Abstract
Activation of genes promoting aerobic glycolysis and suppression of mitochondrial oxidative phosphorylation is one of the hallmarks of cancer. The RUNX2 transcription factor mediates breast cancer (BC) metastasis to bone and is regulated by glucose availability. But, the mechanisms by which it regulates glucose metabolism and promotes an oncogenic phenotype are not known. RUNX2 expression in luminal BC cells correlated with lower estrogen receptor-α (ERα) levels, anchorage-independent growth, expression of glycolytic genes, increased glucose uptake, and sensitivity to glucose starvation, but not to inhibitors of oxidative phosphorylation. Conversely, RUNX2 knockdown in triple-negative BC cells inhibited mammosphere formation and glucose dependence. RUNX2 knockdown resulted in lower LDHA, HK2, and GLUT1 glycolytic gene expression, but upregulation of pyruvate dehydrogenase-A1 (PDHA1) mRNA and enzymatic activity, which was consistent with lower glycolytic potential. The NAD-dependent histone deacetylase, SIRT6, a known tumor suppressor, was a critical regulator of these RUNX2-mediated metabolic changes. RUNX2 expression resulted in elevated pAkt, HK2, and PDHK1 glycolytic protein levels that were reduced by ectopic expression of SIRT6. RUNX2 also repressed mitochondrial oxygen consumption rates (OCR), a measure of oxidative phosphorylation (respiration). Overexpression of SIRT6 increased respiration in RUNX2-positive cells, but knockdown of SIRT6 in cells expressing low RUNX2 decreased respiration. RUNX2 repressed SIRT6 expression at both the transcriptional and post-translational levels and endogenous SIRT6 expression was lower in malignant BC tissues or cell lines that expressed high levels of RUNX2. These results support a hypothesis whereby RUNX2-mediated repression of the SIRT6 tumor suppressor regulates metabolic pathways that promote BC progression.
Keywords: BREAST CANCER, METABOLISM, GLYCOLYSIS, MITOCHONDRIAL RESPIRATION, TRANSCRIPTION
Breast cancer (BC) progression is the result of a complex interplay of tumor cell mutational and microenvironmental events that promote tumor heterogeneity and metastasis [Berman et al., 2010]. Specific transcription factors regulate the tumor’s response to oxygen or nutritional requirements and mediate complex metabolic events such as angiogenesis or glycolysis [Herman and Kahn, 2006; Semenza, 2009] by reducing the expression of genes that inhibit mitochondrial respiration or promoting glycolytic switching [Vander Heiden et al., 2009]. SIRT6 is a mammalian NAD+-dependent histone deacetylase that functions in cellular stress resistance, genomic stability, and aging and is a tumor suppressor that controls cancer metabolism [Sebastian et al., 2012]. Sirt6-deficient mice exhibit profound phenotypes including osteoporosis, loss of subcutaneous fat and severe metabolic hypoglycemia causing death after one month of age [Zhong et al., 2010]. SIRT6 prevents these defects by acting as a negative regulator of Hif1α and deacetylating histone H3K9 on Hif1α target glycolytic genes [Zhong et al., 2010]. SIRT6 also affects the balance of glycolysis and oxidative phosphorylation by acting as a transcriptional repressor to inhibit expression of pyruvate dehydrogenase kinase-1 (PDHK1), which phosphorylates pyruvate dehydrogenase (PDH) and inhibits its activity [Zhong et al., 2010]. PDH is the rate-limiting step in conversion of pyruvate to acetyl-CoA, which enters the tricarboxylic acid (TCA) cycle and increases oxygen consumption and ATP production in the mitochondria. Since SIRT6 represses PDHK1 expression, activation of mitochondrial oxygen utilization through PDH increases. SIRT6 also exhibits non-transcriptional functions, which include a role in DNA repair through its ADP ribosylation activity and protein processing through the proteasome pathway in response to cell stress [Mao et al., 2011].
The RUNX2 gene is a regulator of mammary gland differentiation [Ferrari et al., 2013] and plays a role in metastatic cancer development [Martin et al., 2011; Pratap et al., 2006]. RUNX2 is a DNA-binding transcription factor that can regulate cell transformation [Vitolo et al., 2007], epithelial-mesenchymal transition and tumor suppressor activity [Chimge et al., 2011], and osteolytic metastases in mouse models of breast cancer [Barnes et al., 2004; Pratap et al., 2006]. It also regulates the expression of genes associated with tumor growth, migration, and invasion and is associated with ERα expression [Das et al., 2009]. Some studies have found a negative correlation between RUNX2 expression and ERα status in BC specimens [Onodera et al., 2010]. A subset of cancers expressing both RUNX2 and ERα exhibit opposing effects on cell proliferation [Chimge et al., 2012]. However, the mechanisms through which RUNX2 might promote dedifferentiation and BC progression are not completely clear. Transcription factors respond to environmental nutrients to mediate cellular survival and adaptation [Sellick and Reece, 2005]. We have found that glucose metabolism regulates RUNX2 DNA-binding and transcriptional activity through phosphorylation of a specific cyclin-dependent kinase-1 phosphorylation site on RUNX2 [D’Souza et al., 2009; Pierce et al., 2012]. Since many genes that are the target of glucose metabolism also regulate glucose utilization [Lee and Karsenty, 2008], we examined whether RUNX2 might regulate cellular metabolism and alter the energy balance in BC cells. To test this hypothesis, an inducible RUNX2 breast cancer model in luminal BC cells that do not express RUNX2 and an shRNA-targeted RUNX2 knockdown model in RUNX2-positive, triple-negative BC cells were used. RUNX2 increased glucose uptake and utilization by lowering the levels of SIRT6 protein at both the transcriptional and posttranslational levels. While SIRT6 increased mitochondrial oxygen consumption (oxidative phosphorylation), RUNX2 repressed the expression of SIRT6 and reduced oxygen consumption. These results reveal new mechanisms through which RUNX2 promotes BC progression: regulation of the SIRT6 tumor suppressor and cellular metabolism.
MATERIALS AND METHODS
CELL CULTURE AND CLONAL SELECTION
Derivation of BC cell lines with inducible RUNX2 expression (ER+ MCF7) using the BD™ Tet-Off System was described [Underwood et al., 2012]. RUNX2− MCF7 cells are ER+ and express wild type p53, PTEN, c-myc, and ras, but do not express p16. MCF7 cells containing tTA (Tetracycline-controlled transactivator) regulatory vector (G418 resistant) were purchased from Clontech (Takara Bio, Mountain View, CA), infected with retroviral vectors expressing RUNX2, and selected with 200μg/ml hygromycin B. Cells were frozen within three passages and maintained in DMEM containing 10% FBS and the antibiotics G418 (100μg/ml), hygromycin B (200μg/ml), and doxycycline (2μg/ml) to repress RUNX2 expression. Cells were sub-cloned and grown under similar conditions in the presence or absence of doxycycline for 72 hours to induce RUNX2 expression.
RUNX2+ Hs578t cells are ER-negative, express wild type PTEN, overexpress c-myc, and contain mutant p53 and ras. RUNX2+ Hs578t cells were obtained from ATCC (Manassas, VA) and RUNX2 knockdown clones were selected after lentiviral shRNA infection as recommended by the manufacturer (Sigma-Aldrich; Mission in vivo shRNA system). Briefly, Hs578t parental cells were infected with lentiviral shRNA expressing viruses targeting five different regions of the RUNX2 coding sequence (Sigma-Aldrich, St Louis MO), selected, and frozen within three passages. Cells were maintained in media containing DMEM + 10% FBS with 1μg/ml puromycin. Low-RUNX2 expressing, glucose deprivation-resistant clones of Hs578t cells (Hs578t.LG) were generated by replacing culture media with low-glucose (0.5mM) + 5%FBS for 4 days. Standard culture media was added back to the surviving cells. After 10 days, expanded cells were characterized for RUNX2 expression. SIRT6 overexpressing cells were prepared after transfection of parental Hs578t tumor cells with a Flag-tag SIRT6 cDNA expression plasmid (Genecopoeia, Inc., Rockville, MD) and brief selection with G418 (1 week). SIRT6 protein expression was confirmed by Western blot. For measurement of SIRT6 expression, cells were either starved in the absence of glucose for the indicated time or different concentrations of glucose were added. Treatments with pyruvate (0, 0.1mM, 1 mM) or cycloheximide (10μg/ml) were performed in 5% or 2% FBS, respectively. The proteasome inhibitor MG132 (20 μM; Sigma/Aldrich, St Louis, MO) was used to treat MCF7 cells cultured in the absence (RUNX2+) or presence (RUNX2-) of doxycycline (2μg /ml) after 24 hours of glucose starvation.
CO-IMMUNOPRECIPITATION (CO-IP) AND WESTERN BLOTS
MCF7 cells cultured in the presence or absence of 2μg/ml doxycycline were processed for nuclear extract isolation with NucBuster (Novagen). Briefly, cell extracts were pre-cleared with Protein G-Sepharose (GE Healthcare) for 1 hour at 4°C and the supernatant was incubated with anti-RUNX2 antibody (MBL) overnight. Protein-G-Sepharose was added for 1 hour and the precipitated complexes were washed with 50mM Tris-HCL, 150mM NaCl, 1mM EDTA, 1mM EGTA, 1% Triton X-100, 0.5% NP-40. Proteins were eluted from the beads using 0.1M Glycine buffer (pH 2.5), treated with 1X SDS loading buffer containing β-mercapto-ethanol, and heated at 97°C for 10min. Samples were resolved on a 4–12% Bis-Tris polyacrylamide gel (Invitrogen) and transferred to PVDF membranes (Millipore). Immunoblots were probed with anti- SIRT6 (polyclonal rabbit; Cell Signaling) or anti-FLAG (CY Lin, Georgetown University) antibodies followed by development with enhanced chemiluminescence, ECL (Millipore). Anti-mouse IgG was used as a control antibody. For Western blots, cells were washed with PBS and cytoplasmic and nuclear lysates were obtained using NucBuster (Novagen/EMD4 Biosciences) [Pierce et al., 2012]. Lysates were fractionated by SDS-PAGE with 4–12% gradient gels purchased from Invitrogen. Western analysis was carried out using antibodies recognizing specific peptides or proteins: Flag (CY Lin, Georgetown University), RUNX2 (MBL International Corp., Woburn, MA), GLUT1 (Abcam), SIRT6 (Cell Signaling), pAkt (Cell Signaling), ERα (Santa Cruz), Hif1α (Santa Cruz), Ubiquitin (Santa Cruz), and β-actin (Sigma/Aldrich). ERα protein levels were quantified by scanning gels (n=3) using the generic gray gamma-2.2 profiler in the Apple ColorSync Utility (version 4.6.2) program or NIH Image-J, as indicated. MCF7 cells grown in the presence or absence of doxycycline were starved (no glucose) for 24 hours. 5mM glucose was then added for 4 hours and protein levels of GLUT1 and pAkt were analyzed using cytoplasmic extracts. For SIRT6 expression, cells were starved in the absence of glucose for 16hr followed by glucose treatment at the indicated concentrations (0.5 – 25mM). Cells were also treated with pyruvate (0, 0.1mM, 1mM) and SIRT6 levels were measured by Western blot.
MEASUREMENT OF GLUCOSE UPTAKE
For the assessment of glucose uptake values, MCF7 cells cultured in the presence (RUNX2-) or absence (RUNX2+) of doxycycline were plated and grown in DMEM containing 10% FBS for 24 hours. Cells were then glucose starved for 24 hours followed by incubation with 100μM of 2-NBDG (fluorescently-labeled D-glucose analog 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose) for 30min as recommended by the manufacturer (Invitrogen/Life Technologies Corporation). Cells were harvested and fluorescence intensity was measured by FACS analysis using excitation/emission maxima of 465/540nm and fluorescein optical filters. Results were expressed as relative fluorescence intensity.
CELL VIABILITY AND PROLIFERATION ASSAYS
MCF7 cells were plated on 24-well plates (250,000 cells/well) for 24 hours to allow recovery from trypsinization and treated with either 2-deoxy-glucose (2-DG) (2mM) to inhibit glycolysis or oligomycin (2.5μM) or rotenone (0.2μM) to inhibit ox/phos for 2 days. For 2-DG treatment, the cell media contained 5% serum + 2mM glucose. For oligomycin and rotenone treatment, 5% serum + 5mM glucose was used. Hs578t parental and Hs578t/55.5 RUNX2 knockdown cells were plated and treated with different glucose concentrations (25, 5, 0.5mM). Cell growth was monitored with the crystal violet assay as previously described [Pierce et al., 2012]. In brief, after the indicated time, media and dead cells were removed and attached cells were fixed in 10% formalin, washed with water, and stained with crystal violet (0.5% in 25% methanol; Sigma/Aldrich C3886). After 3 washes with PBS, crystal violet was solubilized in a 0.1M citric acid/50% ethanol solution (Sigma/Aldrich C7254) and the absorbance was measured in a 96-well plate reader at 540nm.
ANCHORAGE-INDEPENDENT GROWTH
MCF7 cells were tested for clonogenic growth in soft agar as described [Qiao et al., 2006; Vitolo et al., 2007]. Briefly, 40,000 cells for each well of a 6-well plate were mixed with 0.33% soft agar in DMEM containing 10% fetal bovine serum and added to a base layer of 0.5% soft agar in media containing 10% fetal bovine serum. Colonies were photographed after 14 days at 37°C. Hs578t cells were tested for clonogenic growth (mammosphere formation) in suspension cultures [Charafe-Jauffret et al., 2009]. Briefly, 6X104 cells were cultured in 6-well ultra-low adhesion plates (Costar 3471) in DMEM media supplemented with 1%FBS, 5mM glucose, bovine pituitary extract (10ng/ml), heparin (4μg/ml), and gentamycin (20μg /ml) in the presence (2ng/ml) or absence of TGFβ (inducer of the mesenchymal phenotype). Mammosphere volume (mean ± SD) was calculated from the formula for an ellipsoid (width2 X length)/2) from duplicate wells and n = 4–9 fields per treatment; spheres > 60 μm3 were used for statistical comparison of control vs treatment groups. Significance was determined using Student’s t-test or Tukey’s post-hoc adjustment for 2-by-2 comparisons following ANOVA with p < 0.05 indicating significant differences.
REAL-TIME QUANTITATIVE RT-PCR
Total RNA was extracted using TRIzol (Life Technologies). cDNA was synthesized using 1 μg of total RNA that was reverse transcribed with oligo-(dT) primer using the SuperScript first-strand synthesis system (Invitrogen). Q-PCR was performed using gene-specific primers. PCR primers used to examine expression of each gene are listed as forward (F) or reverse (R) primers. hHK2: F- GAGCCACCACTCACCCTACT, R-ACCCAAAGCACACGGAAGTT; hLDHA1: F-CAGCCC GAACTGCAAGTTGCTTAT, RTCAGGTAACGG AATCGGGCTGAAT; hSIRT6 F: AAGTTCGACACCACCTTTGAGAGC, R: ACGTACTGCGTCTTACACT TGGCA; hGLUT1: F-AAGGTGATCGAGGAGTTCTACA, R-ATGCCCCCAACAGAAAAGATG; hPDHA1: F-ATGCAGACTGTACGCCGAATG, R-GGGTGAAAGTAAAGCCGTGAG.
CHROMATIN IMMUNEPRECIPITATION (CHIP)
The 5′ flanking genomic region of the Sirt6 gene precedes the Sirt6 open reading frame and contains a TATA- and CCAAT-box less promoter, a 300-bp CpG island, and GATA-x and RUNX/AML transcription factor binding sites [Mahlknecht et al., 2006]. ChIP assays were performed as described [Pierce et al., 2012]. Hs578t cells grown in either full media (FM; 25mM glucose) or low glucose starvation media (5mM glucose) were cross-linked with 1% formaldehyde for 10 min at 37°C. After two washes with cold PBS, cells were collected and lysed with warm SDS lysis buffer (1% SDS, 10 mM EDTA pH 8, 50 mM Tris-HCl pH 8) containing protease inhibitors followed by sonication (5 pulses of 10min, 30sec; Branson Sonifier). The DNA fragmentation was confirmed by agarose gel electrophoresis. After centrifugation for 10 min at 13,000 rpm at 4°C, supernatants were diluted 1/10 in dilution buffer (0.01% SDS,1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl) followed by pre-clearing with magnetic beads (Dynabeads, Invitrogen). RUNX2-specific antibody (MBL International Corp., Woburn, MA; Catalog #D130-3) or IgG non-specific control was added overnight at 4°C on a rotation platform. Magnetic beads were added and the mixture incubated for 1 hour at 4°C with rotation. Beads were collected and washed sequentially for 20min each with high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl pH 8.1, 500 mM NaCl), low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl pH 8.1, 150 mM NaCl), LiCl wash buffer (0.25M LiCl, 1% IGEPAL-CA630, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl pH 8.1), and TE buffer (10mM Tris-HCL, 1mM EDTA). The beads were then eluted with elution buffer (1% SDS, 0.1 M NaHCO3) and 5M NaCl was added and the mixture incubated for 6hr at 65°C to reverse the cross-links. DNA was recovered using the PCI (phenol/chloroform/isoamyl alcohol) extraction method and real-time PCR was performed for analysis. RUNX/AML site a: Forward 5′-CATCCCCTCCTCCAGGAAGCCCT-3′; Reverse 5′AATGGGGC TGGTGGCCTGGAGGA-3′. RUNX/AML sites a and b: Forward 5′-CATCCCCTCCTCCAGGAAGCCCT-3′; Reverse 5′-TTGCCCAGGCTGGAGTGCAGTGG-3′.
LUCIFERASE PROMOTER REPORTER ASSAYS
The Secrete Pair Luminescence Assay Kit and the human promoter reporter clone for SIRT6 (HPRM16920 Target Gene Accession NM_016539) from Genecopoeia (Rockville, MD) were used to measure SIRT6 promoter activity in response to RUNX2 and SIRT6. This method utilizes the SIRT6 Gaussian luciferase promoter reporter clone (pEZX-PG04-GLuc-ON™), which contains two consensus Runx binding sites 952bp and 589bp upstream of the ATG start site. This promoter drives expression of the GLuc gene and a CMV promoter controls expression of the secreted alkaline phosphatase (SEAP) gene. The manufacturer’s recommendations were used to transfect the Flag.tag RUNX2 and SIRT6 expression plasmids (Genecopoeia, Inc. Rockville, MD) into Hs578t, Hs578t.55.5 or HEK293 cells using Lipofectamine-2000 reagent (Life Technologies Inc. Grand Island, NY). After transfection, cells were incubated in low glucose (5mM) for either 24hr (Hs578t cells) or 6hr (HEK293 cells) prior to collection and analysis of conditioned media containing luciferase and alkaline phosphatase.
SMALL-INTERFERING (SI)RNA TARGETING SIRT6
SIRT6 knockdown was performed in Hs578t/55.5 cells (low RUNX2 expression) using the Trilencer-27 Human siRNA oligonucleotides (Origene, Inc., Rockville, MD) containing a Universal Scrambled Negative Control siRNA Duplex and 3 unique 27mer siRNA duplexes (Catalog No. SR309840): siRNAA: rCrG rArGrGrArUrGrUrCrGrGrUrGrArArUrUrArCrGrCrGGC; siRNA B: rCrGrGrArA rGrCrGrGrCrCrUrCrA rArCrArArGrGrGrArAAC; siRNA C: rArGrCrGrGrArArGrGrUrGrUrGrGrG rArArCrUrGrGrCrGAG.
MITOCHONDRIAL OXYGEN CONSUMPTION
Respiration was measured in live cells in a physiologically intact system [Clerc and Polster, 2012] using the Seahorse XF24 system (Seahorse Bioscience, North Billerica, MA), which allows for simultaneous measurement of OCR (O2-consumption rate), a result of oxidative phosphorylation, and ECAR (extracellular acidification rate), largely due to glycolytic lactic acid production [Dranka et al., 2010]. Established protocols were employed to assess oxygen consumption in response to glucose or pyruvate [Dranka et al., 2010]. Cells were pretreated with glucose-free media. Basal OCR was measured, followed by sequential oligomycin to inhibit ATP synthase (2.5nM), Trifluoromethoxy carbonylcyanide phenylhydrazone to uncouple oxidative phosphorylation by dissipating the electrochemical gradient or membrane potential that drives ATP synthesis (FCCP; 0.4μM – 0.75μM), pyruvate as carbon source (10mM) or antimycin-A to inhibit complex III (1μM) as in protocol 2. The difference between basal OCR and oligomycin-insensitive OCR estimates the amount of O2 consumption that is linked to ATP synthesis.
PDH ENZYMATIC ACTIVITY ASSAY
The Pyruvate dehydrogenase (PDH) Enzyme Activity Microplate Assay Kit from Abcam (ab109902) was used to measure cellular PDH activity in MCF7 TetOFF or Hs578t/55.5 RUNX2 knockdown (RUNX2−) compared with Hs578t/54.4 control knockdown (RUNX2+) cells. Briefly, the PDH enzyme is detected in the wells of a 96-well microplate using a specific antibody already bound to the plate and activity is determined from the reduction of NAD+ to NADH, which is coupled to the reduction of a reporter dye. The colored reaction product is detected by measuring the Absorbance at 450nm. Relative activity is expressed as a change in optical density per minute per mg of protein (ΔOD450/min/mg).
TISSUE MICROARRAYS (TMA) AND STATISTICAL ANALYSES
Invasive ductal carcinoma (IDC) samples from the University of Maryland Greenebaum Cancer Center were obtained under IRB approval and processed for histological analysis using a standard protocol with H&E staining. Specific antibodies recognizing RUNX2 or GLUT1 (1:200 dilution) were used to analyze serial sections (5μm) for protein expression. To confirm the specificity of each antibody, specimens were also stained with secondary antibody alone and tissues not expressing RUNX2 or GLUT1 were used as negative controls. SIRT6 expression was detected using previously published IHC protocols [Sebastian et al., 2012] using antibody LS-B5589 from Lifespan Biosciences at 1:200 dilution. The Biomax BR1503 and BR483b breast cancer tissue arrays (http://www.biomax.us/tissue-arrays/Breast/), which included TNM staging, pathology grade, and ER/PR/Her-2/P53 IHC results from 150 breast carcinoma tissue microarray cores were evaluated by two independent pathologists (XFZ and SL). Each core was 1mm in diameter with a thickness of 5μm. Immunostaining for cytokeratin (CK) confirmed the epithelial origin of the tumors. To estimate whether RUNX2, ER, or GLUT1 were elevated in BC patient TMA, tumor specimens were dichotomized (RUNX2 high versus low staining). A Fisher’s exact test with a 0.05 two-sided significance level was used, which exhibited above 90% power to detect the difference between a proportion of tissues with high RUNX2 level of 0.78 and a proportion of 0.27 in ERα-positive tissue, when the number of samples was 18 and 37, respectively. A zero-inflated Poisson model was used to analyze the BR1503 data. Results for the overall test of differences in marker expression were expressed as boxplots displaying the distribution of each protein (GLUT1 cytoplasmic, GLUT1 membrane, RUNX2 cytosol, RUNX2 nuclear) across 5 groups (Normal, fibroadenoma/ductal carcinoma in situ (FA/DCIS), Stage II, Stage II/III, and stage IV). The Spearman correlation coefficient was used to estimate relationships in each group of patients. Quantitation of SIRT6 and RUNX2 in the Biomax BR483b TMAs used a previously published method, with some modifications [Sebastian et al., 2012]. In brief, two independent scorers assigned a weighted score to the staining, which was categorized as negative (0), weak (1), moderate (2) or strong (3). Each of these scores was calculated using the following formula: SIRT6 Labeling Index = 0 x % negative cells + 1 x % weakly stained cells + 2 x % moderately stained cells + 3 x % strongly stained cells. If both observers could not evaluate the tissue section (because of poor fixation, lack of glandular material or low tumor content) it was eliminated from consideration. The Labeling Index was plotted across different categories of tumor specimens, with the number of specimens in each category indicated in parentheses: Normal (7); NAT (1); Stage I (1); Stage II (22); Stage III (15); Stage IV (1). NAT = normal adjacent tumor. Diagnoses included invasive ductal carcinomas (35), mucinous carcinoma (3), invasive papillary carcinoma (1), mixed invasive ductal and lobular carcinoma (1). Because of insufficient Stage I or IV specimens, the mean and SD were calculated for Normal/NAT/Stage I; Stage II; Stage III/IV. P-values relative to normal are indicated in the Figure legend.
For all other analyses, results from culture assays were calculated from at least three replicate samples and expressed as the mean (± SD). To determine statistical significance, comparison of measurements relative to control used the Student’s t-test. For comparison of multiple measurements, Tukey’s post-hoc adjustment for 2-by-2 comparisons following ANOVA was used for data analysis. p values < 0.05 were considered significant.
RESULTS
RUNX2, METABOLIC BC FUNCTION, AND ONCOGENIC POTENTIAL
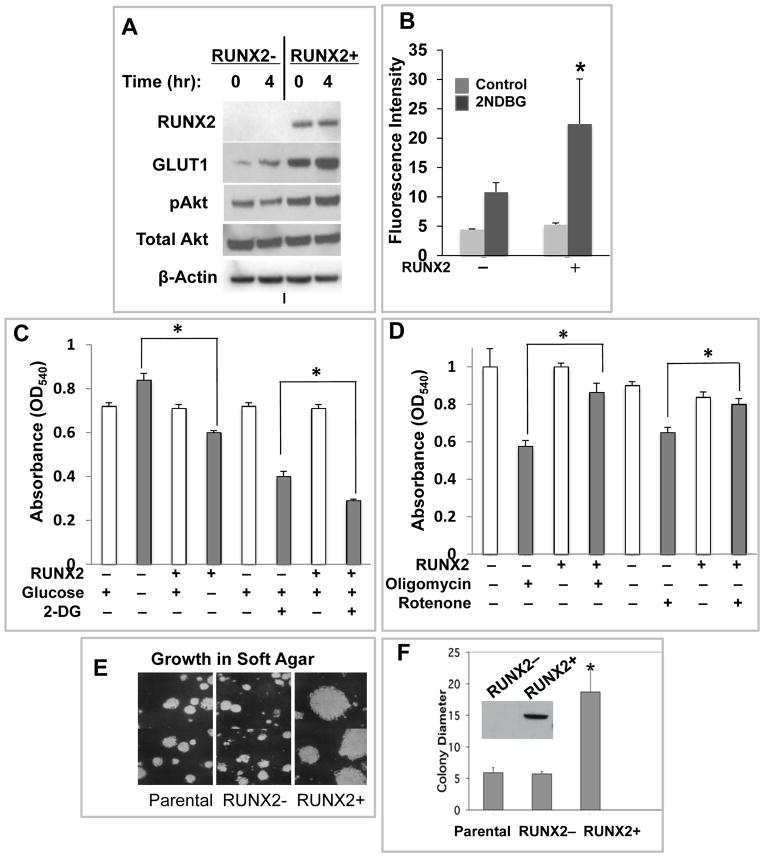
The luminal subtype and ERα+ BC cell line MCF7 (RUNX2-) [Blick et al., 2008; Lau et al., 2006; Neve et al., 2006], was engineered to express RUNX2 in the absence of doxycycline (tet.OFF) [Underwood et al., 2012]. Stable clones expressing RUNX2 were isolated (Supp Fig S1A). Three different RUNX2-negative clonal variants expressed higher levels of ERα than cells expressing RUNX2 (p < 0.018). RUNX2+ MCF7 cells exhibited increased GLUT1 expression and pAkt activation (Fig 1A) relative to RUNX2− cells. Two-fold higher glucose uptake was also observed in RUNX2+ cells (Fig 1B; p < 0.05), which were more sensitive to glucose deprivation or starvation induced by the non-metabolic glucose analog 2-deoxyglucose (2DG) (Fig 1C), but resistant to mitochondrial inhibitors (Fig 1D). Changes in glucose dependence and/or mitochondrial drug sensitivity in RUNX2+ cells might be related to altered glucose metabolism associated with an oncogenic phenotype and a growth advantage [Vander Heiden et al., 2009]. MCF7 cells expressing RUNX2 formed 4-fold larger colonies than parental or RUNX2− cells when cultured in soft agar (Fig 1E, 1F; p < 0.005), consistent with a growth advantage under anchorage-independent conditions.
FIGURE 1. RUNX2 expressing luminal BC cells exhibit a glycolytic phenotype and decreased dependence on mitochondrial oxidative metabolism.
(A) MCF7 cells were starved in 0.5mM glucose for 16hr and treated with 25mM glucose for 0 or 4hr, as shown. GLUT1 glucose transporter expression in RUNX2+ cells and activation of pAkt were determined by Western blot. (B) Glucose uptake in MCF7 RUNX2 +/− cells was measured after starvation in 0.5mM glucose for 16 hours followed by the addition of 100μM of the D-glucose analog 2-NBDG (2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-D-glucose) for 30min. Fluorescence was measured by FACS analysis (control, no 2NBDG (light gray); +2NBDG (dark gray). *p < 0.05 relative to RUNX2− cells (ANOVA). (C) Dependence on glucose for survival. MCF7 cells were glucose starved (no glucose) and treated with 2mM glucose or with 2mM non-metabolic 2-deoxy-glucose (2DG) in the presence of 2mM glucose for 72hr. Surviving cells were measured by crystal violet staining (* indicates p < 0.05 compared to untreated RUNX2− cells without glucose or with 2DG). (D) Dependence on oxidative phosphorylation for survival. Oligomycin (2.5nM) ATP synthase inhibitor or rotenone (0.1 μM) complex I inhibitor was used to treat RUNX2+ or RUNX2− MCF7 cells. Surviving cells were determined by crystal violet staining (*p < 0.05 compared to RUNX2− cells treated with oligomycin or rotenone). (E) Parental MCF7, MCF7 RUNX2 negative (RUNX2-) cells or cells expressing RUNX2 (RUNX2+) protein using a Tet.OFF system were induced for 48hr and clonal growth was measured in soft agar for 2 weeks. Parental cells are shown for comparison at equal magnification. (F) Photographs were taken after 14 days and relative colony diameter measured (mm scale of magnified photos). Quantitation of mean colony diameter +/− SEM was determined with NIH Image-J software from n = 9–10 data points each (*p < 0.005 relative to parental cells). Inset shows Western blot with Flag.tag antibody. RUNX2 = 60kDa.
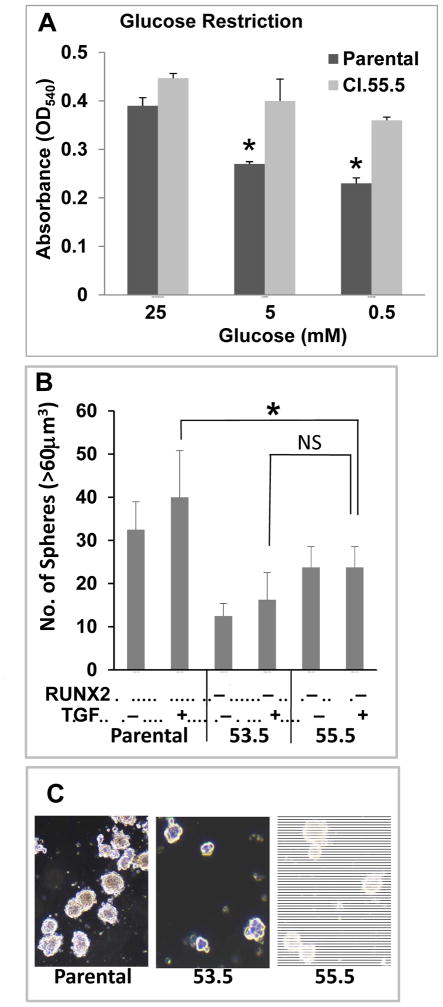
Conversely, endogenous RUNX2 levels were reduced by 90% in a triple negative BC cell line (Hs578t/53.5) or >99% in Hs578t/55.5 selected cells after stable knockdown with two independent lentiviral vectors expressing shRNA targeting RUNX2 (Supp Fig S1B). Control knockdown (Hs578t/54.5) cells expressed levels of RUNX2 that were equivalent to parental Hs578t cells. Glucose restriction reduced the growth of Hs578t parental cells, but RUNX2 knockdown cells were resistant to low glucose conditions (5mM or 0.5mM glucose) relative to parental cells (Fig 2A). Hs578t parental and RUNX2 knockdown clones (53.5 and 55.5) were cultured in low-adhesive tissue culture plates (in suspension) under anchorage-independent conditions that have been used to reveal the tumor initiating, stem-like phenotype of tumor cells [Dimri et al., 2005]. As was observed for MCF7 cells, high RUNX2 expression was associated with the formation of 2–3-fold higher numbers of large (> 60μm3) tumorspheres (Fig 2B), which were denser than those formed by cells with low RUNX2 expression (Fig 2C). Although 55.3 exhibited some residual RUNX2 expression, they appeared to form the fewest tumorspheres, but this trend was not significant in the presence of TGFβ (p > 0.05). MCF7 cells grew at similar rates as adherent cells in media containing 25mM glucose and 10% FBS regardless of RUNX2 expression. Adherent Hs578t parental cells grew at a faster rate after day 2 relative to RUNX2 knockdown cells, which grew at a slower rate as they approached confluence (Supp Fig S1C, S1D).
FIGURE 2. RUNX2 regulates glucose dependence and triple negative BC anchorage-independent growth.
(A) Cell growth in response to glucose availability was determined using Hs578t Parental (RUNX2+) and Hs578t/55.5 (RUNX2 knockdown) cells after 3 days in culture. Culture media contained 10%FBS plus high glucose (25mM) or low glucose (5mM, 0.5mM). Cell growth was assessed by crystal violet assay. *Significant reduction in cell growth compared to 25mM glucose from triplicate samples (p < 0.05). NS, not significant. (B) Triple negative Hs578t cells and the RUNX2 knockdown clones (53.5; 55.5) were harvested from attached cultures and incubated for 6 days in low-adhesion tissue culture plates. The number of spheres greater than 60μm3 from duplicate wells and n = 4–9 fields are reported for each cell line. *p < 0.05. (C) Photos show representative tumorspheres from TGFβ-treated cultures.
GLYCOLYTIC GENE EXPRESSION AND ACTIVITY IN RESPONSE TO RUNX2
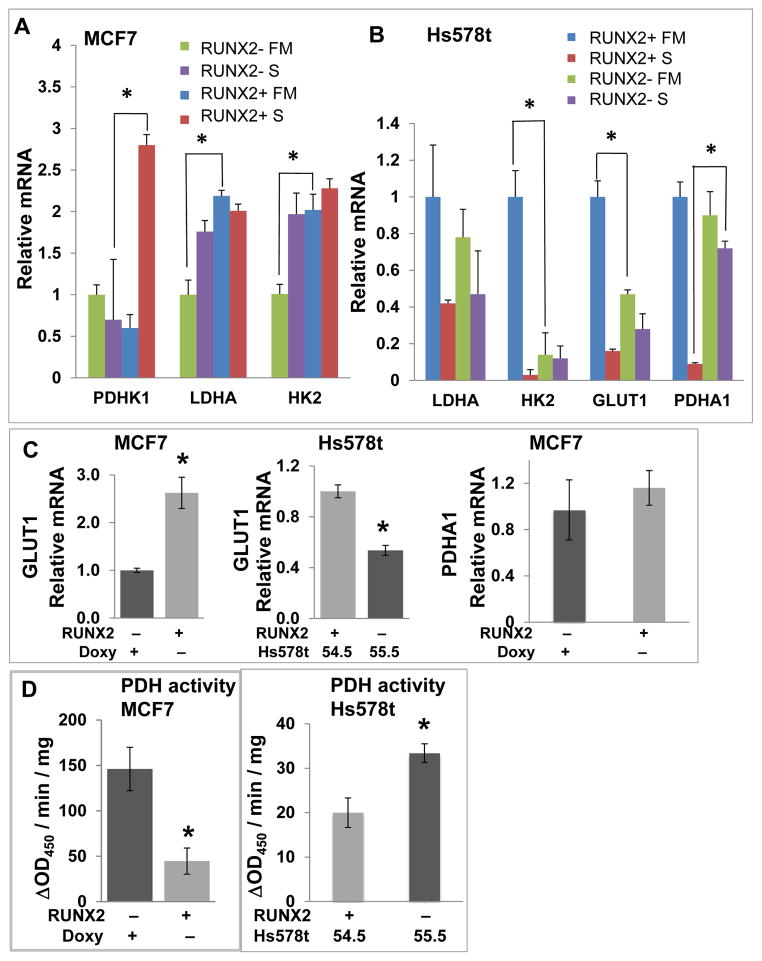
Tumor cell dependence on glycolysis is associated with reduced mitochondrial oxidative phosphorylation (ox/phos) [Vander Heiden et al., 2009] and lower SIRT6 levels [Zhong et al., 2010]. Levels of the pro-glycolytic genes PDHK1, LDHA, and HK2 were significantly elevated upon RUNX2 induction in MCF7 cells (Fig 3A; *p < 0.05). The levels of GLUT1 and HK2 (but not LDHA) were significantly higher in RUNX2+ Hs578t parental cells relative to Hs578t/55.5 knockdown cells (Fig 3B; p < 0.05) while the levels of PDHA1, which is essential for pyruvate utilization and generation of acetyl-CoA for mitochondrial ox/phos, were significantly higher in RUNX2 knockdown cells relative to parental cells cultured in low glucose (S) media (Fig 3B; p < 0.05). In further analysis of GLUT1 and PDHA1 expression, MCF7 cells with inducible RUNX2 expression showed relatively higher levels of GLUT1 mRNA than un-induced cells, as did RUNX2+ Hs578t/54.5 control knockdown cells compared to Hs578t//55.5 knockdown cells (Fig 3C). The relative levels of mRNA expression for PDHA1 in MCF7 cells did not change with RUNX2 expression under any growth conditions (Fig 3C). However, RUNX2 expression was associated with about 3-fold lower PDH enzymatic activity in MCF7 cells compared to RUNX2− cells (Fig 3D). PDH activity in Hs578t RUNX2 knockdown cells (55.5) was increased about 1.7-fold compared with RUNX2 control knockdown (54.5) cells (Fig 3D), consistent with higher PDHA1 mRNA (Fig 3B).
FIGURE 3. Expression of glycolytic or mitochondrial oxidative phosphorylation genes and PDH activity in response to RUNX2.
(A) MCF7 (RUNX2− or RUNX2+) or (B) Hs578t (RUNX2+) or shRNA RUNX2 knockdown (RUNX2-) cells were cultured in either full media (FM; 25mM glucose) or low glucose starvation media (S; 5mM glucose) for 4hr. Levels of mRNA were determined by q-RTPCR with specific primers to detect pyruvate dehydrogenase kinase-1 (PDHK1), lactate dehydrogenase isoform A1 (LDHA1), hexokinase-2 (HK2), glucose transporter-1 (GLUT1), or pyruvate dehydrogenase isoform A1 (PDHA1). (C) GLUT1 or PDHA1 expression was measured by q-RTPCR in MCF7 cells expressing RUNX2 or control cells (RUNX2−). GLUT1 expression in Hs578t cells (55.5 knockdown compared with 54.5 control knockdown; Suppl Fig S1B; Fig S3) is shown for comparison. Significant differences are indicated (*p < 0.05). (D) PDH enzymatic activity was determined in MCF7 TetOFF cells or Hs578t/55.5 (shRNA knockdown; RUNX2 −) and Hs578t/54.5 (control knockdown; RUNX2 +) cells using an antibody-specific microtiter plate assay and expressed as the change in Absorbance at 450nm per minute per mg protein (ΔOD450/min/mg). Significant differences for MCF7 cells and between control knockdown (Hs578t/54.5 cells; Supp Fig S1B) and shRNA RUNX2 knockdown cells (Hs578t/55.5; Supp Fig S1B) are indicated (*p < 0.05).
RUNX2-REGULATED METABOLIC RESPONSE AND SIRT6 EXPRESSION
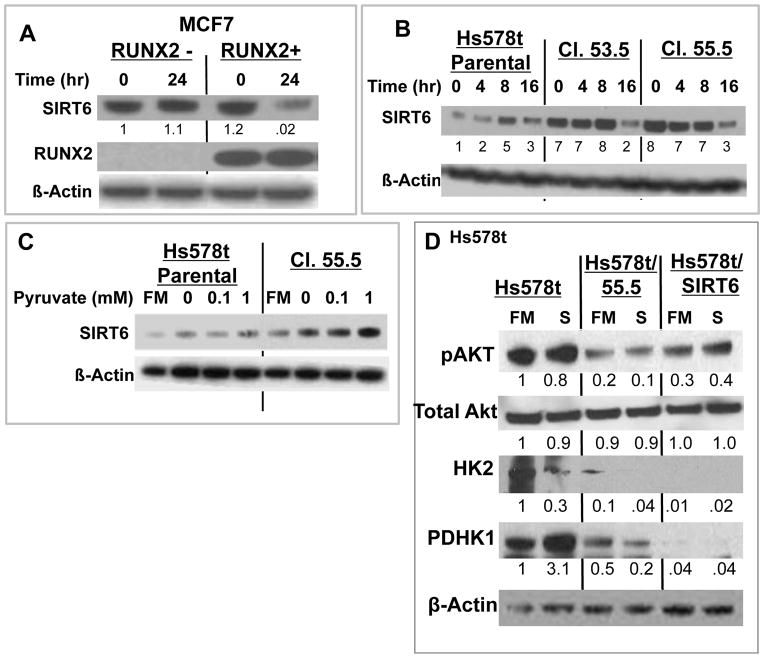
SIRT6 controls glucose homeostasis, is responsive to nutrient conditions [Kanfi et al., 2008], and negatively regulates the energy and anabolic needs of tumor cells [Sebastian et al., 2012]. To determine whether SIRT6 might be responsible for changes in metabolic response after RUNX2 expression, MCF7 were grown in the absence of glucose and Hs578t were grown in low (5mM) glucose. RUNX2-positive MCF7 cells that were glucose starved for 24hr expressed lower levels of SIRT6, relative to RUNX2 negative cells (Fig 4A). The level of SIRT6 in RUNX2 knockdown Hs578t (clones 53.5, 55.5) relative to parental cells was higher over the first 8hr of starvation and declined only after 16hr starvation (Fig 4B; p < 0.04 for 53.5 cells; p < 0.01 for 55.5 cells). Similarly, like Hs578t parental cells, control knockdown cells (Hs578t/54.5 RUNX2+ clone) expressed lower SIRT6 after glucose starvation than RUNX2 knockdown (Hs578t/55.5) cells (Supp Fig S2A). To determine whether the higher levels of SIRT6 in RUNX2 knockdown cells were independent of glycolysis, Hs578t/55.5 cells were treated with pyruvate (in the absence of glucose). Levels of SIRT6 increased with pyruvate treatment and were higher in RUNX2 knockdown cells (Hs578t/55.5) relative to parental cells (p < 0.007) (Fig 4C). Levels of the glycolytic activators pAkt, HK2, and PDHK1 were also lower in RUNX2 knockdown cells relative to parental cells (Fig 4D; lanes 3,4). Overexpression of SIRT6 in Hs578t parental cells (Hs578t/SIRT6; Supp Fig S2B) significantly reduced active pAkt and expression of HK2 and PDHK1 compared with Hs578t parental cells (Figure 4D; lanes 5,6) consistent with a role for SIRT6 in negatively regulating a RUNX2-mediated glycolytic phenotype. Hs578t parental cells were then cultured in 0.5mM glucose for 4 days, which resulted in greater than 90% cell death. Surviving cells were rescued with full culture media (25mM glucose). Three low glucose (LG)-surviving clones (LG1, LG2, LG3) expressed lower endogenous RUNX2 (p < 0.01) and slightly lower levels of GLUT1 (p < 0.06) than parental cells, consistent with a reduced glycolytic requirement in low RUNX2 expressing cells (Supp Fig S2C). Hs578t.LG2 cells also maintained higher SIRT6 levels than parental cells when cultured in low glucose (Supp Fig S2D), confirming the inverse relationship between RUNX2 and SIRT6 levels observed in RUNX2 inducible MCF7 and Hs578t RUNX2 knockdown cells.
FIGURE 4. RUNX2 expression (MCF7 cells) or knockdown of endogenous RUNX2 (Hs578t cells) alters SIRT6 levels.
(A) MCF7 cells grown in 25mM glucose were treated with 0.5mM glucose in media containing 5% serum for 24 hours. Cells were harvested, nuclear extracts were prepared, and SIRT6 and RUNX2 levels were analyzed by immunoblotting. Data were normalized to β-Actin as a loading control. (B) Glucose starvation time course. Hs578t cells were cultured in full media (25mM glucose) and 10% FBS for 24hr and media was replaced with 0.5mM glucose for 0, 4, 8, or 16hr. Significant differences between RUNX2 knockdown cells and parental cells were noted (*p < 0.04 for 53.5 cells; *p < 0.01 for 55.5 cells) over the first 8hr. (C) Glycolysis bypass with pyruvate. Hs578t cells were cultured for 24hr and the media was replaced with conditioned media containing pyruvate (0, 0.1, 1mM) + 10% FBS for 4hr. Full media (FM) = 25mM glucose. SIRT6 levels were detected by Western blot. All samples were nuclear extracts. SIRT6 protein expression (0–1 mM pyruvate) normalized to actin (arbitrary units) = 49.6 ± 5.5 (parental) and 85.6 ± 2.1 (Cl. 55.5 cells); p < 0.007, t-test. (D) Expression of pAkt, total Akt, HK2, and PDHK1 in RUNX2+ (Hs578t), RUNX2 knockdown (Hs578t/55.5), or RUNX2+ cells overexpressing SIRT6 (Hs578t/Sirt6). Cells were cultured in full media (FM, 25mM glucose) or low glucose starvation media (S, 5mM glucose) for 24hr. Protein expression normalized to actin (NIH Image-J) from scanned blots is indicated under each lane relative to Hs578t in FM.
RUNX2 INHIBITS MITOCHONDRIAL RESPIRATION THROUGH SIRT6 REPRESSION
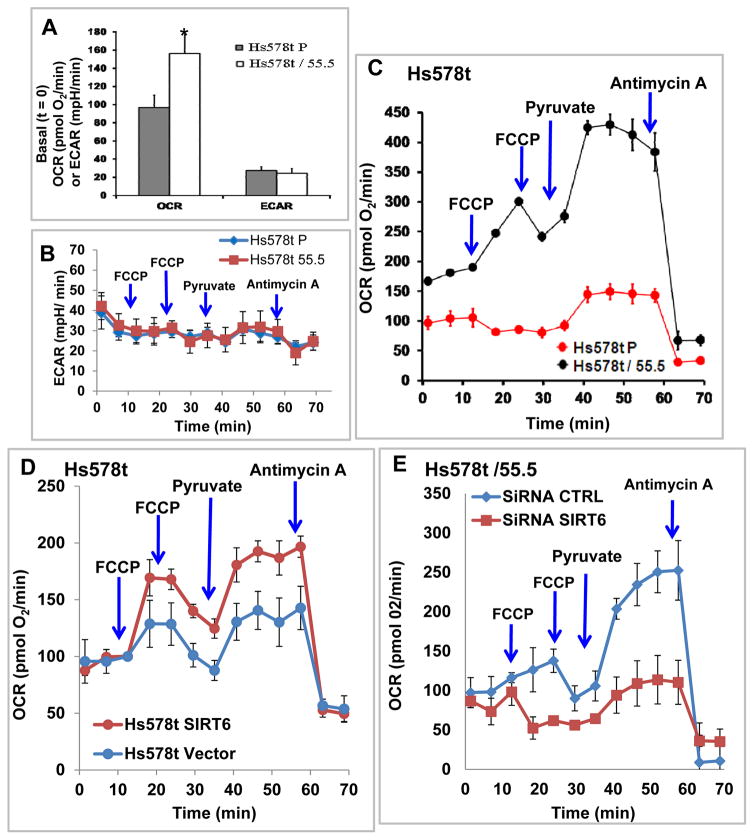
High PDHK1 (Fig 3A; Fig 4D) and low PDHA1 expression (Fig 3B) or activity (Fig 3D) in RUNX2+ cells may indicate that these cells cannot utilize pyruvate for ox/phos very efficiently. Measurement of mitochondrial respiration (OCR) in real time in live cells showed that RUNX2 knockdown Hs578t/55.5 cells exhibited 60% higher basal OCR (t = 0) than parental cells (Fig 5A). However, extracellular acidification rates (ECAR) were not significantly affected by RUNX2 expression in Hs578t cells (Fig 5A, 5B) nor were OCR responses in MCF7 cells (Supp Fig S3A). To test whether maximal electron flux through the electron transport chain was altered by RUNX2 expression in Hs578t cells, FCCP was used to uncouple electron transport from ox/phos and allow the maximum amount of oxygen consumption. With FCCP treatment, Hs578t/55.5 cells showed 3-fold higher maximal OCR relative to parental cells (Fig 5C). When pyruvate was added as a mitochondrial substrate to ensure that substrate availability was not limiting, Hs578t parental cells exhibited about 40% increased OCR from their basal level whereas RUNX2 knockdown cells exhibited a dramatic increase in OCR (4-fold higher than parental cells), consistent with pyruvate utilization through PDH and the mitochondrial TCA cycle rather than through LDH. These observations are consistent with the higher SIRT6 levels observed after pyruvate treatment in RUNX2 knockdown cells (Fig 5C) and with a preference for oxidative phosphorylation in cells expressing low RUNX2. Non-mitochondrial oxygen consumption after antimycin A treatment (inhibitor of complex III) was low in both cell lines. When the FCCP concentration was titrated to depolarize the mitochondrial membrane, both Hs578t parental or control knockdown (54.5) cells (expressing equivalent levels of RUNX2) exhibited relatively similar OCR (Supp Fig S3B), although their response to pyruvate was still much less pronounced when compared with Hs578t RUNX2 knockdown cells (Fig 5C).
FIGURE 5. RUNX2 and SIRT6 regulate mitochondrial oxygen consumption rates (OCR).
(A) Basal OCR and ECAR were measured at t=0 (in 5mM glucose) using Hs578t Parental (RUNX2+) and Hs578t/55.5 RUNX2 knockdown cells. Results are mean ± SD from n=11 separate wells for each cell type (*p < 0.05 relative to Hs578t P). (B) Measurement of changes in ECAR (mpH/min) in Hs578tP (RUNX2+) and Hs578t/55.5 (RUNX2−) cells in real-time. FCCP = 0.75μM; Pyruvate = 10mM; Antimycin A = 1μM. Mean ± SD from n = 2–3 wells/cell. (C) OCR in Hs578tP parental (RUNX2+) and Hs578t/55.5 knockdown cells were measured in real-time. FCCP = 0.75μM; Pyruvate = 10mM; Antimycin = 1μM. Mean ± SD from n = 2–3 wells/cell type. The increase in OCR in shRNA treated cells (55.5) compared to Hs578tP cells was significant (p < 0.05 from 0–60min). (D) OCR were measured in Hs578tP or Hs578t SIRT6 (Supp Fig S2B) overexpressing cells in real-time. FCCP = 0.75μM; Pyruvate = 10mM; Antimycin A = 1μM. Mean ± SD from n = 2–3 wells/cell type. (E) OCR were measured in RUNX2 knockdown Hs578t/55.5 cells (SiRNA CTRL; low RUNX2) or Hs578t/55.5 cells in which SIRT6 levels were also reduced (siRNA SIRT6) using siRNA C (Supp Fig S3C). FCCP = 1.0μM; Pyruvate = 10mM; Antimycin A = 1μM. Mean ± range from n = 2 wells/cell type. Significant differences in OCR in SIRT6 knockdown compared to control cells in response to pyruvate added after FCCP were noted (p < 0.05 from 30–60min).
SIRT6 is known to repress glycolysis and promote mitochondrial respiration [Zhong et al., 2010]. To determine whether RUNX2 was repressing OCR through lower SIRT6 protein levels, Hs578t parental cells (expressing high RUNX2, but low SIRT6) were transfected with a cDNA vector encoding SIRT6 (Supp Fig S2B). SIRT6-transfected cells exhibited higher OCR than vector controls, especially after the addition of pyruvate (Fig 5D). Therefore, the ability of RUNX2 to repress mitochondrial oxygen consumption can be partially reversed by overexpression of SIRT6. To determine if RUNX2 knockdown cells exhibited higher OCR than RUNX2+ cells because of higher SIRT6 levels, SIRT6 knockdown in Hs578t/55.5 cells was tested with three different siRNA oligonucleotides (siRNA A, siRNA B, siRNA C) (Supp Fig S3C). siRNA C treatment resulted in about a 10-fold reduction in SIRT6 levels compared to control. Although RUNX2 levels in Hs578t/55.5 cells were low, overexposure of the Western blot showed that RUNX2 levels did not increase with siRNA C treatment but instead were reduced by 3-fold (Supp Fig S3D). siRNA C-mediated knockdown of SIRT6 in Hs578t/55.5 cells resulted in reduced OCR relative to siRNA control (Fig 5E) suggesting that SIRT6 mediated the increased OCR in RUNX2 knockdown cells.
Since the hypoxia-inducible transcription factor, Hif1α, is a master regulator of glycolysis [Semenza, 2009] and is negatively regulated by the NAD-dependent sirtuin, SIRT6 [Zhong et al., 2010], Hif1α expression was examined in RUNX2+ or RUNX2− MCF7 cells. Hif1α was not detectable in Hs578t cells. Hif1α levels were higher in RUNX2+ (relative to RUNX2-) cells after starvation (t = 0) and increased modestly after glucose treatment (Supp Fig S4A). Conversely, SIRT6 levels were low in RUNX2+ cells that had been starved and the relatively higher SIRT6 levels in RUNX2− cells declined with glucose treatment. The general SIRT inhibitor sirtinol reduced SIRT6 levels even further at 8hr in RUNX2− cells while having modest effects on Hif1α. In cells exposed to hypoxia (1% O2), Hif1α levels increased within 4hr in RUNX2− and RUNX2+ cells, but declined by 24hr (Supp Fig S4B). Hypoxia-inducible levels of Hif1α remained higher in RUNX2+ (relative to RUNX2-) cells even after 24hr. Combined, these data suggest that BC cells expressing RUNX2 upregulate expression of glycolytic genes and the glycolytic factor Hif1α while downregulating SIRT6 levels and mitochondrial pyruvate utilization for respiratory activity.
POST-TRANSLATIONAL AND TRANSCRIPTIONAL REGULATION OF SIRT6 BY RUNX2
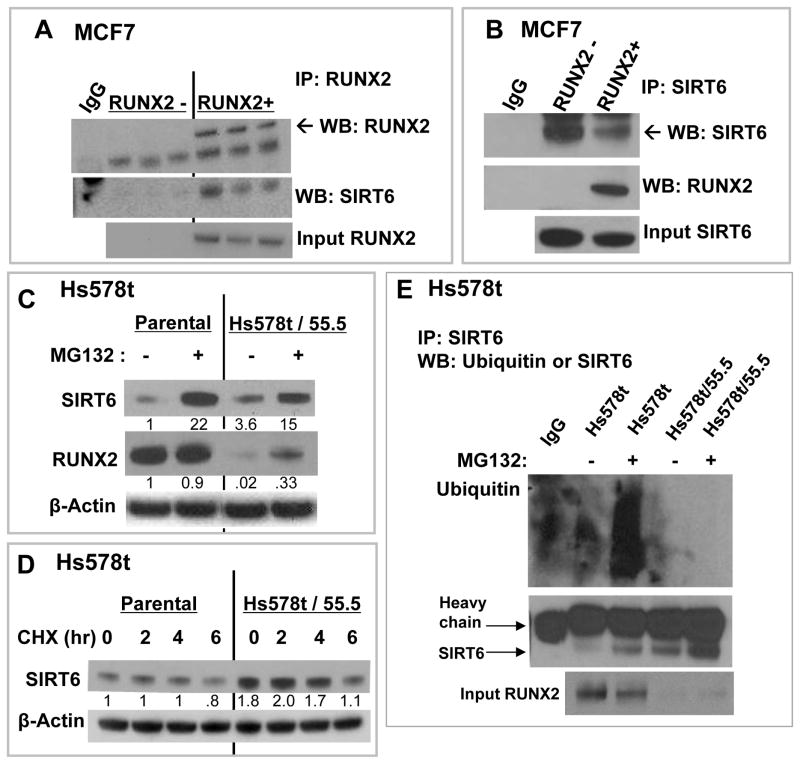
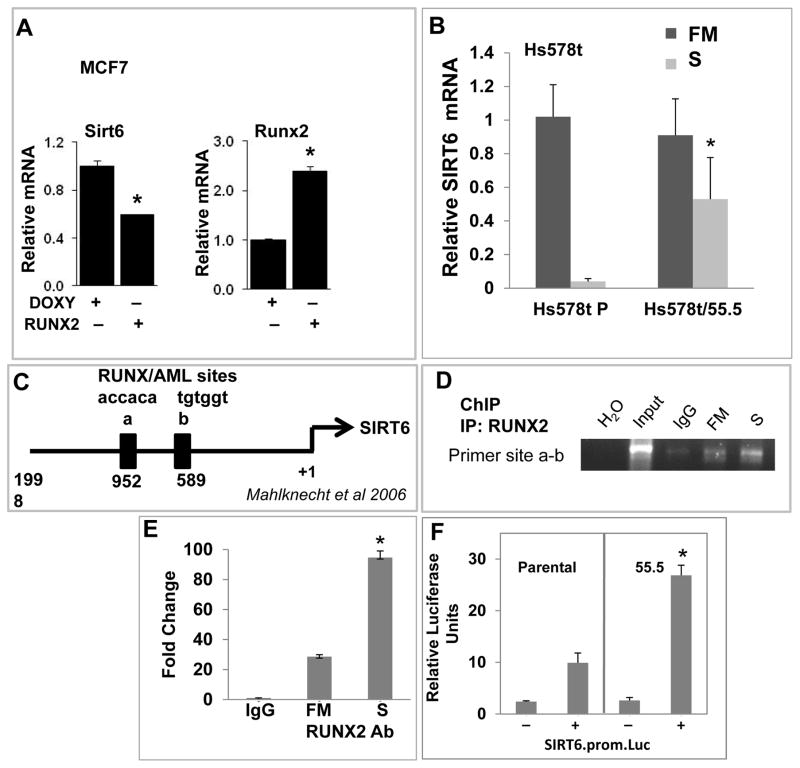
RUNX2 and SIRT6 are regulated through post-transcriptional mechanisms involving nutrient availability, cell cycle kinases, and proteasomal degradation [D’Souza et al., 2009; Kanfi et al., 2008; Pierce et al., 2012; Zhao et al., 2003]. SIRT6 associated with RUNX2 in unstarved MCF7 cells after immunoprecipitation (Fig 6A) and reciprocal co-IP confirmed these results (Fig 6B). Proteasome inhibition with MG132 increased the levels of SIRT6 in Hs578t or 55.5 cells (Fig 6C), but SIRT6 protein stability appeared to be similar in both cells after treatment with the protein synthesis inhibitor cycloheximide (CHX), suggesting that post-translational events might regulate SIRT6 regardless of RUNX2 expression (Fig 6D). Overexpression of SIRT6 in HEK293T cells resulted in high levels of Ubiquitin/SIRT6 only when RUNX2 was co-expressed in the same cells (Supp Fig S5A). This was confirmed in Hs578t cells for endogenous SIRT6 and RUNX2 (Fig 6E). Ubiquitin/SIRT6 complexes were not detectable in RUNX2 knockdown Hs578t cells (Hs578t/55.5), even in the presence of MG132, consistent with higher levels of SIRT6 protein in the immuneprecipitates (Fig 6E), suggesting that SIRT6 levels depend on RUNX2 expression and proteasomal processing. SIRT6 expression is also regulated at the transcriptional level [Zhong et al., 2010]. In MCF7 cells, expression of RUNX2 was associated with lower SIRT6 protein levels (Fig 4; Supp Fig S4A) and SIRT6 mRNA levels (Fig 7A). SIRT6 mRNA expression in Hs578t parental and RUNX2 knockdown cells was measured in full media (FM; 25mM glucose) or in low glucose starvation media (S; 5mM). Culture in low glucose (S) resulted in dramatic (>90%) inhibition of SIRT6 expression in Hs578t parental cells (Fig 7B). However, after RUNX2 knockdown (Hs578t/55.5) only a 50% decrease in SIRT6 mRNA levels was observed. RUNX2 is a known transcriptional repressor when associated with histone deacetylases [Schroeder et al., 2005] and the human SIRT6 promoter contains two consensus RUNX-binding sites (Fig 7C) [Mahlknecht et al., 2006]. Using ChIP analysis, RUNX2 associated with the SIRT6 promoter sites “a and b” in cells cultured in FM (Fig 7D). This association increased in low glucose (S) cultured cells, consistent with the repression of SIRT6 mRNA levels in RUNX2+ cells under starvation (S) conditions (Fig 7B). Results were confirmed quantitatively by qRTPCR (Fig 7E), which showed a 20-fold higher association of RUNX2 with “sites a-b” in FM (relative to IgG control) and a 90-fold increase in low glucose (S). RUNX2 showed negligible association with SIRT6 promoter “site a” alone and the ability to design primers to amplify only site “b” was limited because of the sequences and close proximity of these RUNX/AML sites. Therefore, we conclude that RUNX2 appears to associate with one or both of the RUNX/AML sites on the SIRT6 promoter. Consistent with these observations, SIRT6 promoter-luciferase activity in RUNX2 knockdown Hs578t.55.5 cells was 2.7-fold higher relative to RUNX2+ Hs578t (Parental) cells (Fig 7F). RUNX2 also repressed SIRT6 promoter-luciferase activity about 2-fold in HEK293 cells transfected with RUNX2 and SIRT6 but not with RUNX2 alone (Supp Fig S5B). These combined results suggest that RUNX2 association with the SIRT6 promoter results in active repression of SIRT6.
FIGURE 6. RUNX2 association and post-translational ubiquitination regulate SIRT6 protein levels.
(A) RUNX2 and SIRT6 interactions were measured by co-IP from MCF7 cell nuclear extracts. Cells were cultured in 25mM glucose and 5%FBS as in Figure 3A, with doxycycline to repress RUNX2 expression or in the absence of doxycycline to induce RUNX2. Three separate determinations are shown. (B) MCF7 cells expressing Flag.RUNX2 and endogenous SIRT6 were cultured as in Figure 5A. Nuclear extracts were used for reciprocal co-IP. (C) Parental Hs578t or RUNX2 knockdown (Hs578t/55.5) cells were grown in media containing the proteasomal inhibitor MG132 (20μM) for 16hr. Expression of SIRT6 and RUNX2 were normalized to β-actin as indicated (NIH Image-J). (D) Translational inhibitor (cycloheximide, CHX) was used to determine whether RUNX2 regulated SIRT6 protein stability under starvation (0.5mM glucose) conditions. Hs578t parental and Hs578t/55.5 cells were plated and grown in DMEM (25mM glucose) + 10%FBS. After 24hr, media was replaced with 5% FBS containing 0.5mM glucose and cells were treated with CHX (10 μg/ml) for 0, 2, 4 or 6 hr. Nuclear extracts were analyzed for SIRT6 expression and normalized to β-actin as indicated. (E) Hs578t cells expressing endogenous RUNX2 or cells in which RUNX2 had been reduced by shRNA (Hs578t/55.5) were treated with MG132 for 4hr prior to IP with SIRT6 antibody.
FIGURE 7. SIRT6 mRNA expression is regulated at the transcriptional level by RUNX2 and SIRT6.
(A) SIRT6 and RUNX2 gene expression were measured in MCF7 cells by q-RTPCR with specific primers. Increased RUNX2 expression was associated with lower SIRT6 expression. Significant differences are indicated (*p < 0.05). (B) SIRT6 gene expression in Hs578t BC cells or after RUNX2 knockdown (Hs578t/55.5) was measured either in full media (FM; 25mM glucose) or low glucose starvation media (S; 5mM glucose) for 4hr. Levels of mRNA in RUNX2+ (Parental, Hs578t P) or RUNX2 knockdown (Hs578t/55.5) cells were determined by q-RTPCR. Significant differences are indicated (*p < 0.05). (C) Location of putative RUNX-binding (RUNX/AML a; RUNX/AML b) sites in the human SIRT6 promoter [Mahlknecht et al., 2006]. (D) Chromatin immunoprecipitation (ChIP) assays were performed with Hs578t cells. Protein-DNA complexes from Hs578t cells cultured in full media (FM) or low glucose starvation media (S; 5mM Glucose) for 4hr were immunoprecipitated with RUNX2-specific (MBL) antibody. Agarose gels were used to visualize PCR products specific for sites “a + b”. H2O lane = no DNA template; input = prior to immunoprecipitation; IgG = non-specific, isotype-matched IgG control; FM = full media (25mM glucose) + RUNX2 Ab; S = low glucose starvation (5mM) + RUNX2 Ab. (E) ChIP analysis using quantitative PCR was performed after immunoprecipitation of protein/DNA complexes with non-specific IgG or RUNX2-specific antibody in cells cultured in full media (FM) or low glucose starvation (S) for 4hr. Specific PCR primers were used to amplify sites “a + b”. Significant differences between FM and S media are indicated (*p < 0.05; ANOVA). (F) SIRT6 promoter-luciferase (prom.luc) activity was determined with the dual Gaussian luciferase (GLuc) normalized to secreted alkaline phosphatase (SEAP) in Hs578t (Parental) or RUNX2 knockdown (55.5) cells transfected with or without the SIRT6.prom.Luc vector. After transfection, cells were incubated in low glucose (5mM) for 24hr prior to collection of conditioned media. Significant differences in SIRT6 promoter activity are indicated (*p < 0.05; ANOVA).
RUNX2/SIRT6 EXPRESSION AND METABOLIC POTENTIAL IN HUMAN BC
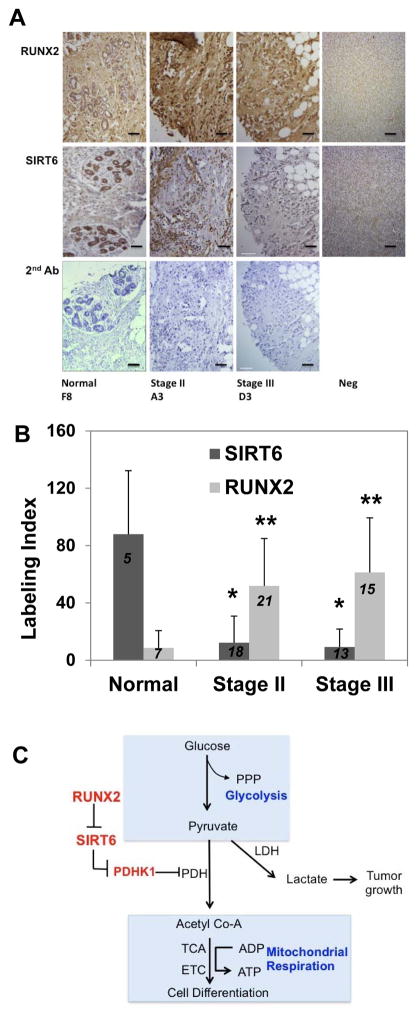
Expression of RUNX2 in a limited group of invasive ductal carcinoma (IDC) lesions from BC patients at the University of Maryland Greenebaum Cancer Center was determined by IHC. RUNX2 expression was inversely correlated with ER+/PR+ status (Supp Fig S6) and directly correlated with proliferative index (Ki67) and standard uptake values (SUV; estimated from FDG-PET scans), indicative of increased glucose metabolism. The glucose transporter, GLUT1, was expressed in proliferative, ERα-negative and RUNX2+ tumors. ERα is a marker of differentiated mammary epithelia that is lost during the epithelial-mesenchymal transition (EMT) [Guttilla et al., 2012a; Guttilla et al., 2012b]. ERα expression has been reported to inversely correlate with RUNX2 expression [Chimge et al., 2012; Onodera et al., 2010]. RUNX2 and GLUT1 expression were therefore examined in an independent commercially available tumor microarray (TMA), which included normal tissue, FA/DCIS (stage I), stage II, stage II/III, and stage III cancers. These tumors were validated for ERα, Ki67, and p53 status. GLUT1 was used as a marker of glycolysis and ERα as a marker of the differentiated phenotype. Two blinded and independent clinical anatomic pathologists assessed IHC staining. Normal glandular tissue (H & E) exhibited low RUNX2 and high GLUT1 staining while stage III cancers were poorly differentiated (H & E) with high RUNX2 and GLUT1 staining (Supp Fig S7A). Cytoplasmic GLUT1 (Glut1C) was detected in normal, stage II/III, and stage III tumors (Supp Fig S7B). Plasma membrane-bound GLUT1 (Glut1M) was not found in normal tissue, but increased with tumor grade. RUNX2 was not expressed in normal tissue, but cytoplasmic RUNX2 (RUNX2C) increased with tumor grade (80% of stage II tumors expressed RUNX2) and was found in the nucleus of 68% of stage III tumors (RUNX2N). Cytoplasmic GLUT1 expression was different across all groups (p < .0001) as was plasma membrane bound GLUT1 (p < .008) and total RUNX2 (p < .0001) (Supp Fig S7B). There was also a negative correlation (r = −0.48) between total RUNX2 and ERα both in normal tissue (high ER; low RUNX2) and in stage III tumors (low ER; high RUNX2) (Supp Fig S7C). ERα expression was high (40–80% of all tissues were positive) at all stages except stage III (12% positive). RUNX2 expression was high (40–80% of all tissues were positive) at all stages, except normal tissue (12.5% positive). Generally, RUNX2 and ERα were co-expressed at all tumor stages, except stage III. Interestingly, there was a possible negative correlation between RUNX2 (low) and cytosolic GLUT1 (high) in normal tissues (r = −0.75). However, there was a possible positive correlation between RUNX2 and membrane-bound GLUT1 in stage III cancers (r = 0.42). Examination of serial-sectioned primary breast tumor specimens from a similar tumor microarray (Fig 8A, 8B) showed that the incidence of SIRT6 expression was high in normal tissue (mean labeling index = 88) while stage II (mean labeling index = 12.2) or III (mean labeling index = 9.2) malignant tissue exhibited lower SIRT6 expression (p < 0.05). The incidence of RUNX2 in normal tissue (mean labeling index = 8.6) was significantly lower than stage II (mean labeling index = 51.9) or stage III (mean labeling index = 61.3) tumors, thus supporting an inverse relationship between RUNX2 and SIRT6 expression in human BC.
FIGURE 8. RUNX2 and SIRT6 expression in human BC: TMA analysis.
(A) IHC for SIRT6 and RUNX2 from normal human mammary glands or adjacent normal tissue and stage II and III breast cancers. Some stromal elements stained with SIRT6 antibody in stage II cancers. Controls using secondary antibody alone (2nd Ab; H&E stain) and negative tissue stained with either RUNX2 or SIRT6 antibody are shown. Scale bar = 100μm. (B) RUNX2 or SIRT6 Labeling Index for samples from panel A was calculated as described from IHC of human BC tumor microarrays (TMA) across 3 groups of tumors: Normal/NAT/Stage I (n = 5); Stage II (n = 18); Stage III/IV (n = 13). Shown are the mean ± SD for each stage; *p < 0.005 or **p < 0.05 compared to normal tissue. (C) Model for RUNX2 regulation of breast cancer glucose metabolism. When RUNX2 is expressed, SIRT6 levels decline, which in turn maintains high active PDHK1 to inhibit PDH. Low PDH activity results in reduced conversion of pyruvate to acetyl CoA (low OCR and mitochondrial respiration). Conversely, when RUNX2 levels are low, SIRT6 levels are high and so PDHK1 expression is repressed. This results in active PDH (TCA cycle and ETC), consistent with higher OCR. SIRT6 may restrict growth by also reducing histone acetylation and protein synthesis. (PPP, pentose-phosphate pathway; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PDHK1, pyruvate dehydrogenase kinase-1; TCA, tricarboxylic acid; ETC, electron transport chain.
DISCUSSION
Metabolic adaptations during cancer progression allow tumor cells to use oxygen and nutrients from the microenvironment [Fukumura and Jain, 2007; Semenza, 2009] to generate energy, support the biosynthetic burdens of proliferation, adapt to hypoxia and oxidative stress, and support tumor growth [Cairns et al., 2011]. Glycolysis is a major driver of tumorigenicity, but the mechanisms through which tumor cells suppress mitochondrial oxidative phosphorylation are not completely understood [Vander Heiden et al., 2009]. RUNX2-expressing BC cells metabolized glucose and were dependent on glucose for survival. The results extend our earlier findings that glucose can activate RUNX2 DNA-binding [D’Souza et al., 2009; Pierce et al., 2012] and show that RUNX2 can regulate BC metabolic activity. Further, RUNX2 may promote BC progression in some cells by suppressing SIRT6-mediated mitochondrial respiration. RUNX2 inhibited SIRT6 mRNA expression, interacted with the SIRT6 promoter, repressed SIRT6 promoter activity, and was associated with lower levels of SIRT6 protein. These results reveal a novel mechanism through which RUNX2 regulates cellular metabolism and promotes tumor progression in BC cells in part by altering glycolytic gene expression or mitochondrial respiration (Fig 8C).
RUNX2 has been shown to exhibit tumor suppressor properties by antagonizing ERα oncogenic activity [Chimge et al., 2012]. However, RUNX2 can also increase growth in soft agar of NIH323 fibroblasts, an oncogenic function [Qiao et al., 2006; Vitolo et al., 2007]. Current results show that RUNX2 increased anchorage-independent growth in soft agar (Fig 1) and mammosphere formation (Fig 2), a cancer stem cell phenotype [Kim et al., 2012] and an indicator of oncogenic potential [Dimri et al., 2005]. Triple negative Hs578t are part of the NCI60 panel of cell lines that express putative tumor stem cell markers [Stuelten et al., 2010], oncogenic (mutant) p53, mutant ras, and c-myc, but lack p16 [Ikediobi et al., 2006]. Others have reported that Hs578t cells also express high levels of RUNX2 protein [Lau et al., 2006], but do not grow well in vivo in mice or as anchorage-independent colonies in soft agar [Hughes et al., 2008]. However, we found that these cells grow readily as mammospheres in suspension, which might reflect their claudin-low phenotype, expression of EMT markers, and stem-like characteristics [Prat et al., 2010]. RUNX2 is known to cooperate with TGFβ through RUNX/Smad transcriptional regulation of target genes [Pratap et al., 2006]. TGFβ displays a dual role in breast cancer, inhibiting growth at early stages while increasing growth in late stages of tumor progression [Massague, 2008]. Although the addition of TGFβ did not augment metabolic gene expression in these cells, it is possible that constitutive Smad activation may still cooperate with RUNX2 to alter BC metabolism. Our results are consistent with reciprocal effects of RUNX2 and ER [Chimge et al., 2012] and with a role for RUNX2 in promoting a BC cell oncogenic phenotype. Luminal breast cancers are heterogeneous [Berman et al., 2010; Eroles et al., 2012] and a subpopulation of MCF7 cells was found to exhibit stem cell properties and express RUNX2 [Kim et al., 2012]. BC cells that gain RUNX2 expression may acquire a survival advantage mediated by GLUT1 expression, glucose uptake, and glucose utilization. Our results are consistent with the hypothesis that RUNX2 supports a glycolytic phenotype in cells with Hif1α, but low SIRT6 expression. Other studies have found that SIRT6-deficient cells express higher Hif1α, with increased glycolysis and diminished mitochondrial respiration [Zhong et al., 2010]. RUNX2 interacts with Hif1α through the RUNX DNA-binding domain, leading to stabilization of Hif1α protein [Kwon et al., 2011], as observed in RUNX2 expressing MCF7 cells (Supp Fig S4). Extended starvation leads to reduction in Hif1α levels perhaps because of depletion of metabolic intermediates (such as succinate and fumarate) that normally inhibit prolyl hydroxylases to maintain higher levels of Hif1α. It is also possible that RUNX2/Hif1α cooperation could regulate metabolic reprogramming through PI3K/Erk or PI3K/Akt-mediated RUNX2 phosphorylation [Ge et al., 2009; Pande et al., 2013; Qiao et al., 2004].
RUNX2 expression was associated with elevated PDHK1, HK2, GLUT1, and lower PDHA1, which are known to promote glycolysis. Higher PDHA1 levels and activity in RUNX2 knockdown cells (relative to RUNX2+ cells) are indicative of the potential to divert glucose towards mitochondrial oxidative phosphorylation. SIRT6 and RUNX2 play important roles in several common biological processes: metabolism, senescence, aging, bone formation, and insulin secretion. But, so far, no link between these transcriptional regulators has been reported. The association of RUNX2 and SIRT6 may contribute to ubiquitination and proteasomal degradation of SIRT6 (Fig 6; Supp Fig S5). In addition, when cultured in low glucose, SIRT6 mRNA expression was low in RUNX2+ cells but was higher in RUNX2 knockdown cells (Fig 7). RUNX2 associated with the SIRT6 promoter (Fig 7) and RUNX2+ cells, which express low SIRT6, exhibited lower SIRT6 promoter-luciferase activity than RUNX2 knockdown cells, which express higher SIRT6. The ability of RUNX2 to repress SIRT6 promoter activity in parental cells that express low SIRT6 may be due to the presence of sufficient endogenous SIRT6 cofactor that cooperates with RUNX2. However, it is possible that other co-repressors (such as HDAC6) that are known to interact with RUNX2 could also cooperate with RUNX2 to inhibit SIRT6 expression. The data suggest that RUNX2 can be actively recruited to SIRT6 promoter sites and that it may be a regulatory factor to repress SIRT6 gene expression in BC cells. RUNX2 expression may not only promote a glycolytic phenotype, but may also suppress mitochondrial respiration (Fig 5). Knockdown of endogenous SIRT6 in Hs578t/55.5 cells (low RUNX2) inhibited mitochondrial respiration (Fig 5E) without elevating RUNX2 (Supp Fig S3), suggesting that the increased OCR observed upon lowering RUNX2 levels was dependent on SIRT6. Similarly, MEFs from SIRT6 KO mice, which exhibit defects in bone formation and a lethal hypoglycemia [Mostoslavsky et al., 2006], exhibit reduced OCR relative to wild type MEFs. In these cells, SIRT6 promoted glucose utilization through mitochondrial oxidative phosphorylation [Zhong et al., 2010]. Our results reveal for the first time that the changes in metabolism in response to RUNX2 involve inhibition of oxygen utilization because of SIRT6 repression. However, it is possible that RUNX2 may regulate other mitochondrial factors that control oxygen consumption.
Glucose uptake as measured by 18F-2DG PET scans has been used clinically for many years to diagnose primary tumors and metastatic lesions [Wu and Gambhir, 2003]. Our data reveal that in tumors with high GLUT1 expression, membrane-bound GLUT1 increased with tumor grade and was associated with RUNX2 expression, which is consistent with higher glucose uptake values and a glycolytic phenotype in poorly-differentiated tumors [Cairns et al., 2011; Zhang et al., 2011]. Expression of RUNX2 in cancer tissues was observed in the cytoplasm and nucleus (Supp Fig S7). Previous work reported RUNX2 cytoplasmic staining in epithelial and stromal cells of prostate tumors [Yun et al., 2012] where nuclear staining correlated with metastatic disease. RUNX2 expression was also observed in the cytoplasm of cells in culture [Deepak et al., 2011; Kim et al., 2003] and taxol treatment led to the accumulation of RUNX2 in the cytosol because of failure to translocate to the nucleus after synthesis [Pockwinse et al., 2006]. Low SIRT6 but high RUNX2 staining was observed in late stage II–III BC tissue with high Ki67 (proliferative index) but low ERα expression (Fig 8), which is consistent with a tumor suppressor role for SIRT6 in BC progression. A recent study has also shown that SIRT6 acts as a tumor suppressor in gastro-intestinal cancers [Sebastian et al., 2012]. In summary, RUNX2 appears to attenuate SIRT6 expression, which is a critical regulator of glucose homeostasis, especially under glucose restrictive conditions that are common in the tumor microenvironment.
Supplementary Material
Supplemental Figure S1: Breast cancer cell models expressing RUNX2. (A) Inducible RUNX2 expression in ERα+ MCF7 cells was established using the BD™-Tet-Off System. RUNX2 and ERα expression were measured with specific antibodies. RUNX2+ MCF7 cells expressed lower levels of ERα (mean = 0.56 ± 0.04) relative to RUNX2− cells (mean = 0.74 ± 0.01; p < 0.018; t-test). (B) Triple negative Hs578t cells were infected with lentiviral shRNA targeting endogenous RUNX2. After stable selection, clones with reduced RUNX2 expression relative to parental cells were analyzed (Hs578t/53.5; Hs578t/55.5) while another clone (Hs578t/54.5) served as a control knockdown clone that did not show a decrease in RUNX2 expression. MCF7.TetOFF (RUNX2−) or cells expressing RUNX2 (RUNX2+) (C) or Hs578t parental or shRNA knockdown cells (D) were grown for 3 days in complete media. Cell growth was measured using crystal violet staining (Absorbance 540nm). Each point is the mean ± SD of triplicate samples; p > 0.05).
Supplemental Figure S2: SIRT6 expression in Hs578t knockdown clones. (A) Comparison of SIRT6 expression in parental (Hs578t), control knockdown 54.5, or Runx2 knockdown (55.5) Hs578t cells. Cells were culture in full media containing 10%FBS and 25mM glucose (FM) or in glucose starvation media containing 2%FBS for 8hr (S). Nuclear extracts were examined for RUNX2 and SIRT6 expression by Western blot. Relative SIRT6 protein expression normalized to actin (NIH Image-J) from scanned blots is indicated under each lane. (B) Overexpression of SIRT6 in breast cancer cells. Hs578t cells (RUNX2+) were transfected with empty vector (Vector) or cDNA expression vector encoding human SIRT6. After a brief selection (1 week), nuclear extracts were isolated and analyzed for SIRT6 and RUNX2 expression by Western blotting. SIRT6 relative density in arbitrary units (AU) normalized to actin was calculated from three determinations using NIH Image-J; * indicates p < 0.05 relative to Vector. (C) Glucose starvation-resistant cells (Hs578t.LG) from Hs578t cells were obtained in low glucose (0.5mM with 5% FBS) for 4 days. Surviving cells were expanded in standard cell culture media (DMEM+10% FBS) for 10–14 days. Cells were analyzed for RUNX2 and GLUT1 expression. Significant differences in RUNX2 (p < 0.01; t-test) and GLUT1 (p < 0.06; t-test) expression for LG clones compared to parental cells were calculated by Image-J. (D) Hs578t parental and LG2 cells were treated with different concentrations of glucose as indicated and analyzed for SIRT6 expression after 16hr. Relative SIRT6 protein expression normalized to actin (NIH Image-J) from scanned blots is indicated.
Supplemental Figure S3: Mitochondrial OCR in MCF7 and Hs578t cells . (A) MCF7 RUNX2 + or RUNX2 − cells were compared for OCR using the Seahorse metabolic flux analyzer. FCCP was used to depolarize the inner mitochondrial membrane and inhibitors of mitochondrial ETC were used as in Figure 5. Treatments include: FCCP + pyruvate (Pyr) ( 0.4μM + 10mM), FCCP (0.1μM), FCCP (0.1μM), and antimycin-A (1μM). Cell protein was extracted after determination of OCR and was similar for RUNX2+ (10.68 ± 0.17 μg) and RUNX2− (10.10 ± 2.07 μg) from n = 11 wells per sample (p > 0.05)
(B) Hs578tP (Parental) or control knockdown (Hs578t/54.5) cells were compared for OCR using the Seahorse metabolic flux analyzer. As in Figure 5, FCCP (0.75μM), pyruvate (10mM), and antimycin-A (1μM) were used to treat cells. Oligomycin (2.5nM) inhibition indicates that in these cells >95% of the oxygen consumption was linked to mitochondrial ATP synthesis. Inset shows RUNX2 expression in Hs578t/54.5 (control knockdown) and Hs578t/55.5 (RUNX2 knockdown) cells compared to parental cells. (C) Knockdown of SIRT6 in breast cancer cells. Hs578t/55.5 (low RUNX2 expression) cells were transfected with scrambled (Control) siRNA or three different SIRT6-specific siRNA oligonucleotides (siRNA A, siRNA B, and siRNA C) from Origene. Levels of SIRT6 were determined by Western blot with specific SIRT6 antibody. Relative density in arbitrary units (AU) represents the mean from three determinations normalized to actin (NIH Image-J); * indicates p < 0.05 relative to Control. (D) Hs578t/55.5 cells were transfected with control or siRNA C targeting SIRT6. SIRT6 and RUNX2 protein levels are shown and compared relative to actin. The Western blot was overexposed to visualize the low levels of RUNX2 in 55.5 knockdown cells. Band density relative to control is indicated.
Supplemental Figure S4: Hif1α and SIRT6 expression in response to RUNX2. (A) MCF7 cells cultured in the presence (+Dox; RUNX2-) or absence of doxycycline (−Dox; RUNX2+) were starved in the absence of glucose for 16hr to lower Hif1α levels and then treated with 5mM glucose for the indicated times. Some cells were treated with the SIRT inhibitor, sirtinol (Sirt; 10μM) and 5mM glucose for 8hr. Cell lysates were prepared and the expression of Hif1α, SIRT6, or RUNX2 was measured by Western blotting. Gels were stripped and reprobed for β-actin (loading control). Relative density of scanned Western blot (NIH Image-J) was normalized to actin and expressed as arbitrary units (AU). (B) MCF7 cells cultured in +Dox or −Dox to induce RUNX2 were cultured in 5mM glucose, placed in a hypoxia chamber, and exposed to 1%O2 for the indicated times. Cells were lysed and the expression of Hif1α was measured by Western blotting. Relative band intensities were determined with NIH Image-J software from these Western blots. Essentially similar results were obtained from two additional determinations
Supplemental Figure S5: Ubiquitination of SIRT6 and transcriptional activity of RUNX2 in HEK293T cells. (A) Cells were transfected with SIRT6 and/or RUNX2 expression vectors for 24hr and treated with MG132 for 4hr. Nuclear extracts were isolated and immunoprecipitated with SIRT6-specific antibody followed by Western blotting using anti-Ubiquitin or SIRT6 antibodies. (B) HEK293 cells were transfected with the indicated plasmids (Flag empty vector, Flag.RUNX2, Flag.SIRT6, or SIRT6.prom.Luc) for 24hr prior to incubation in low glucose (5mM) for 6hr. Relative GLuc:SEAP luciferase units were measured from conditioned media. Differences in SIRT6 promoter activity after RUNX2 transfection compared to Flag vector are indicated (NS; ANOVA). Differences in activity from RUNX2 + SIRT6 expression relative to SIRT6 + Flag are indicated (*p < 0.05; ANOVA).
Supplemental Figure S6: Clinical correlates, RUNX2, and metabolic profiling in BC patients with IDC. Tissues from patients admitted to the University of Maryland Greenebaum Cancer Center with invasive ductal carcinoma were analyzed for GLUT1 and RUNX2 expression by immunohistochemistry (IHC). Scale bar = 400μm for all panels. Inset magnification shows areas with cytoplasmic, membrane, nuclear, or negative staining. Tumor profiling for patients with IDC was performed by the Department of Anatomic Pathology (University of Maryland School of Medicine) and included ER/PR status, proliferative index (Ki67), and standard uptake values (SUV) from 18F-DG positron emission tomography (FDG-PET) scans.
Supplemental Figure S7: Expression of RUNX2, GLUT1, and ERα in human BC. (A) Panels show IHC representative photos of normal or stage III tissue stained with H & E or specific antibodies recognizing RUNX2 or Glut1. Controls used secondary antibody alone (2° Ab) and negative tissue stain for GLUT1 or RUNX2. Scale bar = 100μm. (B) Results from TMA of normal, stage I (fibroadenoma/ductal carcinoma in situ – FA/DCIS), stage II, stage II/III, and stage III patients. Panels show IHC representative photos of tissue expressing RUNX2 or GLUT1 vs stage: cytoplasmic GLUT1 (GLUT1C), membrane-localized GLUT1 (GLUT1M), cytoplasmic RUNX2 (RUNX2C), and nuclear RUNX2 (RUNX2N). The Spearman correlation coefficient was used to determine whether total RUNX2 expression was different across the five groups (normal, FA/DCIS, stage II, stage II/III, and stage III). Significant differences between RUNX2 and GLUT1 across these groups were noted (p < 0.0001). A negative correlation between RUNX2 and cytosolic GLUT1 in normal tissues (r = −0.75) and a possible positive correlation between RUNX2 and membrane-bound GLUT1 in stage III cancers (r = 0.42) were also noted. Scale bar = 100μm. Inset magnification shows cytoplasmic, membrane, or nuclear localization. (C) Association between total RUNX2 and ERα. Using the Spearman correlation coefficient, RUNX2 total and ERα were assessed for each group: Normal (n = 16); FA/DCIS (n = 7); Stage II (n = 88); Stage II/III (n = 14); Stage III (n = 16). A negative correlation between RUNX2 and ERα in normal tissue and in stage III tumors was noted (r = −0.48).
Acknowledgments
We appreciate the advice and support of other members of the laboratory during the preparation of this manuscript including Maria Mochin, Karen Underwood, Adam Pierce, Haeyoung Cho, and Helen Zhao. We also thank Dr. CY Lin (Georgetown University, Washington, DC) for his generous gift of monoclonal anti-Flag antibody.
This work was supported by:
Contract grant sponsor: VA Research and Development Service
Contract grant number: Merit award I01 BX002205A (AP, MC)
Contract grant sponsor: State of Maryland University of Maryland Greenebaum Cancer Center
Contract grant number: Cigarette Restitution Fund (AP, MC, GDG)
Contract grant sponsor: Department of Pathology
Contract grant number: Institutional Funds (XFZ, SL)
Contract grant sponsor: NIH
Contract grant number: R01NS064978 (BMP)
Contract grant sponsor: NIH
Contract grant number: P01HD16596-26A1 (GMF, BMP)
ABBREVIATIONS
- BC
breast cancer
- 2-DG
2-deoxy-glucose
- ECAR
extracellular acidification rate
- ECL
enhanced chemiluminescence
- ER-α
estrogen receptor-α
- FCCP
Trifluoromethoxy carbonylcyanide phenylhydrazone
- GLUT1
glucose transporter-1
- HK2
hexokinase-2
- IHC
immunohistochemistry
- LDHA1
lactate dehydrogenase isoform A1
- OCR
oxygen consumption rate
- Ox/phos
oxidative phosphorylation
- PDHA1
pyruvate dehydrogenase isoform A1
- PDHK1
pyruvate dehydrogenase kinase-1
- SEAP
secreted alkaline phosphatase
- TCA
tricarboxylic acid
- TMA
tissue microarray
Footnotes
Authors’ Contributions
MC and AP designed the experiments. MC, BP, GF, and AP developed the methodology. MC, SC, JB, XFZ, SL, LB, MSK, and BP acquired the data, provided specimens, or analyzed specimens. MC, AP, MSK, and OG analyzed and interpreted the data. MC, AP, BP, and GG wrote and edited the manuscript. GG and AP supervised the study.
The authors declare that they have no conflicts of interest.
Contributor Information
Moran Choe, Email: mchoe001@umaryland.edu.
Jessica L. Brusgard, Email: jbenn003@umaryland.edu.
Sara Chumsri, Email: schumsri@umm.edu.
Lekhana Bhandary, Email: lekhanabhandary@umaryland.edu.
X. Frank Zhao, Email: Frank.Zhao2@va.gov.
Song Lu, Email: song_lu@hotmail.com.
Olga G. Goloubeva, Email: goloubeva@som.umaryland.edu.
Brian M. Polster, Email: bpolster@anes.umm.edu.
Gary M. Fiskum, Email: gfiskum@anes.umm.edu.
Geoffrey D. Girnun, Email: Geoffrey.Girnun@stonybrookmedicine.edu.
Myoung Sook Kim, Email: MKim@som.umaryland.edu.
Antonino Passaniti, Email: apass001@umaryland.edu.
References
- Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64:4506–13. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- Berman HK, Gauthier ML, Tlsty TD. Premalignant breast neoplasia: a paradigm of interlesional and intralesional molecular heterogeneity and its biological and clinical ramifications. Cancer Prev Res (Phila) 2010;3:579–87. doi: 10.1158/1940-6207.CAPR-10-0073. [DOI] [PubMed] [Google Scholar]
- Blick T, Widodo E, Hugo H, Waltham M, Lenburg ME, Neve RM, Thompson EW. Epithelial mesenchymal transition traits in human breast cancer cell lines. Clin Exp Metastasis. 2008;25:629–42. doi: 10.1007/s10585-008-9170-6. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–13. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Little GH, Chen YB, Kahn M, Tripathy D, Borok Z, Frenkel B. Regulation of breast cancer metastasis by Runx2 and estrogen signaling: the role of SNAI2. Breast Cancer Res. 2011;13:R127. doi: 10.1186/bcr3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge NO, Baniwal SK, Luo J, Coetzee S, Khalid O, Berman BP, Tripathy D, Ellis MJ, Frenkel B. Opposing effects of Runx2 and estradiol on breast cancer cell proliferation: in vitro identification of reciprocally regulated gene signature related to clinical letrozole responsiveness. Clin Cancer Res. 2012;18:901–11. doi: 10.1158/1078-0432.CCR-11-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Polster BM. Investigation of mitochondrial dysfunction by sequential microplate-based respiration measurements from intact and permeabilized neurons. PLoS One. 2012;7:e34465. doi: 10.1371/journal.pone.0034465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza DR, Salib MM, Bennett J, Mochin-Peters M, Asrani K, Goldblum SE, Renoud KJ, Shapiro P, Passaniti A. Hyperglycemia Regulates RUNX2 Activation and Cellular Wound Healing through the Aldose Reductase Polyol Pathway. J Biol Chem. 2009;284:17947–55. doi: 10.1074/jbc.M109.002378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Leong DT, Gupta A, Shen L, Putti T, Stein GS, van Wijnen AJ, Salto-Tellez M. Positive association between nuclear Runx2 and oestrogen-progesterone receptor gene expression characterises a biological subtype of breast cancer. Eur J Cancer. 2009;45:2239–48. doi: 10.1016/j.ejca.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepak V, Zhang Z, Meng L, Zeng X, Liu W. Reduced activity and cytoplasmic localization of Runx2 is observed in C3h10t1/2 cells overexpressing Tbx3. Cell Biochem Funct. 2011;29:348–50. doi: 10.1002/cbf.1753. [DOI] [PubMed] [Google Scholar]
- Dimri G, Band H, Band V. Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res. 2005;7:171–9. doi: 10.1186/bcr1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radic Biol Med. 2010;48:905–14. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Ferrari N, McDonald L, Morris JS, Cameron ER, Blyth K. RUNX2 in mammary gland development and breast cancer. J Cell Physiol. 2013;228:1137–42. doi: 10.1002/jcp.24285. [DOI] [PubMed] [Google Scholar]
- Fukumura D, Jain RK. Tumor microvasculature and microenvironment: targets for anti-angiogenesis and normalization. Microvasc Res. 2007;74:72–84. doi: 10.1016/j.mvr.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C, Xiao G, Jiang D, Yang Q, Hatch NE, Roca H, Franceschi RT. Identification and functional characterization of ERK/MAPK phosphorylation sites in the Runx2 transcription factor. J Biol Chem. 2009;284:32533–43. doi: 10.1074/jbc.M109.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttilla IK, Adams BD, White BA. ERalpha, microRNAs, and the epithelial-mesenchymal transition in breast cancer. Trends Endocrinol Metab. 2012a;23:73–82. doi: 10.1016/j.tem.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res Treat. 2012b;132:75–85. doi: 10.1007/s10549-011-1534-y. [DOI] [PubMed] [Google Scholar]
- Herman MA, Kahn BB. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J Clin Invest. 2006;116:1767–75. doi: 10.1172/JCI29027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes L, Malone C, Chumsri S, Burger AM, McDonnell S. Characterisation of breast cancer cell lines and establishment of a novel isogenic subclone to study migration, invasion and tumourigenicity. Clin Exp Metastasis. 2008;25:549–57. doi: 10.1007/s10585-008-9169-z. [DOI] [PubMed] [Google Scholar]
- Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O’Meara S, Santarius T, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Hunter C, Jenkinson A, Jones D, Kosmidou V, Lugg R, Menzies A, Mironenko T, Parker A, Perry J, Raine K, Richardson D, Shepherd R, Small A, Smith R, Solomon H, Stephens P, Teague J, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Reinhold W, Weinstein JN, Stratton MR, Futreal PA, Wooster R. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of SIRT6 protein levels by nutrient availability. FEBS Lett. 2008;582:543–8. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Villadsen R, Sorlie T, Fogh L, Gronlund SZ, Fridriksdottir AJ, Kuhn I, Rank F, Wielenga VT, Solvang H, Edwards PA, Borresen-Dale AL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Tumor initiating but differentiated luminal-like breast cancer cells are highly invasive in the absence of basal-like activity. Proc Natl Acad Sci U S A. 2012;109:6124–9. doi: 10.1073/pnas.1203203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, Karsenty G, Taniguchi T, Takayanagi H. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–91. doi: 10.1101/gad.1119303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon TG, Zhao X, Yang Q, Li Y, Ge C, Zhao G, Franceschi RT. Physical and functional interactions between Runx2 and HIF-1alpha induce vascular endothelial growth factor gene expression. J Cell Biochem. 2011;112:3582–93. doi: 10.1002/jcb.23289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, Bhalla KN, Zhu T, Ito Y, Sukumar S. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66:6512–20. doi: 10.1158/0008-5472.CAN-06-0369. [DOI] [PubMed] [Google Scholar]
- Lee NK, Karsenty G. Reciprocal regulation of bone and energy metabolism. Trends Endocrinol Metab. 2008;19:161–6. doi: 10.1016/j.tem.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Ho AD, Voelter-Mahlknecht S. Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int J Oncol. 2006;28:447–56. [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–6. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Zielenska M, Stein GS, van Wijnen AJ, Squire JA. The Role of RUNX2 in Osteosarcoma Oncogenesis. Sarcoma. 2011;2011:282745. doi: 10.1155/2011/282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta in Cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–29. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Miki Y, Suzuki T, Takagi K, Akahira J, Sakyu T, Watanabe M, Inoue S, Ishida T, Ohuchi N, Sasano H. Runx2 in human breast carcinoma: its potential roles in cancer progression. Cancer Sci. 2010;101:2670–5. doi: 10.1111/j.1349-7006.2010.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande S, Browne G, Padmanabhan S, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Oncogenic cooperation between PI3K/Akt signaling and transcription factor Runx2 promotes the invasive properties of metastatic breast cancer cells. J Cell Physiol. 2013;228:1784–92. doi: 10.1002/jcp.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AD, Anglin IE, Vitolo MI, Mochin MT, Underwood KF, Goldblum SE, Kommineni S, Passaniti A. Glucose-activated RUNX2 phosphorylation promotes endothelial cell proliferation and an angiogenic phenotype. J Cell Biochem. 2012;113:218–92. doi: 10.1002/jcb.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockwinse SM, Rajgopal A, Young DW, Mujeeb KA, Nickerson J, Javed A, Redick S, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Doxsey SJ. Microtubule-dependent nuclear-cytoplasmic shuttling of Runx2. J Cell Physiol. 2006;206:354–62. doi: 10.1002/jcp.20469. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Qiao M, Shapiro P, Fosbrink M, Rus H, Kumar R, Passaniti A. Cell cycle-dependent phosphorylation of the RUNX2 transcription factor by cdc2 regulates endothelial cell proliferation. J Biol Chem. 2006;281:7118–28. doi: 10.1074/jbc.M508162200. [DOI] [PubMed] [Google Scholar]
- Qiao M, Shapiro P, Kumar R, Passaniti A. Insulin-like Growth Factor-1 Regulates Endogenous RUNX2 Activity in Endothelial Cells Through a PI3K/ERK-dependent and Akt-independent Signaling Pathway. J Biol Chem. 2004;279:42709–18. doi: 10.1074/jbc.M404480200. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–25. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, Cosentino C, Greenson JK, Macdonald AI, McGlynn L, Maxwell F, Edwards J, Giacosa S, Guccione E, Weissleder R, Bernstein BE, Regev A, Shiels PG, Lombard DB, Mostoslavsky R. The Histone Deacetylase SIRT6 Is a Tumor Suppressor that Controls Cancer Metabolism. Cell. 2012;151:1185–99. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Reece RJ. Eukaryotic transcription factors as direct nutrient sensors. Trends Biochem Sci. 2005;30:405–12. doi: 10.1016/j.tibs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of cancer cell metabolism by hypoxia-inducible factor 1. Semin Cancer Biol. 2009;19:12–6. doi: 10.1016/j.semcancer.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Stuelten CH, Mertins SD, Busch JI, Gowens M, Scudiero DA, Burkett MW, Hite KM, Alley M, Hollingshead M, Shoemaker RH, Niederhuber JE. Complex display of putative tumor stem cell markers in the NCI60 tumor cell line panel. Stem Cells. 2010;28:649–60. doi: 10.1002/stem.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood KF, D’Souza DR, Mochin MT, Pierce A, Kommineni S, Choe M, Bennett J, Gnatt A, Habtemariam B, Mackerell AD, Jr, Passaniti A. Regulation of RUNX2 transcription factor-DNA interactions and cell proliferation by Vitamin D3 (cholecalciferol) prohormone activity. J Bone Min Res. 2012;27:913–25. doi: 10.1002/jbmr.1504. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitolo MI, Anglin IE, Mahoney WM, Jr, Renoud KJ, Gartenhaus RB, Bachman KE, Passaniti A. The RUNX2 Transcription Factor Cooperates with the YES-associated Protein, YAP65, to Promote Cell Transformation. Cancer Biol Ther. 2007;6:856–863. doi: 10.4161/cbt.6.6.4241. [DOI] [PubMed] [Google Scholar]
- Wu D, Gambhir SS. Positron emission tomography in diagnosis and management of invasive breast cancer: current status and future perspectives. Clin Breast Cancer. 2003;4(Suppl 1):S55–63. doi: 10.3816/cbc.2003.s.016. [DOI] [PubMed] [Google Scholar]
- Yun SJ, Yoon HY, Bae SC, Lee OJ, Choi YH, Moon SK, Kim IY, Kim WJ. Transcriptional repression of RUNX2 is associated with aggressive clinicopathological outcomes, whereas nuclear location of the protein is related to metastasis in prostate cancer. Prostate Cancer Prostatic Dis. 2012;15:369–73. doi: 10.1038/pcan.2012.31. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, Jung HJ, McCaffery JM, Kurland IJ, Reue K, Lee WN, Koehler CM, Teitell MA. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. Embo J. 2011;30:4860–73. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem. 2003;278:27939–44. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–93. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Breast cancer cell models expressing RUNX2. (A) Inducible RUNX2 expression in ERα+ MCF7 cells was established using the BD™-Tet-Off System. RUNX2 and ERα expression were measured with specific antibodies. RUNX2+ MCF7 cells expressed lower levels of ERα (mean = 0.56 ± 0.04) relative to RUNX2− cells (mean = 0.74 ± 0.01; p < 0.018; t-test). (B) Triple negative Hs578t cells were infected with lentiviral shRNA targeting endogenous RUNX2. After stable selection, clones with reduced RUNX2 expression relative to parental cells were analyzed (Hs578t/53.5; Hs578t/55.5) while another clone (Hs578t/54.5) served as a control knockdown clone that did not show a decrease in RUNX2 expression. MCF7.TetOFF (RUNX2−) or cells expressing RUNX2 (RUNX2+) (C) or Hs578t parental or shRNA knockdown cells (D) were grown for 3 days in complete media. Cell growth was measured using crystal violet staining (Absorbance 540nm). Each point is the mean ± SD of triplicate samples; p > 0.05).
Supplemental Figure S2: SIRT6 expression in Hs578t knockdown clones. (A) Comparison of SIRT6 expression in parental (Hs578t), control knockdown 54.5, or Runx2 knockdown (55.5) Hs578t cells. Cells were culture in full media containing 10%FBS and 25mM glucose (FM) or in glucose starvation media containing 2%FBS for 8hr (S). Nuclear extracts were examined for RUNX2 and SIRT6 expression by Western blot. Relative SIRT6 protein expression normalized to actin (NIH Image-J) from scanned blots is indicated under each lane. (B) Overexpression of SIRT6 in breast cancer cells. Hs578t cells (RUNX2+) were transfected with empty vector (Vector) or cDNA expression vector encoding human SIRT6. After a brief selection (1 week), nuclear extracts were isolated and analyzed for SIRT6 and RUNX2 expression by Western blotting. SIRT6 relative density in arbitrary units (AU) normalized to actin was calculated from three determinations using NIH Image-J; * indicates p < 0.05 relative to Vector. (C) Glucose starvation-resistant cells (Hs578t.LG) from Hs578t cells were obtained in low glucose (0.5mM with 5% FBS) for 4 days. Surviving cells were expanded in standard cell culture media (DMEM+10% FBS) for 10–14 days. Cells were analyzed for RUNX2 and GLUT1 expression. Significant differences in RUNX2 (p < 0.01; t-test) and GLUT1 (p < 0.06; t-test) expression for LG clones compared to parental cells were calculated by Image-J. (D) Hs578t parental and LG2 cells were treated with different concentrations of glucose as indicated and analyzed for SIRT6 expression after 16hr. Relative SIRT6 protein expression normalized to actin (NIH Image-J) from scanned blots is indicated.
Supplemental Figure S3: Mitochondrial OCR in MCF7 and Hs578t cells . (A) MCF7 RUNX2 + or RUNX2 − cells were compared for OCR using the Seahorse metabolic flux analyzer. FCCP was used to depolarize the inner mitochondrial membrane and inhibitors of mitochondrial ETC were used as in Figure 5. Treatments include: FCCP + pyruvate (Pyr) ( 0.4μM + 10mM), FCCP (0.1μM), FCCP (0.1μM), and antimycin-A (1μM). Cell protein was extracted after determination of OCR and was similar for RUNX2+ (10.68 ± 0.17 μg) and RUNX2− (10.10 ± 2.07 μg) from n = 11 wells per sample (p > 0.05)
(B) Hs578tP (Parental) or control knockdown (Hs578t/54.5) cells were compared for OCR using the Seahorse metabolic flux analyzer. As in Figure 5, FCCP (0.75μM), pyruvate (10mM), and antimycin-A (1μM) were used to treat cells. Oligomycin (2.5nM) inhibition indicates that in these cells >95% of the oxygen consumption was linked to mitochondrial ATP synthesis. Inset shows RUNX2 expression in Hs578t/54.5 (control knockdown) and Hs578t/55.5 (RUNX2 knockdown) cells compared to parental cells. (C) Knockdown of SIRT6 in breast cancer cells. Hs578t/55.5 (low RUNX2 expression) cells were transfected with scrambled (Control) siRNA or three different SIRT6-specific siRNA oligonucleotides (siRNA A, siRNA B, and siRNA C) from Origene. Levels of SIRT6 were determined by Western blot with specific SIRT6 antibody. Relative density in arbitrary units (AU) represents the mean from three determinations normalized to actin (NIH Image-J); * indicates p < 0.05 relative to Control. (D) Hs578t/55.5 cells were transfected with control or siRNA C targeting SIRT6. SIRT6 and RUNX2 protein levels are shown and compared relative to actin. The Western blot was overexposed to visualize the low levels of RUNX2 in 55.5 knockdown cells. Band density relative to control is indicated.
Supplemental Figure S4: Hif1α and SIRT6 expression in response to RUNX2. (A) MCF7 cells cultured in the presence (+Dox; RUNX2-) or absence of doxycycline (−Dox; RUNX2+) were starved in the absence of glucose for 16hr to lower Hif1α levels and then treated with 5mM glucose for the indicated times. Some cells were treated with the SIRT inhibitor, sirtinol (Sirt; 10μM) and 5mM glucose for 8hr. Cell lysates were prepared and the expression of Hif1α, SIRT6, or RUNX2 was measured by Western blotting. Gels were stripped and reprobed for β-actin (loading control). Relative density of scanned Western blot (NIH Image-J) was normalized to actin and expressed as arbitrary units (AU). (B) MCF7 cells cultured in +Dox or −Dox to induce RUNX2 were cultured in 5mM glucose, placed in a hypoxia chamber, and exposed to 1%O2 for the indicated times. Cells were lysed and the expression of Hif1α was measured by Western blotting. Relative band intensities were determined with NIH Image-J software from these Western blots. Essentially similar results were obtained from two additional determinations
Supplemental Figure S5: Ubiquitination of SIRT6 and transcriptional activity of RUNX2 in HEK293T cells. (A) Cells were transfected with SIRT6 and/or RUNX2 expression vectors for 24hr and treated with MG132 for 4hr. Nuclear extracts were isolated and immunoprecipitated with SIRT6-specific antibody followed by Western blotting using anti-Ubiquitin or SIRT6 antibodies. (B) HEK293 cells were transfected with the indicated plasmids (Flag empty vector, Flag.RUNX2, Flag.SIRT6, or SIRT6.prom.Luc) for 24hr prior to incubation in low glucose (5mM) for 6hr. Relative GLuc:SEAP luciferase units were measured from conditioned media. Differences in SIRT6 promoter activity after RUNX2 transfection compared to Flag vector are indicated (NS; ANOVA). Differences in activity from RUNX2 + SIRT6 expression relative to SIRT6 + Flag are indicated (*p < 0.05; ANOVA).
Supplemental Figure S6: Clinical correlates, RUNX2, and metabolic profiling in BC patients with IDC. Tissues from patients admitted to the University of Maryland Greenebaum Cancer Center with invasive ductal carcinoma were analyzed for GLUT1 and RUNX2 expression by immunohistochemistry (IHC). Scale bar = 400μm for all panels. Inset magnification shows areas with cytoplasmic, membrane, nuclear, or negative staining. Tumor profiling for patients with IDC was performed by the Department of Anatomic Pathology (University of Maryland School of Medicine) and included ER/PR status, proliferative index (Ki67), and standard uptake values (SUV) from 18F-DG positron emission tomography (FDG-PET) scans.
Supplemental Figure S7: Expression of RUNX2, GLUT1, and ERα in human BC. (A) Panels show IHC representative photos of normal or stage III tissue stained with H & E or specific antibodies recognizing RUNX2 or Glut1. Controls used secondary antibody alone (2° Ab) and negative tissue stain for GLUT1 or RUNX2. Scale bar = 100μm. (B) Results from TMA of normal, stage I (fibroadenoma/ductal carcinoma in situ – FA/DCIS), stage II, stage II/III, and stage III patients. Panels show IHC representative photos of tissue expressing RUNX2 or GLUT1 vs stage: cytoplasmic GLUT1 (GLUT1C), membrane-localized GLUT1 (GLUT1M), cytoplasmic RUNX2 (RUNX2C), and nuclear RUNX2 (RUNX2N). The Spearman correlation coefficient was used to determine whether total RUNX2 expression was different across the five groups (normal, FA/DCIS, stage II, stage II/III, and stage III). Significant differences between RUNX2 and GLUT1 across these groups were noted (p < 0.0001). A negative correlation between RUNX2 and cytosolic GLUT1 in normal tissues (r = −0.75) and a possible positive correlation between RUNX2 and membrane-bound GLUT1 in stage III cancers (r = 0.42) were also noted. Scale bar = 100μm. Inset magnification shows cytoplasmic, membrane, or nuclear localization. (C) Association between total RUNX2 and ERα. Using the Spearman correlation coefficient, RUNX2 total and ERα were assessed for each group: Normal (n = 16); FA/DCIS (n = 7); Stage II (n = 88); Stage II/III (n = 14); Stage III (n = 16). A negative correlation between RUNX2 and ERα in normal tissue and in stage III tumors was noted (r = −0.48).