | 1.68 |
| CUFF.839.1 | 0.01 | pol polyprotein [Nosema bombycis] | 2.29 | 0 |
| CUFF.1025.1 | 0.01 | hypothetical protein NBO_29g0030 [Nosema bombycis CQ1] | 10.9 | 4.64 |
| CUFF.1841.1 | 0.01 | Hypothetical protein NCER_102284 [Nosema ceranae BRL01] | 0.932 | 0 |
| CUFF.1912.1 | 0.01 | Hypothetical spore wall protein [Nosema bombycis] | 8.46 | 0.566 |
| CUFF.1897.1 | 0.01 | hypothetical protein NBO_546g0002 [Nosema bombycis CQ1] | 8.19 | 3.40 |
| CUFF.1151.1 | 0.02 | Microtubule-associated protein 1A [Nosema bombycis CQ1] | 2.08 | 0.194 |
| CUFF.2402.1 | 0.02 | No annotation | 2.49 | 0 |
| CUFF.1781.1 | 0.02 | Nuclear transcription factor Y subunit B-2 [Nosema bombycis CQ1] | 7.51 | 2.25 |
| CUFF.2140.1 | 0.02 | Hypothetical protein NBO_613g0002 [Nosema bombycis CQ1] | 1.22 | 0 |
| CUFF.2377.1 | 0.02 | No annotation | 1.45 | 0 |
| CUFF.1341.1 | 0.02 | No annotation | 1.54 | 0 |
| CUFF.2503.1 | 0.02 | Pol polyprotein [Nosema bombycis] | 6.67 | 0 |
| CUFF.2331.1 | 0.02 | Polar tube protein 2 [Nosema bombycis CQ1] | 3.88 | 0.305 |
| CUFF.2008.1 | 0.02 | Transketolase 1, partial [Nosema bombycis CQ1] | 2.51 | 0.570 |
| CUFF.1536.1 | 0.03 | Vacuolar ATP synthase catalytic subunit A [Nosema bombycis CQ1] | 0.884 | 0 |
| CUFF.456.1 | 0.03 | Hypothetical protein TcasGA2_TC003734 [Tribolium castaneum] | 1.48 | 0 |
| CUFF.909.1 | 0.03 | Spore wall protein 5 [Nosema bombycis CQ1] | 4.54 | 0.831 |
| CUFF.2082.1 | 0.03 | Hypothetical protein NBO_6g0002 [Nosema bombycis CQ1] | 1.96 | 0.399 |
| CUFF.1162.1 | 0.03 | No annotation | 1.05 | 0 |
| CUFF.1687.1 | 0.03 | Ricin B lectin [Nosema bombycis CQ1] | 0.866 | 0 |
| CUFF.1778.1 | 0.03 | DNA-directed RNA polymerase [Nosema bombycis CQ1] | 1.21 | 0 |
| CUFF.2029.1 | 0.03 | Ricin B lectin [Nosema bombycis CQ1] | 0.691 | 0 |
| CUFF.525.1 | 0.03 | No annotation | 1.79 | 0 |
| CUFF.872.1 | 0.03 | Hypothetical protein NBO_27g0003 [Nosema bombycis CQ1] | 9.72 | 0.622 |
| CUFF.753.1 | 0.04 | Mannose-1-phosphate guanyltransferase 2 [Nosema bombycis CQ1] | 1.93 | 0.335 |
| CUFF.687.1 | 0.04 | Chromosome segregation protein [Nosema bombycis CQ1] | 20.7 | 7.47 |
| CUFF.1326.1 | 0.04 | Hypothetical protein [Plasmodium yoelii yoelii 17XNL] | 17.4 | 0 |
| CUFF.1490.1 | 0.04 | Transcription factor steA [Nosema bombycis CQ1] | 2.15 | 0 |
| CUFF.1884.1 | 0.04 | No annotation | 1.55 | 0 |
| CUFF.1340.1 | 0.05 | Hypothetical protein NBO_38g0001 [Nosema bombycis CQ1] | 2.76 | 0 |
| CUFF.256.1 | 0.05 | General transcription factor II-I repeat domain-containing protein 2A-like [Oreochromis niloticus] | 1.34 | 0 |
| CUFF.56.1 | 0.05 | Hypothetical protein NCER_102516 [Nosema ceranae BRL01] | 4.67 | 0 |
| CUFF.72.1 | 0.05 | Hypothetical protein NBO_1040g0001 [Nosema bombycis CQ1] | 1.06 | 0 |
| CUFF.935.1 | 0.05 | Pol polyprotein [Nosema bombycis] | 1.93 | 0 |
| CUFF.688.1 | 0.05 | Structural maintenance of chromosomes protein 1 [Nosema bombycis CQ1] | 40.5 | 16.8 |
| CUFF.1981.1 | 0.05 | Alanyl-tRNA synthetase, mitochondrial [Nosema bombycis CQ1] | 1.85 | 0.760 |
| CUFF.448.1 | 0.001 | Nucleoporin NUP170 [Nosema bombycis CQ1] | 41.9 | 108 |
| CUFF.1882.1 | 0.007 | Hypothetical protein NBO_54g0012 [Nosema bombycis CQ1] | 9.01 | 20.9 |
| CUFF.2629.1 | 0.01 | No annotation | 9.50 | 27.6 |
| CUFF.885.1 | 0.01 | Hypothetical protein NBO_399g0001 [Nosema bombycis CQ1] | 11.6 | 23.3 |
| CUFF.631.1 | 0.02 | No annotation | 2.61 | 7.94 |
| CUFF.100.1 | 0.02 | No annotation | 185 | 377 |
| CUFF.2608.1 | 0.03 | Hypothetical protein NBO_857g0001 [Nosema bombycis CQ1] | 3.63 | 7.66 |
| CUFF.2474.1 | 0.04 | No annotation | 3.30 | 8.49 |
| CUFF.1525.1 | 0.05 | Protein phosphatase PP2-A regulatory subunit A [Nosema bombycis CQ1] | 1.18 | 2.73 |
Figure 3.
The relative expression levels of 15 randomly selected genes in two groups (UGS and GS) measured by qRT-PCR and RNA-seq The results of all the selected genes are similar to those measured by RNA-seq. The internal control gene is β-tubulin. For qRT-PCR, the fold change of each gene was calculated by the 2−ΔΔCT method. For RNA-seq, the fold change of each gene was calculated by the FPKM. The fold change for each gene was determined by three independent quantitative PCR amplifications of RNA extracted independently and three independent RNA-seqs. Data were presented as mean ± SD from three independent biological replications. PTP2, polar tube protein 2; PTP3, polar tube protein 3; MAP1, microtubule-associated protein 1A; TKL1, Transketolase 1; NUP170, Nucleoporin NUP170; GNRP, glutamate NMDA receptor-associated protein 1; SWP5, spore wall protein 5; G6PI, glucose-6-phosphate isomerase; PP2A, protein phosphatase PP2-A regulatory subunit A; M1PG, mannose-1-phosphate guanyltransferase 2; NTFY, nuclear transcription factor Y subunit B-2; ATRS: alanyl-tRNA synthetase; PP, protein peanut; 60RP, 60S ribosomal protein L6; MADS, MADS domain containing protein.
Clustering and functional enrichment of DEGs
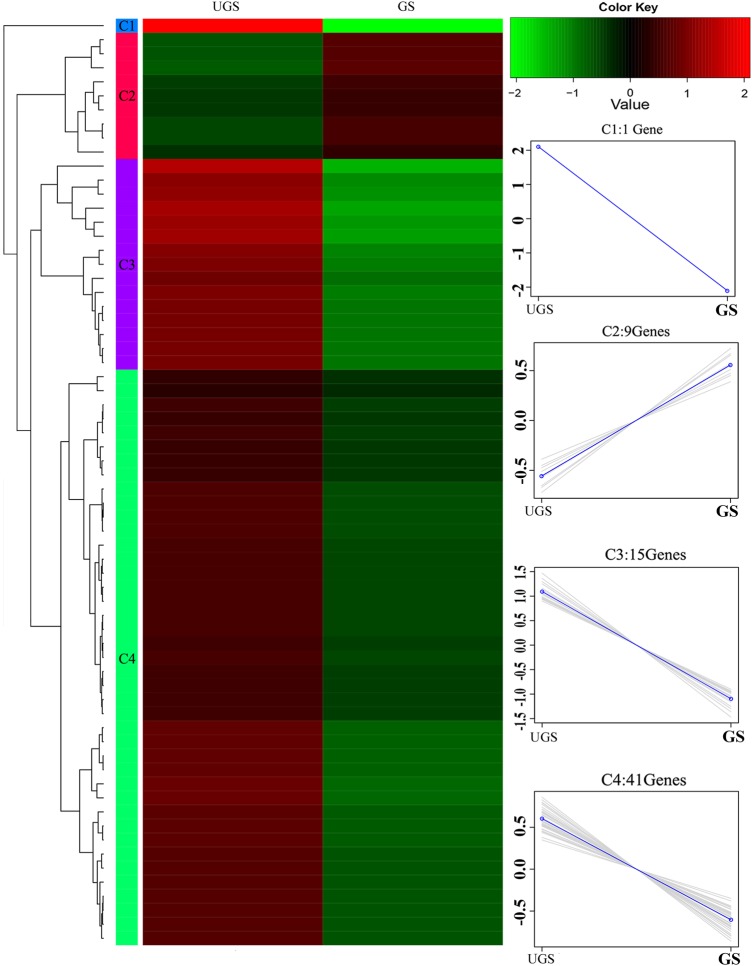
Hierarchical clustering of the 66 DEGs using the cluster distance method associated with complete linkage was performed to further illustrate the relationships between DEGs with various expression patterns (Fig. 4). These DEGs were mainly divided into four clusters. The red color represents the up-regulated genes, and the green color represents the down-regulated genes. The linear tendency of the four clusters was obtained to summarize the expression patterns. According to the expression patterns, cluster 1 (C1) which includes the genes that exhibited the most significant difference in expression was annotated as hypothetical proteins with unknown function, however unigenes in this cluster (CUFF.1326.1) might represent a valuable subject because of their marked down-regulation after germination; cluster 2 (C2) contains nine DEGs that were mainly up-regulated after germination; whereas the other two clusters were all down-regulated.
Figure 4.
Hierarchical clustering of DEGs Heatmap of DEGs The four main clusters are shown, the expression values for the UGS and GS sets are presented, and the DEGs with decreased (green) and increased (red) expression in the different groups are shown. Expression patterns of genes in the four main clusters, namely C1–C4, corresponding to the hierarchical heatmap are shown on the right.
To identify the functional category of the annotated genes, GO was employed to classify the transcripts annotated as known proteins (Supplementary Tables S3 and S4). The mapping of 2756 unigenes in GO Consortium (http://geneontology.org/) is shown in Supplementary Table S3. Supplementary Table S4 is the summary of GO level 2 classification. In total, 841 of the 2756 (31%) unigenes were assigned one or more GO terms for a total of 4400 GO assignments. The distribution of the unigenes in different GO categories is shown in Fig. 5. It was found that 1650 records were annotated with a cellular component, whereas 1034 were annotated with a molecular function, and 1716 were annotated with a biological process. Among the various biological process categories, ‘Binding’ and ‘Catalytic Activity’ were dominantly represented within the ‘Molecular Function’ category, up to 56% and 51%, respectively. The ‘Cellular Process’ and ‘Metabolic Process’ categories accounted for 59% and 60% within the ‘Biological Process’ category, respectively. Within the ‘Cellular Component’ category, 48% of the genes were located in the ‘Cell and Cell Part’ categories. In the ‘Cellular Process’ (GO:0009987) and ‘Catalytic Activity’ (GO:0003824) categories, some important genes were found, such as trehalose-phosphate synthase, glucose transporter, and trehalase. These three proteins which usually play a key role in the synthesis and transport of carbohydrate molecules may contribute to the intrasporal osmotic pressure. From the GO assignments for the ‘Membrane’ (GO:0016020) categories, the aquaporin protein was identified. The aquaporin protein transports water through the member and regulates the intrasporal osmotic pressure. The change of intrasporal osmotic pressure will affect the spore germination process. These proteins were detected in the GS, suggesting their potential role in germination of N. bombycis.
Figure 5.
Gene ontology classification of UGS and GS transcriptome
To further investigate the functional classification and pathway assignments, pathway-based analysis was conducted based on the KEGG Pathway database using Blastx/Blastp 2.2.24+. The mapping of 2756 unigenes in Kyoto Encyclopedia of Genes and Genomes (http://www.kegg.jp/) are shown in Supplementary Table S5. The summary of unigenes mapped in each pathway are presented in Supplementary Table S6. The results showed that a total of 925 genes from the UGS and GS transcriptome were assigned to 202 KEGG pathways. After KEGG enrichment analysis (Supplementary Table S7), the most enriched pathways, i.e. the pentose phosphate pathway as well as the amino sugar and nucleotide sugar metabolism pathway, had the lowest P-values. Some glycometabolism-associated pathways, such as fructose and mannose metabolism, were also identified (P> 0.05). Some interesting and potentially valuable pathways involved in calcium regulation were also found in the transcriptome. Calcium regulation was considered to be involved in the response to osmotic pressure change. All the assigned KEGG pathway diagrams are shown in Supplementary Dataset 02.
Prediction of candidate genes involved in biological changes during germination
At present, the knowledge of the biological changes that occur during germination in microsporidia, particularly in N. bombycis, remains limited. Furthermore, germination is the most critical process in N. bombycis spore invasion because it is the first step of the process in which an infective sporoplasm enters the host cell. In contrast to other molecular biology experimental tools, transcriptome analysis and selection which are bioinformatics tools will provide some new information that may help clarify this process. Some interesting and valuable candidates underlying the biological changes during germination are described in the following paragraphs.
Protein phosphorylation is the major mechanism for the regulation of protein activity. Most proteins in the cell can be directly or indirectly regulated by this mechanism. In our DEGs analysis, a unigene (CUFF.1525.1) that was annotated as protein phosphatase PP2-A regulatory subunit A was found to be significantly up-regulated after germination (P< 0.05), reflecting an increase in the overall protein dephosphorylation levels. The total proteins from UGS and GS were subject to western blot analysis using anti-phosphoserine antibody. The results showed that the phosphorylated protein levels with a molecular weight of ∼120 kDa were markedly decreased after germination (Fig. 6). This western blot analysis results further confirmed that the level of dephosphorylation was indeed increased significantly (P< 0.05).
Figure 6.
Detection of phosphorylated protein levels in the UGS and GS samples of N. bombycis The level of dephosphorylation was increased significantly after germination (*P< 0.05). The β-actin protein and target protein were quantified using Quantify One software. Data were presented as the mean ± SD from three independent biological replications. Statistical analyses were performed using the Statistical Package for the Social Sciences, version 13.0. Results were analyzed using one-way ANOVA followed by Student's t-test. A P-value <0.05 was considered statistically significant.
It has been reported that the increase in osmotic potential caused by carbohydrate catabolism appears to play a role in the germination of N. algerae [33]. In this study, three annotated glycometabolism-associated unigenes (CUFF.1848.1, CUFF.2008.1, and CUFF.753.1) were found to be significantly down-regulated after germination. The P-value of one unigene (CUFF.1848.1) was found to be 0.0076. The detailed annotation and statistical information of these three unigenes are shown in Table 2. The DEGs analysis revealed that eight cytoarchitecture-associated unigenes (Table 2) which were mainly annotated as SWPs and polar tube proteins were significantly down-regulated after germination. Three of these were significantly down-regulated compared with the UGS group (P <0.01). Other interesting unigenes (Table 2) that were annotated as ricin B lectin were also significantly down-regulated after germination.
Discussion
The investigation of changes in transcription during germination, a key turning point from the dormant to the reproduction state, may provide a deeper understanding of the invasion mechanism and make a contribution to the development of parasite control strategies in the silkworm industry. Most studies have shown that some biological changes during germination are associated with AQP [24,25], SWP [29–31], polar tube protein [30,39,40], protease [26–28], carbohydrate conversion [33,34,41], and so on. Biological control against microsporidia depending on the mechanism of spore germination is far from successful because of the lack of knowledge on the key proteins involved in germination. High-throughput mRNA sequencing (RNA-Seq) offers the possibility to measure the transcript expression levels in different samples using a single assay [42–44]. Compared with the transcriptome analysis of Encephalitozoon cuniculi conducted in 2013 [10], these researchers obtained ∼9–17 million reads at three post-infection time-points with a genome size of 2.9 Mbp. In this study, we produced an N. bombycis reference transcriptome consisting of ∼30 million high-quality reads with a genome size of 15.7 Mbp. All of these reads were then assembled into 2756 transcripts, and information of N. bombycis transcriptome in the germination process could be obtained.
As a process that is equally important as phosphorylation, dephosphorylation is also considered as a major protein-regulation mode [45–49]. A unique protein phosphatase with Kelch-like domains (PPKL) in Plasmodium was demonstrated to modulate ookinete morphology, motility, and invasion [50,51], and ‘Shewanella-like’ phosphatases (Shelphs) in Plasmodium falciparum were found to be located in the apical complex of maturing schizonts [52], which is necessary for invasion [53]. In Trichomonas vaginalis, protein phosphatase 1 gamma (TvPP1c) was proven to play a critical role in the proliferation and attachment of the parasite to a mammalian cell [54]. Evidence of the roles of protein phosphatases in the invasion and reproduction of parasite in the host cells has been reported, but the mechanism remains unknown, particularly for microsporidia. Our study showed that protein phosphatase PP2-A of N. bombycis functions after spore germination. The change of overall protein dephosphorylation levels may function in the spore germination process. Protein phosphatase may take part in the invasion of N. bombycis because germination of N. bombycis is regarded as the first step in host cell infection. Some proteins that regulate and control spore germination may be dephosphorylated by protein phosphatase PP2-A and lose their biological function in maintaining spore dormancy. Our results provide some new insights into the mechanism underlying spore germination.
According to the results from the DEGs analysis and KEGG enrichment analysis, three genes (CUFF.1848.1, CUFF.2008.1, and CUFF.753.1) and one glycometabolism-associated pathway (pentose phosphate pathway) were identified. Some changes in sugars may occur during germination. In some other microsporidia, such as Nosema algerae, Edhazardia aedis, and Vavraia culicis, the total and reducing sugars displayed a marked change during the germination process, and trehalose was converted into reducing sugars. The osmotic potential increase caused by the catabolism of trehalose appears to be sufficient for germination [33,34]. Sugar metabolism may directly or indirectly affect N. bombycis spore germination. SWPs have been proven to be associated with germination and invasion [29–31]. Polar tube proteins (PTPs) are regarded as the most critical cell structural proteins involved in germination and invasion. They act as channels that allow the release of the infectious materials (sporoplasm) into the host cell [55]. Our results showed that SWP5 and three PTPs are significantly down-regulated after germination. Genes encoding PTPs and SWPs in Encephalitozoon cuniculi were remarkably up-regulated from 24 to 48 h and from 24 to 72 h post-infection, respectively. These results indicated that mRNAs which were produced in higher quantity during the sporogony stage are then accumulated in the spores [10]. Our result is in agreement with this research. We infer that these proteins may be accumulated to sufficient quantity in the immature stage and consumed during germination. Thus, the expression level of these genes may not be maintained at a high level.
According to the results of transcriptome analysis, the gene expression level of NbRBL was significantly down-regulated after germination. Two ricin B lectin-like secreted proteins were also identified in the supernatant of germinated Spraguea lophii spores [9]. Some research showed that lectins play diverse roles in parasites such as mediating parasite adhesion to host cell during infection, mediating immune evasion, and triggering different developmental pathways in the parasite [56,57]. Some microsporidian spores could utilize glycosaminoglycans (GAGs) as host cell receptors for adhesion and infection, possibly through the interaction with a major SWP named EnP1 [58]. In Antonospora (Paranosema) locustae, fat bodies from experimentally infected and controlled Locusta migratoria locusts were not decorated with anti-ricin B antibodies as revealed by immunoblotting [59]. Based on these findings, we infer that the gene expression level of NbRBL might be high in dormant state to accumulate NbRBL protein, and that this functional protein might play a role in spore invasion after germination.
In summary, the results of this study provide an overview of the regulations and changes of transcriptome in spore germination and pave the way for future studies on the molecular mechanisms of the complex interactions for microsporidia invasion.
Supplementary Data
Funding
This work was supported by the grants from the Modern Agricultural Industrial Technology System (No. CARS-22-ZJ0202), Zhejiang Sericultural Sci-Tech Innovation Team (No. 2011R50028), and the National Natural Science Foundation of China (No. 31302033).
Supplementary Material
Acknowledgements
We thank Dr Changwei Liu (College of Life Science, Zhejiang University) and Dr Jing Zhao (College of Animal Sciences, Zhejiang University) for their excellent advice in this research.
References
- 1.Keeling P. Five questions about microsporidia. PLoS Pathog 2009, 5: e1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat SA, Bashir I, Kamili AS. Microsporidiosis of silkworm, Bombyx mori L. (Lepidoptera— bombycidae): a review. Afr J Agr Res 2009, 4: 1519–1523. [Google Scholar]
- 3.Becnel JJ, Andreadis TG. Microsporidia in insect. In: Wittner M, Weiss LM eds. The Microsporidia and Microsporidiosis. Washington, DC: ASM Press, 1999, 447–501. [Google Scholar]
- 4.Vavra J, Larsson JIR. Structure of the microsporidia. In: Wittner M, Weiss LM eds. The Microsporidia and Microsporidiosis. Washington, DC: ASM Press, 1999, 7–84. [Google Scholar]
- 5.Haro M, Izquierdo F, Henriques-Gil N, Andres I, Alonso F, Fenoy S, del Aquila C. First detection and genotyping of human-associated microsporidia in pigeons from urban parks. Appl Environ Microbiol 2005, 71: 3153–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schottelius J, Schmetz C, Kock NP, Schuler T, Sobottka I, Fleischer B. Presentation by scanning electron microscopy of the life cycle of microsporidia of the genus Encephalitozoon. Microbes Infect 2000, 2: 1401–1406. [DOI] [PubMed] [Google Scholar]
- 7.Chen YP, Pettis JS, Zhao Y, Liu XY, Tallon LJ, Sadzewicz LD, Li R et al. Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics 2013, 14: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001, 414: 450–453. [DOI] [PubMed] [Google Scholar]
- 9.Campbell SE, Williams TA, Yousuf A, Soanes DM, Paszkiewicz KH, Williams BA. The Genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS Genet 2013, 9: e1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grisdale CJ, Bowers LC, Didier ES, Fast NM. Transcriptome analysis of the parasite Encephalitozoon cuniculi: an in-depth examination of pre-mRNA splicing in a reduced eukaryote. BMC Genomics 2013, 14: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue YJ, Tang XD, Xu L, Yan W, Li QL, Xiao SY, Fu XL et al. Early responses of silkworm midgut to microsporidium infection—a digital gene expression analysis. J Invertebr Pathol 2015, 124: 6–14. [DOI] [PubMed] [Google Scholar]
- 12.Pan G, Xu J, Li T, Xia Q, Liu SL, Zhang G, Li C et al. Comparative genomics of parasitic silkworm microsporidia reveal an association between genome expansion and host adaptation. BMC Genomics 2013, 14: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu HD, Pan G, Luo B, Li T, Yang Q, Vossbrinck CR, Debrunner-Vossbrinck BA et al. Intraspecific polymorphism of rDNA among five Nosema bombycis isolates from different geographic regions in China. J Invertebr Pathol 2013, 113: 63–69. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Pan G, Li T, Huang W, Luo B, Zhou Z. Ultrastructure, chromosomal karyotype, and molecular phylogeny of a new isolate of microsporidian Vairimorpha sp. BM (Microsporidia, Nosematidae) from Bombyx mori in China. Parasitol Res 2012, 110: 205–210. [DOI] [PubMed] [Google Scholar]
- 15.Jyothi N, Patil C, Dass C. Action of carbendazim on the development of Nosema bombycis Naegeli in silkworm Bombyx mori L. J Appl Entomol 2005, 129: 205–210. [Google Scholar]
- 16.Ravikumar G, Urs SR, Prakash NBV, Rao CGP, Vardhana KV. Development of a multiplex polymerase chain reaction for the simultaneous detection of microsporidians, nucleopolyhedrovirus, and densovirus infecting silkworms. J Invertebr Pathol 2011, 107: 193–197. [DOI] [PubMed] [Google Scholar]
- 17.Xiang H, Pan G, Zhang R, Xu J, Li T, Li W, Zhou Z et al. Natural selection maintains the transcribed LTR retrotransposons in Nosema bombycis. J Genet Genomics 2010, 37: 305–314. [DOI] [PubMed] [Google Scholar]
- 18.Lin L, Pan G, Li T, Dang X, Deng Y, Ma C, Chen J et al. The protein import pore Tom40 in the microsporidian Nosema bombycis. J Eukaryot Microbiol 2012, 59: 251–257. [DOI] [PubMed] [Google Scholar]
- 19.De Graaf DC, Masschelein G, Vandergeynst F, De Brabender H, Jacobs F. In vitro germination of Nosema apis (Microspora: Nosematidae) spores and its effect on their alpha-alpha-trehalose/d-glucose ratio. J Invertebr Pathol 1993, 62: 220–225. [Google Scholar]
- 20.Undeen A, Becnel JJ. Longevity and germination of Edhazardia aedis (Microspora, Amblyosporidae) spores. Biocontrol Sci Techn 1992, 2: 247–256. [Google Scholar]
- 21.Undeen A. A proposed mechanism for the germination of microsporidian (Protozoa: Microspora) spores. J Theor Biol 1990, 142: 223–235. [Google Scholar]
- 22.Undeen A, Frixione E. The role of osmotic pressure in the germination of Nosema algerae spores. J Protozool 1990, 37: 561–567. [DOI] [PubMed] [Google Scholar]
- 23.Undeen A, Vander Meer R. The effect of ultraviolet radiation on the germination of Nosema algerae Vavra and Undeen (Microsporida: Nosematidae) spores. J Protozool 1990, 37: 194–199. [DOI] [PubMed] [Google Scholar]
- 24.Vivarès CP, Gouy M, Thomarat F, Méténier G. Functional and evolutionary analysis of a eukaryotic parasitic genome. Curr Opin Microbiol 2002, 5: 499–505. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh K, Cappiello CD, McBride SM, Occi JL, Cali A, Takvorian PM, McDonald TV et al. Functional characterization of a putative aquaporin from Encephalitozoon cuniculi, a microsporidia pathogenic to humans. Int J Parasitol 2006, 36: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sironmani TA. Biochemical characterization of the microsporidian Nosema bombycis spore proteins. World J Microb Biot 1999, 15: 239–248. [Google Scholar]
- 27.Dang X, Pan G, Li T, Lin L, Ma Q, Geng L, He Y et al. Characterization of a subtilisin-like protease with apical localization from microsporidian Nosema bombycis. J Invertebr Pathol 2013, 112: 166–174. [DOI] [PubMed] [Google Scholar]
- 28.Millership JJ, Chappell C, Okhuysen PC, Snowden KF. Characterization of aminopeptidase activity from three species of microsporidia: Encephalitozoon cuniculi, Encephalitozoon hellem, and Vittaforma corneae. J Parasitol 2002, 88: 843–848. [DOI] [PubMed] [Google Scholar]
- 29.Cai S, Lu X, Qiu H, Li M, Feng Z. Identification of a Nosema bombycis (Microsporidia) spore wall protein corresponding to spore phagocytosis. Parasitology 2011, 138: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Pan G, Li T, Huang W, Chen J, Geng L, Yang DL et al. SWP5, a spore wall protein, interacts with polar tube proteins in the parasitic microsporidian Nosema bombycis. Eukaryot Cell 2012, 11: 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Wu Z, Pan G, He W, Zhang R, Hu J, Zhou Z. Identification of a novel spore wall protein (SWP26) from microsporidia Nosema bombycis. Int J Parasitol 2009, 39: 391–398. [DOI] [PubMed] [Google Scholar]
- 32.Zhu F, Shen Z, Hou J, Zhang J, Geng T, Tang X. Identification of a protein interacting with the spore wall protein SWP26 of Nosema bombycis in a cultured BmN cell line of silkworm. Infect Genet Evol 2013, 17: 38–45. [DOI] [PubMed] [Google Scholar]
- 33.Undeen AH, Vander Meer RK. Conversion of intrasporal trehalose into reducing sugars during germination of Nosema algerae (Protista: Microspora) spores: a quantitative study. J Eukaryot Microbiol 1994, 41: 129–132. [Google Scholar]
- 34.Undeen AH, Vander Meer RK. Microsporidian intrasporal sugars and their role in germination. J Invertebr Pathol 1999, 73: 294–302. [DOI] [PubMed] [Google Scholar]
- 35.Lu XM, Huang SK. Comparative study on the infectivity and spore surface protein of Nosema bombycis and its morphological variant strain. Agr Sci China 2005, 4: 475–480. [Google Scholar]
- 36.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2009, 38: 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc 2008, 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 39.Bouzahzah B, Nagajyothi F, Ghosh K, Takvorian PM, Cali A, Tanowitz HB, Weiss LM. Interactions of Encephalitozoon cuniculi polar tube proteins. Infect Immun 2010, 78: 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Weiss LM. The microsporidian polar tube: a highly specialised invasion organelle. Int J Parasitol 2005, 35: 941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dijksterhuis J, van Driel K, Sanders M, Molenaar D, Houbraken J, Samson RA, Kets EPW. Trehalose degradation and glucose efflux precede cell ejection during germination of heat-resistant ascospores of Talaromyces macrosporus. Arch Microbiol 2002, 178: 1–7. [DOI] [PubMed] [Google Scholar]
- 42.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008, 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 43.Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 2008, 5: 613–619. [DOI] [PubMed] [Google Scholar]
- 44.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 2007, 12: 169–176. [DOI] [PubMed] [Google Scholar]
- 46.Hunter T. Protein kinases and phosphatases: the Yin and Yang of protein phosphorylation and signaling. Cell 1995, 80: 225–236. [DOI] [PubMed] [Google Scholar]
- 47.Guergnon J, Dessauge F, Dominguez V, Viallet J, Bonnefoy S, Yuste JV, Mercereau-Puijalon O et al. Use of penetrating peptides interacting with PP1/PP2A proteins as a general approach for a drug phosphatase technology. Mol Pharmacol 2006, 69: 1115–1124. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez J. Phosphorylation in eukaryotic cells. Role of phosphatases and kinases in the biology, pathogenesis and control of intracellular and bloodstream protozoa. Rev Med Chile 2000, 128: 1150–1160. [PubMed] [Google Scholar]
- 49.Striepen B. Switching parasite proteins on and off. Nat Methods 2007, 4: 999–1000. [DOI] [PubMed] [Google Scholar]
- 50.Philip N, Vaikkinen HJ, Tetley L, Waters AP. A unique Kelch domain phosphatase in Plasmodium regulates ookinete morphology, motility and invasion. PLoS One 2012, 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guttery DS, Poulin B, Ferguson DJP, Szöo˝r B, Wickstead B, Carroll PL, Ramakrishnan C et al. A unique protein phosphatase with Kelch-like domains (PPKL) in Plasmodium modulates ookinete differentiation, motility and invasion. PLoS Pathogens 2012, 8: 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol 2010, 28: 91–98. [DOI] [PubMed] [Google Scholar]
- 53.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell 2006, 124: 755–766. [DOI] [PubMed] [Google Scholar]
- 54.Muñoz C, Pérez M, Orrego PR, Osorio L, Gutiérrez B, Sagua H. A protein phosphatase 1 gamma (PP1 gamma) of the human protozoan parasite Trichomonas vaginalis is involved in proliferation and cell attachment to the host cell. Int J Parasitol 2012, 42: 715–727. [DOI] [PubMed] [Google Scholar]
- 55.Texier C, Vidau C, Viguès B, El Alaoui H, Delbac F. Microsporidia: a model for minimal parasite–host interactions. Curr Opin Microbiol 2010, 13: 443–449. [DOI] [PubMed] [Google Scholar]
- 56.Petri WA, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin–carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu Rev Microbiol 2002, 56: 39–64. [DOI] [PubMed] [Google Scholar]
- 57.Loukas A, Maizels RM. Helminth C-type lectins and host–parasite interactions. Parasitol Today 2000, 16: 333–339. [DOI] [PubMed] [Google Scholar]
- 58.Southern TR, Jolly CE, Lester ME, Hayman JR. EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukaryot Cell 2007, 6: 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Senderskiy IV, Timofeev SA, Seliverstova EV, Pavlova OA, Dolgikh VV. Secretion of Antonospora (Paranosema) locustae proteins into infected cells suggests an active role of microsporidia in the control of host programs and metabolic processes. PLoS One 2014, 9: e93585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.