Functional development of CD4+ memory T cells features distinct, definable steps
Key words: Bcl6 (B-cell lymphoma 6), CD4 memory T-cell development, cognate interaction with non-GC B cells, memory B-cell recall response, stepwise transcriptional regulation, T-follicular helper cells
Abstract
Memory CD4+ T cells promote protective humoral immunity; however, how memory T cells acquire this activity remains unclear. This study demonstrates that CD4+ T cells develop into antigen-specific memory T cells that can promote the terminal differentiation of memory B cells far more effectively than their naive T-cell counterparts. Memory T cell development requires the transcription factor B-cell lymphoma 6 (Bcl6), which is known to direct T-follicular helper (Tfh) cell differentiation. However, unlike Tfh cells, memory T cell development did not require germinal center B cells. Curiously, memory T cells that develop in the absence of cognate B cells cannot promote memory B-cell recall responses and this defect was accompanied by down-regulation of genes associated with homeostasis and activation and up-regulation of genes inhibitory for T-cell responses. Although memory T cells display phenotypic and genetic signatures distinct from Tfh cells, both had in common the expression of a group of genes associated with metabolic pathways. This gene expression profile was not shared to any great extent with naive T cells and was not influenced by the absence of cognate B cells during memory T cell development. These results suggest that memory T cell development is programmed by stepwise expression of gatekeeper genes through serial interactions with different types of antigen-presenting cells, first licensing the memory lineage pathway and subsequently facilitating the functional development of memory T cells. Finally, we identified Gdpd3 as a candidate genetic marker for memory T cells.
Introduction
During the immune response, activated CD4+ T cells proliferate and differentiate into multiple effector cells with distinct functions, such as helper T-cell subset 1 (Th1), Th2, Th17, regulatory T (Treg) cells and T-follicular helper (Tfh) cells (1). Tfh cells are unique in their capacity, due to expression of the chemokine (C-X-C motif) receptor 5 (CXCR5), to migrate into B-cell follicles where they provide essential help to B cells for the germinal center (GC) reaction (2). B-cell lymphoma 6 (Bcl6) is a transcription factor essential for both Tfh-cell differentiation and GC B-cell development. Bcl6-deficient CD4+ T cells fail to develop into Tfh cells, resulting in impaired GC B-cell development and reduced high-affinity antibody memory responses (3–6).
The majority of CD4+ effector cells generated during an immune response die by apoptosis during the contraction phase that occurs after the peak of the response. However, some survive as memory cells and are maintained for a long period in peripheral tissues (7). These CD4+ memory T cells are heterogeneous in terms of their origin and phenotype. Based on analysis of antigen-specific T cells generated during acute lymphocytic choriomeningitis virus (LCMV) infection, it has been suggested that virus-specific Th1 and Tfh effector cells can differentiate into Th1- and Tfh-like memory cells, respectively (8, 9). Likewise, Listeria monocytogenes (Lm) infection generates Th1 effector memory cells and Tfh-like memory cells expressing CC chemokine receptor 7 (CCR7)+ (10), a characteristic feature of central memory cells as reported by Sallusto et al. (11). Generation of CXCR5+ Tfh-like memory cells in response to protein antigens has been also reported (12, 13).
CD4+ memory T cells are distinguished from naive CD4+ T cells by their longevity and characteristic functions. In response to pathogens, Th1- and Tfh-like CD4+ memory T cells proliferate more extensively than naive T cells, and this is then followed by the production of large quantities of cytokines and the generation of effector cells with Tfh and Th1 signatures (7–10). In response to protein antigens, it has been reported that Tfh-like CD4 memory T cells enhance the GC reaction and class switching in a primary B-cell response more efficiently than the primary responding CD4 T cells (14). However, it remains unclear how effector cells survive the contraction phase and are converted to quiescent memory cells with such unique activities.
In the present study, based on our observation that CD4+ memory T cells play a pivotal role in humoral immunity by controlling the terminal differentiation of memory B cells, we evaluated cellular events directing the fate of effector CD4 T cells differentiating into memory cells in vivo by measuring their longevity and acquisition of functionality to promote memory B-cell recall responses. Using a combination of analyses for cellularity, surface phenotype, function and genetic signatures, our results led to a stepwise developmental model for CD4 memory T cells. It begins with lineage commitment due to Bcl6 expression followed by the expression of high levels of transcripts associated with metabolic pathways and homeostasis, events that are, in part, shared with Tfh cells. Subsequently, through cognate interaction with B cells, mainly non-GC B cells, memory precursor T cells undergo dynamic changes in gene regulation and acquire the capacity to support the memory B-cell recall response. From a general perspective, we propose that such stepwise gene regulation is a fundamental strategy used by the immune system to ensure the proper development of memory T cells with specific functions.
Methods
Mice
Eight to ten-week-old C57BL/6 mice were purchased from Clea Inc. Bcl6-flox mice have been described previously (6). The mb1-cre mice and B1-8hi mice were kindly provided by Drs M. Reth and M. Nussenzweig, respectively. OT-II transgenic mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). Rag-1−/− and CD4-cre mice were obtained from Taconic (Hudson, NY, USA). All experiments were performed in accordance with guidelines established by the RIKEN Animal Safety Committee.
ELISA and ELISPOT assays
ELISA and ELISPOT assays were performed as described previously (15).
Adoptive transfer of carrier-primed CD4 T cells
Splenocytes were prepared from Bcl6+/+/CD4-cre and Bcl6f/f/CD4-cre, or Bcl6+/+/mb1-cre+/− and Bcl6f/f/mb1-cre+/− mice that had been immunized with chicken gamma globulin (CG) 40 days previously, and CD4+ T cells were enriched using the MACS system (Miltenyi Biotech, Gladbach, Germany). (4-hydroxy-3-nitrophenyl)acetyl (NP)-specific IgG1+ memory B cells were purified by FACS sorting from the pooled spleens of CG coupled to NP (NP-CG)-primed C57BL/6 mice at day 40 post-immunization (6). Naive B cells were enriched from C57BL/6 mice as described previously (6). CG-primed CD4 T cells (4×106) and naive B cells (1×106), as filler cells, with or without IgG1 memory B cells (1×103), were transferred into Rag-1−/− mice, followed by immunization with soluble NP-CG. The number of total and high-affinity anti-NP/IgG1+ antibody-secreting cells (ASCs) in the spleen was measured by ELISPOT (15).
Adoptive transfer of naive CD4 T cells
To purify CD4+ T cells, splenocytes from OT-II transgenic mice (with or without of CD4 T cell-specific deletion of Bcl6) were incubated with a mixture of biotinylated mAbs against B220, CD25, CD8α, CD11b, DX5, NK1.1, TER119 and TCRγδ (BioLegend, San Diego, CA, USA), followed by negative MACS selection using streptavidin microbeads (Miltenyi Biotech). Thereafter, the cells were stained with anti-CD90.2FITC (BioLegend), anti-CD62LPE-Cy7 (eBioscience, San Diego, CA, USA), anti-CD44PE or CD44Pacific Blue (BioLegend), and streptavidinPE-TexasRed (BD Pharmingen, San Diego, CA, USA). Cells were resuspended in a staining buffer containing propidium iodide (PI, 1 µg/ml; Sigma, St. Louis, MO, USA), and live PI-, CD90.2+CD25−CD62LhiCD44lo cells were sorted as naive CD4 T cells using a FACS Aria (BD Biosciences, San Jose, CA, USA). 3 x 106 naive CD4 T cells from OT-II transgenic mice (CD45.1 or CD45.2) were adoptively transferred into congenic recipient mice (CD45.2 or CD45.1, respectively), followed by immunization i.v. with 100 µg ovalbumin (OVA)323–339 peptide plus 25 µg LPS (Sigma) on the day after transfer.
Immunohistofluorescence analysis of the secondary antibody response
OVA-specific/CD45.1+ memory T cells (1 x 105) were transferred into CD45.2+ C57BL/6 mice that had been immunized with NP-CG/alum 40 days previously, followed by immunization i.v. with 50 µg of OVA coupled to (4-hydroxy-3-nitrophenyl)acetyl (NP-OVA). Spleens were obtained from the immunized mice at 5–7 days after immunization and embedded in OCT compound after fixation with 4% paraformaldehyde. Frozen sections (8 µm) were stained with anti-IgG1FITC (BD Pharmingen), anti-CD4efluor450 (eBioscience) and anti-IgDAlexaFluor647 (BioLegend). Confocal images were acquired on a Leica SP2AOBS or TCS SP5 confocal microscope using a 20x objective.
Flow cytometric analysis of memory and Tfh cells
This analysis was performed as previously described (6). To prepare single cell suspensions, spleens were minced and incubated with collagenase IV (200 unit/ml; Sigma) and DNase I (20 µg/ml; Roche Diagnostics, Mannheim, Germany) for 30min at 37°C. After washing, splenocytes were depleted of red blood cells and incubated with anti-FcγRII/III mAb (2.4G2; ATCC). For analysis of CD4+ memory and Tfh cells, the cells were incubated with a mixture of biotinylated mAbs against B220, CD8α, CD11b, NK1.1, TER119 and TCRγδ (BioLegend).
Cells were stained with APC-conjugated anti-CXCR5APC (BD Pharmingen), streptavidinPE-TexasRed, anti-CD4V500 (BD Pharmingen), anti-TCRβAPC-eFluor780 (eBioscience), anti-CD44FITC, anti-CD62LPacific Blue (BioLegend) and PE-conjugated mAbs against either PD1, CCR7, ICOS, Ly6c, CD45RB, IL7R (all from BioLegend), CXCR3 (R&D systems, Minneapolis, MN, USA) or PSGL1 (BD Pharmingen). To analyze the expression of intracellular transcription factors, cells were fixed after cell surface staining, permeabilized using the Foxp3 staining buffer set (eBioscience) and stained with anti-Bcl6PE mAb (BD Biosciences).
Cells were washed, resuspended in a staining buffer containing PI for cell surface staining and analyzed using a FACS Aria. Data were analyzed using FlowJo software (Tree Star, San Carlos, CA, USA) as described in Kaji et al. (6).
Purification of CD4 memory T cells from recipients transferred with CD4 T cells
Splenocytes were prepared from the pooled spleens of recipient mice and transferred with OT-II CD4 T cells at the indicated time after immunization. Cells were incubated with a mixture of biotinylated mAbs as described above in the Flow cytometric analysis of memory and Tfh cells section and anti-CD45.1 or CD45.2 Abs to exclude contamination by recipient T cells, followed by negative MACS selection using streptavidin microbeads. Thereafter, the cells were stained with anti-CXCR5APC, streptavidinPE-TexasRed, anti-CD4V500, anti-TCRβAPC-eFluor78, PE-Cy7-conjugated anti-CD45.1 or CD45.2 (anti-CD45.1PE-Cy7 or anti-CD45.2PE-Cy7), anti-CD44FITC, anti-CD62LPacific Blue and anti-PD1PE, followed by sorting into CXCR5+ Tfh cells, CD62Lhi central memory T-cell (Tcm) and CD62Llo effector memory T-cell (Tem) populations for RNA extraction.
For sorting of donor T cells for culture or adoptive transfer experiments, CD4 T cells were MACS enriched and then stained with anti-CD90.2FITC, anti-CD62LPE-Cy7, anti-CD44PE, and anti-CD45.1APC or anti-CD45.2APC, followed by sorting into Tcm and Tem populations.
Tfh-cell and GC B-cell analyses in immunized mice
Analysis of Tfh and GC B cells in NP-CG-immunized or non-immunized Bcl6+/+/mb1-cre+/− or Bcl6f/f/mb1-cre+/− mice was performed as described previously (6).
Analysis of secondary adoptive responses
Naive OT-II CD4 T cells and long-term CD62Lhi and CD62Llo CD4 memory T cells were sorted as described above in the section on adoptive transfer of CD4 OT-II T cells and CD4 memory T cell purification, respectively. Naive B and T cells were enriched from C57BL/6 mice as described previously (6). OT-II T cells, NP-specific IgG1 memory B cells, naive CD4 T cells, and naive B cells were transferred into Rag-1−/− mice, followed by immunization with 50 µg soluble NP-OVA. The number of anti-NP/IgG1+ ASCs in the spleens of adoptive recipients was determined by ELISPOT at day 10 post-immunization.
In vitro secondary responses by co-culture of memory B and T cells
1×103 NP-specific memory B cells and 5×103 to 1×104 OVA-specific memory T cells were co-cultured in a 96-well U-bottomed plate in the presence of NP-OVA (0.3 µg ml–1, Biosearch Technologies, Novato, CA, USA) or OVA (Sigma) in RPMI1640 medium (Wako Pure Chemical, Osaka, Japan) supplemented with 15% FCS (Japan Bioserum, Hiroshima, Japan), 55 μM 2-mercaptoethanol, 2mM l-glutamine, 1% Minimum Essential Media (MEM) non-essential amino acids, 1% MEM sodium pyruvate solution and penicillin-streptomycin (all from Life Technologies, Grand Island, NY, USA). Culture supernatants were replaced with fresh medium (120 µl) at 3 and 5 days after cultivation and harvested on day 7 for ELISA.
Microarray analysis
Total RNA was extracted from cells by using the TRIzol reagent according to the manufacturer’s instructions (Life Technologies) and the microarray analysis was performed with Affymetrix GeneChip Mouse 430 2.0 Arrays. RNA samples were labeled using the Ovation RNA Amplification System V2 and FL-Ovation cDNA Biotin Module V2 Kits (Nugen, San Carlos, CA, USA). Image files were scanned and processed by AGCC (Affymetrix GeneChip Command Console Software) and the microarray data were normalized with GC Robust Multi-array Average (16). Differentially expressed genes between samples were selected with the RankProd method (17) with a q-value < 0.05. The full set of raw data is available from the RefDIC database (18) under the accession numbers RSM14249-14254, RSM14258-14262, RSM14277-14279 and RSM14802.
RNA-seq analysis
Total RNA was extracted from cells by using the TRIzol reagent per the manufacturer’s instructions. The cDNA synthesis and amplification were performed with the SMARTer Ultra Low RNA Kit for Illumina Sequencing (Clontech, Mountain View, CA, USA). The Covaris AFA system (Covaris, Woburn, MA, USA) was used for controlled cDNA shearing, and cDNA preparation was performed with the TruSeq DNA sample prep kit (Illumina, San Diego, CA, USA). The size range of the resulting cDNA was estimated using the 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The molar concentration was estimated using LightCycler 480 (Roche Diagnostics), and the cDNA molecules were used for cluster generation and sequencing on the Illumina HiSeq 1000 (Illumina) instrument. Eight or nine samples per lane were used to generate 100bp single end reads. The raw data were subjected to the CASAVA 1.8.2 (Illumina) to generate FASTQ files. The sequence reads were aligned to the Mus musculus reference genome (Build 37) using TopHat v1.4.0 (19). According to the mapped data, Cufflinks v1.3.0 (20) was used to calculate the FPKM (fragments per kilobase of exon per million reads) with the Mus musculus genome annotation NCBI build 37.2 from its website (http://cufflinks.cbcb.umd.edu/igenomes.html). The statistical significance of the differential gene expression between cell populations was evaluated using the Cuffdiff software (20).
Statistical analysis
Student’s t-test was used with KaleidaGraph 4.0 software (Synergy Software, Reading, PA, USA). A P-value of < 0.05 was considered to indicate a significant difference.
Results
Persisting antigen-specific CD4 T cells in adoptive hosts efficiently promote the secondary antibody response
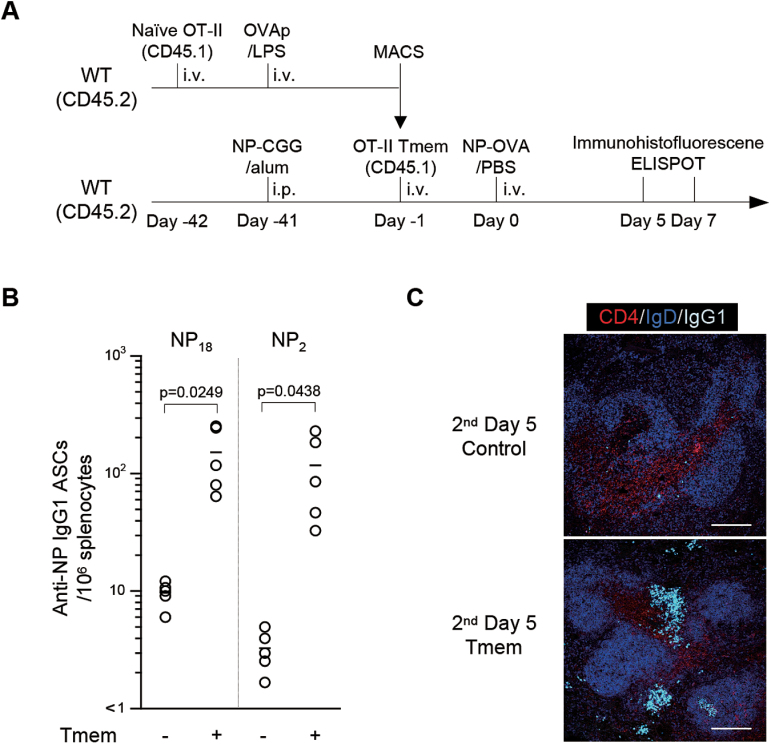
To determine the properties of antigen-specific memory T cells, OVA-specific αβ+CD4+ T cells were prepared from pooled spleens of naive OT-II transgenic mice (21) and transferred into CD45.2+ congenic mice, which were then immunized with OVA (Fig. 1A). Forty days later, the donor OT-II T cells were purified from the recipient mice, and then transferred into CD45.2 congenic recipient mice that had been immunized with NP-CG conjugate in alum. The recipient mice were then challenged with soluble NP-OVA and analyzed for their antibody response. As shown in Fig. 1B, transfer of only 105 OVA-specific memory T cells into the NP-CG-primed recipients was sufficient to induce significant numbers of high-affinity NP-specific/IgG1 ASCs upon NP-OVA challenge.
Fig. 1.
Memory T cells support the secondary antibody response. (A) Diagram of the experimental protocol. Naive CD45.1+CD4+ OT-II T cells were adoptively transferred into CD45.2+ WT mice, followed by immunization with OVA323-339 peptide/LPS. Donor CD45.1+ OT-II memory T cells that developed in CD45.2+ C57BL/6 recipient mice were transferred into C57BL/6 mice that had been immunized with NP-CG 40 days previously in a synchronized manner, followed by immunization with soluble NP-OVA. At 5–7 days after secondary immunization, the recipient mice were sacrificed for ELISPOT (B) and immunofluorescence (C). (B) Anti-NP/IgG1 ASCs in the spleen of NP-CG immunized mice with (+) or without (−) transfer of OVA-specific memory T cells and immunization with NP-OVA. The number of total (NP18) and high-affinity (NP2) anti-NP/IgG1+ ASCs in the spleen was measured by ELISPOT. Combined data from two independent experiments are shown (n = 5). (C) NP-CG immunized recipient mice (CD45.2+) with (memory T cells, Tmem) or without (Control) CD45.1+CD4+ OT-II memory T cells were rechallenged with soluble NP-OVA. Five days later, splenic cryosections were stained with anti-CD4 (red), anti-IgD (blue) and anti-IgG1 (light blue). Representative results of two mice are shown. Scale bar indicates 150 µm.
Consistently, B cells expressing high levels of IgG were detected in both B-cell and T-cell areas at day 5–7 after the secondary challenge (Fig. 1C), and some of them formed clusters with donor memory T cells in the B-cell follicles of recipient mice (data not shown). These results suggest that CD4+ T cells acquired the capacity to promote a memory B-cell response in the adoptive hosts by 40 days after immunization.
CD62Lhi memory T cells support IgG1 memory B-cell responses more efficiently than CD62Llo memory T cells
Persisting CD4+ memory T cells in the adoptive hosts at day 40 after transfer had the phenotype CXCR5dullPD-1−CCR7+, distinct from the Tfh-cell phenotype (CXCR5+PD-1+CCR7−) at day 7 after immunization (Fig. 2A). As for transcription factors, Bcl6 was expressed in Tfh but not in day 40 CD4+ T cells, whereas both CD4+ T-cell populations expressed T-bet at low levels (data not shown). In terms of CD62L expression, Tfh cells were CD62L−, whereas persisting TCRαβ+CD4+ T cells had a similar frequency of CD62Lhi and CD62Llo T-cell subsets (Fig. 2A). We designated CD62Lhi T cells as Tcm and CD62Llo as Tem, according to a previous report (8).
Fig. 2.
Long-term CD62Lhi CD4 Tcm cells have the ability to promote a secondary response by IgG1+ memory B cells. (A) Naive CD45.1+CD4+ OT-II T cells were adoptively transferred into CD45.2+ WT mice, followed by immunization with OVA323-339 peptide/LPS. CXCR5 and PD1 expression by CD45.1+TCRβ+CD4+ T cells was analyzed by flow cytometry at day 7 (left panels) and day 40 (right panels) after immunization. Day 7 CD4+CXCR5hi donor T cells and day 40 CD4+ donor T cells were examined for expression of surface CD62L, CD44 and CCR7 (middle and bottom panels). Numbers in the plots indicate the percent of cells in quadrants. Representative results of three independent experiments with more than three mice per group are shown. (B) Persisting CD4+ OT-II T cells were purified from recipient mice at day 40 after immunization and separated into CD62Lhi (CM) or CD62Llo (EM) cells by FACS. Naive OT-II T cells (Naive) were purified from non-immunized OT-II transgenic mice. Naive or day 40 OT-II T cells (1×105) were transferred into Rag-1−/− mice, together with NP-specific IgG1+ memory B cells (2×103) and naive B cells (1×106) and T cells (1×106) as filler cells (n = 3), followed by immunization with soluble NP-OVA. The number of total (left panel) and high-affinity (right panel) anti-NP/IgG1+ ASCs in the spleen was measured by ELISPOT at day 10 post-immunization using NP18-BSA and NP2-BSA as coating antigens, respectively. Circles represent the number of cells in individual mice. Data are representative of two independent experiments. (C) Naive OT-II T cells (Naive) or CD62Lhi memory T (CM) cells (5×103) and NP-specific memory B cells (1×103) were co-cultured in triplicate with NP-OVA or OVA. The titer of total (left panel) and high-affinity (right panel) anti-NP IgG1 Abs in culture supernatants at day 7 was determined by ELISA. Circles represent the titer in individual wells. Data are representative of two independent experiments.
To characterize the functional capacity of long-term CD62Lhi and CD62Llo CD4+ T cells, these donor-derived T-cell subsets were purified from recipient mice at day 40 after immunization and transferred into RAG-1−/− mice together with NP-specific IgG1+ memory B cells, followed by immunization with soluble NP-OVA. Figure 2B shows that day 40 CD62Lhi T cells could promote IgG antibody production by NP-specific memory B cells, whereas CD62Llo T cells had little such activity, as previously observed in an influenza virus model (22).
To characterize the functional properties of CD62Lhi memory T cells in more detail, we utilized an in vitro assay system that enables us to analyze memory B-cell responses with a small number of CD62Lhi memory T cells (see Methods). To this end, donor-derived OT-II CD62Lhi memory T cells were purified from adoptively transferred recipient mice and co-cultured with NP-specific IgG1+ memory B cells from NP-CG immunized mice in the presence of NP-OVA or OVA. Consistent with the results in adoptive transfer experiments, CD62Lhi memory T cells could promote antibody production by memory B cells in vitro upon stimulation with NP-OVA, but not OVA (Fig. 2C), indicating that the response was mediated by cognate memory T- and B-cell interactions. By contrast, naive OT-II T cells could not trigger a memory B-cell recall response.
Together, these results suggest that CD4 memory T cells able to promote memory B-cell responses are enriched in the CD62LhiCXCR5− CD4+ T-cell population.
Bcl6 deletion in CD4 T cells abrogates the generation of long-term memory T cells
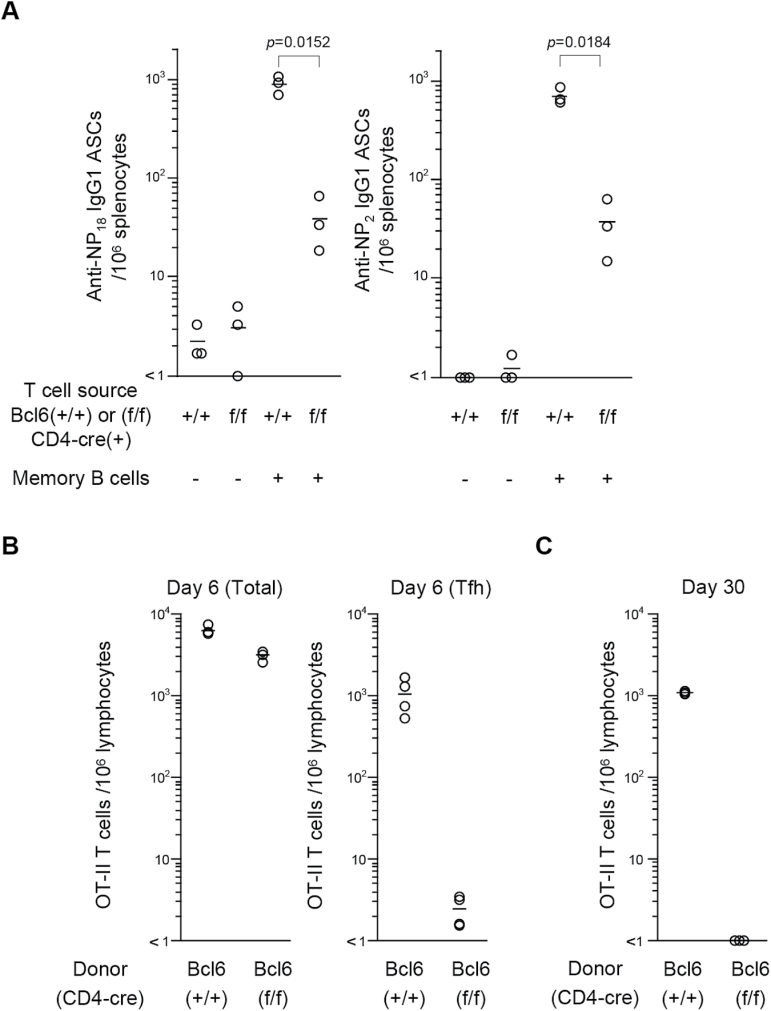
It has been reported that Bcl6 gene deletion impaired the generation of long-term CD4 T cells in adoptive recipients (23). In this context, we analyzed the role of Bcl6 in memory T-cell generation by using mutant mice with a conditional deletion of Bcl6 in CD4 T cells (6). As shown in Fig. 3A, we observed a large number of IgG1 ASCs in adoptive hosts reconstituted with NP-specific memory B cells and CG-primed CD4+ T cells from (Bcl6+/+/CD4-cre) control mice; however, the number of ASCs was significantly reduced in recipients of CG-primed CD4 T cells from the mutant (Bcl6f/f/CD4-cre) mice. These results suggest that the adoptive secondary antibody response was largely dependent on carrier-primed CD4 T cells that had expressed Bcl6.
Fig. 3.
Bcl6-deficient CD4 T cells fail to develop into long-term memory T cells. (A) CD4+ T cells from CG-primed Bcl6+/+/CD4-cre (+/+) and Bcl6f/f/CD4-cre (f/f) and naive B cells with (+) or without (−) NP-specific IgG1 memory B cells (1×103) were transferred into Rag-1−/− mice (n = 3), followed by immunization with NP-CG. The number of total (left panel) and high-affinity (right panel) anti-NP/IgG1+ ASCs in the spleen was determined as in Fig. 2B. Data are representative of two independent experiments. (B and C) Naive CD45.2+CD4+ T cells from OT-II transgenic Bcl6+/+/CD4-cre (+/+) or Bcl6f/f/CD4-cre (f/f) mice were adoptively transferred into CD45.1+ WT mice, followed by immunization with OVA323-339 peptide/LPS. Total CD45.2+CD4+ donor T cells were enumerated from 106 cells within the lymphocyte gate at day 6 (B) and day 30 (C) after immunization. The number of OT-II T cells with a Tfh signature (CXCR5hiPD1hi) is shown in the right panel in B. Circles represents the number of cells in individual mice. Data are representative of two independent experiments.
The conditional deletion of Bcl6 in OT-II T cells did not affect in vitro proliferation and cytokine production by OVA-specific CD4+ T cells in response to OVA (Supplementary Figure 1, available at International Immunology Online), but did impair the generation of Tfh cells and long-term memory CD4 T cells in adoptively transferred hosts after immunization with OVA (Fig. 3B and C). Together, these results suggest that Bcl6 has an essential role in the generation of long-term memory CD4 T cells.
The GC is required for Tfh but not for CD4 memory T-cell development
Since Bcl6 is a master regulator of Tfh differentiation, we asked whether memory T precursor cells are derived from committed Tfh cells. It has been suggested that development of CD4+ T cells into Tfh cells requires GC B cells (24). Indeed, in response to NP-CG, conditional Bcl6 deletion in B cells (Bcl6f/f/mb-1-cre+/−) significantly reduced the number of T cells with a Tfh signature (i.e. CXCR5+–hiPD1+–hi) in the spleen, in association with the loss of GC B cells (Fig. 4A).
Fig. 4.
Loss of GC B cells reduces the number of Tfh cells, but not memory T cells, in the response to NP-CG. (A) Splenocytes were recovered from Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice heterozygous for mb1-cre at day 7 post-immunization with NP-CG in alum, followed by FACS analysis. Dot plots show levels of CXCR5 and PD1 expression by CD4+ T cells from the Bcl6+/+ (+/+, bottom left panel) and Bcl6f/f (f/f, bottom right panel) mice. Naive mice are also shown as controls (top panels). Percentages of CXCR5hiPD1hi cells are indicated. The middle and right panels show the number of Tfh cells and NP-specific/IgG1+ GC B cells, respectively, at day 7 after immunization. Naive mice were used as a control for Tfh cells. Circles represent the number of cells in individual mice. Representative results of two independent experiments with three to four mice per group are shown. (B) CD4 T cells from CG-primed Bcl6+/+/mb1-cre+/− (+/+) and Bcl6f/f/mb1-cre+/− (f/f) and naive B cells with (+) or without (−) NP-specific IgG1 memory B cells (1×103) were transferred into Rag-1−/− mice, followed by immunization with soluble NP-CG. The number of total (left panel) and high-affinity (right panel) anti-NP/IgG1+ ASCs in the spleen was determined as in Fig.2B. Data are representative of two independent experiments.
To determine whether memory T-cell development requires GC B cells, CD4+ T cells were prepared from mutant (Bcl6f/f/mb-1-cre+/−) GC B-cell-deficient mice, which had been primed with CG 40 days before and transferred into adoptive hosts together with NP-specific IgG1 memory B cells. After immunization with NP-CG, adoptively transferred memory B cells produced large numbers of total and high-affinity IgG1 ASCs in the presence of CD4+ T cells from either mutant or control mice (Fig. 4B). These results suggest that CD4 memory T cells can develop even under conditions where Tfh development is not fully supported.
The GC is not required for CD62LhighCD4+ memory T-cell development
To further examine the role of GC B cells in memory T-cell development, CD4+ T cells were purified from pooled spleens of OT-II transgenic mice and transferred into CD45.2+ congenic control (Bcl6+/+/mb-1-cre+/−) or mutant (Bcl6f/f/mb-1-cre+/−) mice with a conditional deletion of Bcl6 in B cells. At day 7 after immunization with OVA, the frequency of CD4+ T cells with a Tfh signature among total CD4+ donor T cells was significantly reduced in mutant mice deficient in GC formation (Fig. 5A). The reduction was particularly significant in Tfh cells expressing the highest levels of CXCR5 and intracellular Bcl6 (Fig. 5A, right panel). In contrast, the frequency of non-Tfh cells was comparable between wild-type (WT) and mutant mice (data not shown). Since the expression levels of CXCR5 and PD1 are highest on T cells physically located within GCs (25), this result suggests that the GC is indeed required for full development of Tfh.
Fig. 5.
GC B cells are not essential for the development of Tcm cells. (A and B) Naive CD45.1+CD4+CD44lo OT-II T cells were adoptively transferred into CD45.2+ Bcl6+/+ (+/+) and Bcl6f/f (f/f) mice heterozygous for mb1-cre, followed by immunization with OVA323-339 peptide/LPS. (A, left panel) the number of donor CD4+ T cells (total) and Tfh cells at day 7 and the percentage of Tfh cells among CD4+ T cells. (A, right panels) CXCR5 and intracellular Bcl6 expression by CD45.1+CD4+ donor T cells (red dots) and CD45.1−CD4+CD62LhiCD44lo recipient T cells (blue dots) were analyzed by flow cytometry at day 7 after immunization. (B) The number of donor CD4+ T cells (total) and CD62Lhi Tcm cells at day 40. The number of cells in (A) and (B) were determined as in Fig. 3. Representative results of two independent experiments with three mice per group are shown. (C) CD62Lhi OT-II Tcm cells (CM) from Bcl6+/+ (+/+) and Bcl6f/f (f/f) recipient mice heterozygous for mb1-cre were prepared as in A at day 40 after immunization. Naive OT-II T cells (Naive) and NP-specific/IgG1+ memory B cells were prepared as in Fig. 2B. T cells and memory B cells were co-cultured in triplicate with NP-OVA and the titer of total (left panel) and high-affinity (right panel) anti-NP IgG1 antibodies in culture supernatants at day 7 was determined as in Fig. 2C. Circles represent the titer in individual wells. Data are representative of two independent experiments. (D) Naive CD45.1+CD4+ OT-II T cells were adoptively transferred into CD45.2+ Bcl6+/+ (red) and Bcl6f/f (blue) mice heterozygous for mb1-cre, followed by immunization with OVA323-339 peptide/LPS. Representative FACS profiles of CD4+CD62Lhi donor Tcm cells at day 40 stained with the indicated mAbs. FACS profiles of recipient naive T cells from control mice (CD45.1–) are also shown (dark shading). Representative results of two independent experiments with three mice per group are shown.
By contrast, CD62LhighCD4+ memory T cells were sustained at comparable levels in the presence or absence of GC B cells at day 40 after immunization (Fig. 5B). Day 40 Tcm cells recovered from GC-deficient mutant and control recipient mice were indistinguishable in their ability to support IgG NP antibody production by memory B cells in an in vitro recall response (Fig. 5C) and by cell-surface phenotype (Fig. 5D). Day 40 CD62LhighCD4+ memory T cells in wild-type mice had the phenotype CXCR5−PD-1−CCR7+ICOSdullPSGL1+, similar to those in the GC-less mice. The CD62Lhigh memory T cells were also Ly6clo, a phenotype that has been reported to preferentially mark antiviral memory cells with greater longevity and proliferative responses to secondary infection (8, 9).
Thus, the GC response is not essential for the functional development of Tcm cells that can promote a memory B-cell recall response. Furthermore, these results led us to conclude that CD62LhighCD4+ memory T cells are not derived from Tfh cells undergoing full development through interaction with GC B cells.
Genetic signatures of Tcm cells that develop in the presence or absence of GC B cells are similar
To elucidate the genetic signatures for CD62LhiCD4+ memory T cells that had developed in the presence or absence of GC B cells, gene expression was assessed using Affymetrix GeneChip technology in three replicates of OVA-specific donor T cells that had been purified from adoptively transferred recipients at day 7 and day 40 after immunization. Hierarchical cluster analysis of all arrays for the expression of 45037 probes resulted in a dendrogram with four major branches (Fig. 6A). CD62Lhi memory T cells were clustered as an independent population next to day 7 CD62Lhi non-Tfh cells and clearly separated from naive and Tfh cells. As shown in Fig. 6B, the gene expression profiles between CD62Lhi memory T cells that developed in control and mutant mice lacking GCs were mostly similar, with the exception of a few genes, including Gdpd3 (26) (see below). These results demonstrate that memory T-cell subsets maintain their capacity to help memory B-cell recall responses, along with a unique gene expression signature, regardless of the presence or absence of GC B cells during their development.
Fig. 6.
A hierarchical clustering of triplicate samples of OT-II T cells. (A) Naive OT-II T cells (Naive) were prepared from the pooled spleens of OT-II transgenic mice (n = 3). CD62LloCXCR5hi (Tfh) and CD62LhiCXCR5lo (CD62Lhi non-Tfh) effector cells were purified from immunized mice (n = 3) at day 7 after immunization with OVA. CD62LhiCXCR5lo Tcm cells were purified from control (Bcl6+/+mb-1-cre+/−) and GC-less mutant (Bcl6f/fmb-1-cre+/−) mice (n = 4–7) that had been transferred with CD4+ OT-II T cells and immunized with OVA 40 days previously as described in Methods. Total RNA was extracted from each sample in three independent experiments. The number for each sample (e.g. Naive 1) annotates each experiment. Colors in the heatmap depict the Pearson’s correlation coefficient between a pair of samples. The heatmap was generated with R package (http://www.r-project.org/). To investigate the cellular state of different cells, a correlation coefficient of gene expression profiles among all sample pairs was calculated and these values were analyzed by hierarchical clustering. The cluster with a value greater than 95 is considered to be strongly supported by the data (82). (B) Heatmap representing the expression profile of the differentially expressed genes between Tcm and GC-less cells with a statistical significance (q-value < 0.05) from the analysis in A.
CD4 T cells develop into functional CD62Lhi Tcm cells through cognate B-cell interactions
In an acute infection with LCMV or Lm, CD4 memory T cell development is diminished in B-cell-deficient µmt−/− mice (9, 10, 27), implying a requirement of B cells for memory T cell development. However, it remains unknown whether memory T-cell development requires cognate or bystander B cells. Furthermore, it is possible that the memory T cell defect in the µmt−/− mutant mice reflects the abnormal T-cell zone architecture observed in these mice rather than a requirement for B cells (28). Therefore, we asked whether CD4 T-cell development into Tcm cells requires cognate B-cell interaction or can be induced by bystander B cells, as was observed in Tfh-cell development (29). We tracked the development of OT-II donor T cells in B1-8hi mice, whose B cells express a BCR encoded by the NP-specific IgH gene, V186.2, with a point mutation resulting in high affinity for the NP hapten (30). In contrast to µmt−/− mice, the histological architecture in lymphoid organs is normal in B1-8hi mice (30). B1-8hi mice do not produce detectable levels of IgM or IgG1 antibodies upon immunization with OVA in alum (Supplementary Figure 2, available at International Immunology Online). Thus, antigen presentation to OVA-specific OT-II T cells in these mice should be restricted to non-B-cell antigen-presenting cells.
Figure 7A shows that the number of OT-II CD4+ T cells with a Tfh signature is reduced in the spleen of B1-8hi recipient mice at 7 days after transfer and subsequent immunization with OVA, in parallel with the reduction in number of OT-II CD4+ T cells. The ratio of Tfh among total donor T cells was comparable in the spleen of wild-type and B1-8hi recipients; however, the frequency of PD1highCXCR5high Tfh cells was less in B1-8hi recipient mice (Fig. 7B). This result suggests that cognate B cells may play a role in sustaining GC Tfh cells. Likewise, the frequency of donor-derived Tcm cells was comparable in the spleen of wild-type and B1-8hi recipient mice at day 40 after immunization (Fig. 7C). To characterize the activity of CD4+ CD62Lhi T cells persisting in hosts lacking cognate B cells, Tcm cells were purified from B1-8hi or control recipient mice that had been primed with OVA 40 days previously and co-cultured with NP-specific IgG1 memory B cells in the presence of NP-OVA. Figure 7D shows that day 40 CD62Lhi T cells promoted a high level of IgG antibody production by NP-specific memory B cells in vitro, whereas CD62Lhi T cells from B1-8hi recipient mice had little such activity. These results suggest that for memory T cells to attain the capacity to promote memory B-cell responses, cognate B-cell interactions are required.
Fig. 7.
B-cell cognate interaction is required for functional development of Tcm. (A-C) Naive CD45.2+CD4+CD44lo OT-II T cells were adoptively transferred into CD45.1+ WT (WT) and B1-8hi (B1-8hi) mice, followed by immunization with OVA323-339 peptide/LPS. The number of donor CD4+ T cells (total) and Tfh cells at day 7 (A and B) and CD4+ T cells (total) and CD62Lhi Tcm cells at day 40 (C) was determined as in Fig. 3. (B) The perecentage of Tfh cells among total CD4 T cells in WT and B1-8high recipients (left panel), together with representative FACS profiles of CXCR5 and PD1 expression by CD45.2+TCRβ+CD4+ T cells in WT and B1-8high recipients (right panels). The numbers in the profiles indicate the percentage of CXCR5+PD1+ cells among total CD4 T cells. Circles represent the number of cells in individual mice. Data are representative of two independent experiments. (D) CD62Lhi OT-II Tcm cells (CM) from WT (WT) and B1-8hi (B1-8hi) mice were prepared as in A. Naive OT-II T cells (Naive) and NP-specific/IgG1+ memory B cells were prepared as in Fig. 2B. T cells and memory B cells were co-cultured in triplicate with NP-OVA and the titer of total (left panel) and high-affinity (right panel) anti-NP IgG1 antibodies in culture supernatants at day 7 was determined as in Fig. 2C. Circles represent the titer in individual wells. Data are representative of three independent experiments.
Functional CD62Lhi Tcm development is associated with expression of a group of transcripts induced by cognate B-cell interaction
To elucidate the molecular basis for functional differences in CD62Lhi memory T cells that developed in the presence or absence of cognate B-cell interaction, we assessed their gene expression profiles by RNA sequence analyses in triplicate. Approximately 30 transcripts were found to be highly expressed in day 40 Tcm to a greater extent than in naive T cells (Supplementary Tables 1 and 2, available at International Immunology Online).
As shown in Fig. 8A, CD62Lhi memory T cells that developed in the absence of cognate B-cell interaction had significantly reduced levels of transcripts that have been reported to be associated with T-cell homeostasis [Il7r (31)], glucose transport [Tbc1d4 (32)], protein stability [Trib2 (33)] and growth and activation, such as Ppic (cyclophilin C) and Plac8 (34). Interestingly, we observed that conventional long-term Tcm cells expressed high levels of transcripts from the Gdpd3 gene, which encodes GroPIns phosphodiesterase, an enzyme that hydrolyzes GroPIns to glycerol and inositol 1-phosphate (26). Expression of this gene could distinguish these Tcm cells from other naive and effector T cells and from Tcm cells that developed in hosts deficient in cognate B-cell interaction (Fig. 8A; Supplementary Tables 3 and 4, available at International Immunology Online). Thus, Gdpd3 is a promising candidate genetic marker for memory T cells. Since GDPD3 in conjunction with another intracellular protein SPRED1 is a recently described biomarker for neoadjuvant platinum chemotherapy response in certain urothelial carcinomas, the necessary reagents for this potentially important analysis should be readily available (35).
Fig. 8.
Transcriptional profiling suggests that functional development of Tcm is a stepwise process. The bar plots depict the mean FPKM value for each transcript from four distinct cell populations. OVA-specific CD4+TCRβ+CD62Lhi Tcm cells were purified by FACS from the pooled spleens (n = 4–7) of adoptively transferred WT (a, green column) or B1-8hi mutant (b, red column) hosts at day 40 after transfer of CD4+CD44lo OT-II T cells and subsequent immunization with OVA323-339 peptide/LPS as in Fig. 7. Naive T cells (c, blue column) were purified from non-immunized OT-II transgenic mice. CD4+TCRβ+CXCR5+ Tfh cells (d, open column) were purified from WT hosts (n = 3) at day 7 post-immunization. RNA-seq was performed with three replicates of each purified cell population as described in Methods. (A) Transcripts, including those associated with survival, that were down-regulated in the day 40 Tcm cells in the absence of cognate B-cell interaction (b, cognate-less) relative to those in the presence of cognate interaction. (B) Transcripts, including those associated with mitochondria activities, homeostasis and immune reactivity, that were significantly up-regulated in Tcm as compared with the levels found in naive T cells (c, Naive) and sustained in the presence or absence of cognate B cells. (C) Transcripts that were significantly up-regulated in Tfh (d), compared with the levels in Tcm cells in the presence (a) and absence of cognate B-cell interaction (b) and naive T cells (c). (D)Transcripts, including those with inhibitory activities for T-cell responses, that were significantly up-regulated in Tcm in the absence of cognate B cells. Error bars represent 95% confidence interval for the means of triplicates.
Curiously, day 40 Tcm cells that had developed in the absence of cognate B-cell interaction had significantly up-regulated expression of ENC1, which suppresses the activity of Nrf2, a transcriptional factor supporting Th2 differentiation (36, 37), as well as a group of genes that encode proteins with inhibitory activities for T-cell response such as Ly6C (38), Hspa1a (39), lfit3, CD200 (40) and Zfp36 (36, 37, 41, 42) (Fig. 8B, see also Supplementary Table 5, available at International Immunology Online). In addition, these cognate-less day 40 Tcm had up-regulated the expression of genes such as Pdlim2 (43), Tbc1d10c (44) and Spry1 (45), which are associated with a negative regulatory loop for the TCR signaling pathway.
Taken together, these results suggest that co-stimulatory signals from B cells generated through cognate interaction may play a critical role in transcriptional regulation in CD4 T cells, resulting in a persisting immune response leading to the development of memory cells.
Genetic signatures in long-term memory T cells and Tfh cells are different. Tfh expressed ~200 transcripts to a greater extent than Tcm and naive T cells, including Batf, a transcriptional factor indispensable for Tfh development (46, 47), and c-maf, which is reported to be regulated directly by Batf and required for IL-21 and CD40L production (48–50) (Fig. 8C; Supplementary Table 3, available at International Immunology Online). Day40 Tcm barely expressed several Tfh-related transcripts (3, 30, 49, 51–54), such as CXCR5, TIGIT, IL21, IFg, PD-1 and POU2AF1. POU2AF1 is a known regulator of CXCR5 expression by B cells (54).
On the other hand, memory T cells and Tfh cells shared expression of a group of transcripts, the majority of which did not change significantly in the absence or presence of cognate B cells (Fig. 8D). These mRNAs encode molecules associated with mitochondria activities, such as NADH dehydrogenase ubiquinone 1 beta subcomplex subunit 7 (Ndufb7), ubiquinol-cytochrome-c reductase (Uqcr), cytochrome P450, family 4 (Cyp4v3), reactive oxygen species modulator 1 (ROMO1), Ncf4 and mitochondrial ribosomal protein L15 (Mrpl15), as well as those associated with metabolism and cellular activity, such as dehydrogenase/reductase (SDR family) member 7(Dhrs7), ADK, Nrp1 (55), Zc3h12d (56), and Lrp10 (57) (Fig. 8A and B).
These results suggest that early during T-cell activation, the transition from quiescence to rapid cell growth and subsequent differentiation into memory T cell and Tfh subsets is accompanied by unique gene expression profiles through interaction with different antigen-presenting cells.
Discussion
Despite the crucial role of CD4+ T cells in B cell help to establish long-term humoral immunity to pathogens, the factors regulating the transition of effector CD4+ T cells to memory CD4+ T cells are largely unknown. Although specific markers for CD4 memory T cells remain unknown at present, these cells are operationally defined as cells capable of long-term survival after immunization and with the capacity for intense proliferation and cytokine production upon re-stimulation, greater than in the primary response (58, 59). In addition, the CD4+ memory T cells can enhance GC formation and immunoglobulin class-switch in the primary B-cell response better than the primary responding T cells (14, 60).
The present study clearly demonstrates that OVA-specific CD4+ memory T cells promote terminal differentiation of NP-specific IgG1+ memory B cells in response to NP-OVA far more effectively than antigen-specific naive CD4+ T cells. CD4 memory T cells are divided into CD62Lhi and CD62Llo memory T-cell subsets (61). We showed that compared with CD62Llo T cells, the CD62Lhi memory T cell subset has a superior capacity to support the IgG1 memory B-cell recall response in adoptive hosts. CD62Lhi memory T cells with this activity were mostly PSGL1hiLy6clo, a T-cell phenotype that is also generated during LCMV infection and characterized by greater longevity and proliferative responses during a secondary infection compared with the PSGL1hiLy6chi Tem phenotype (8, 9).
It has been reported that memory T cells consist of two types of cells derived from CXCR5+ Tfh and non-Tfh effector cells (7–9, 12, 53). The present study demonstrates that Bcl6 has an essential role in the development of CD4+ memory T cells, as predicted from the studies of Lm infection (10). Since Bcl6 is a master regulator for Tfh differentiation, we addressed the question of whether memory T precursor cells are derived from committed Tfh cells. Tfh development begins during dendritic cell (DC) priming, during which ICOS provides a critical signal to induce the transcription factor Bcl6 (62). Bcl6 then induces CXCR5 and reduces CCR7 expression, allowing for migration of the primed T cells into B-cell follicles (62). Within the follicles, Tfh form complexes with GC B cells, which facilitates maintenance of Bcl6 expression and stable commitment to Tfh differentiation (3, 24, 63–65).
Considering the possibility that CD4 memory T cells are derived from cells in GC at a late stage of Tfh development (65), we analyzed CD62Lhigh memory T-cell development in mutant mice lacking GC B cells, wherein Tfh development is grossly inhibited due to the conditional Bcl6 deficiency in B cells. Our results demonstrate that CD4+CD62Lhigh memory T cells develop normally in the absence of GC B cells, GC structures or a substantial number of Tfh cells, and with no apparent alterations in their phenotype, function and genetic signature. These results support the conclusion that CD62Lhigh memory T cells are not derived from Tfh cells undergoing full development through their interaction with GC B cells. Supporting this notion, memory T cells display a phenotype and genetic signatures distinct from CXCR5hiPD1hi Tfh cells.
Previous reports suggested that CD4 T-cell development to memory T cells is diminished in B-cell-deficient µmt−/− mice (9, 10, 27); however, it remained unclear whether the defect reflected the loss of cognate B cells or a bystander effect due to lack of B cells. Furthermore, the possibility remained that memory T cell development was retarded by the aberrant T zone architecture in µmt−/− mutant mice (28). To better explore the role of B-cell cognate interaction in memory T-cell development, we tracked the development of OVA-specific donor T cells in B1-8hi mutant mice, which have normal lymphoid histological architecture but where the B cells express a VH gene encoding a high-affinity NP-specific antibody (30). Under these circumstances, antigen presentation to OVA-specific T cells is limited to non-B-cell antigen-presenting cells. The results showed that the frequency of persisting OVA-specific CD4+ T cells was comparable in B1-8hi mutant and WT mouse hosts. However, long-term memory CD4 T cells in the hosts without cognate B cells had significantly reduced activity to promote a memory B-cell recall response. These results demonstrate that cognate B cells are dispensable for the survival or maintenance of CD4 memory T cells but indispensable for their functional development. Curiously, loss of GC B cells did not affect the number and effector function of persisting memory CD4+ T cells.
Our gene expression profiling suggests that long-term CD4+ T cells persisting in hosts lacking cognate B-cell interaction up-regulated transcripts encoding molecules inhibitory for T-cell responses, but down-regulated expression of genes that have been reported to be associated with activation, homeostasis and metabolism (see Fig. 8). On the other hand, genetic signatures in CD4+ T cells persisting in the hosts in the absence or presence of GC B cells were mostly similar, suggesting a role of non-GC B cells in the acquisition by CD4+ T cells of functional competence to support a memory B-cell recall response. In this situation, prior to their encounter with cognate B cells, CD62Lhigh memory T-cell precursors with such genetic signatures could not have optimal memory T-cell functions without first down-regulating the expression of these negative regulators. Thus, although the mechanism remains obscure, it seems that B-cell cognate interaction releases memory precursors from a paused state to allow their full functional development into memory T cells by down-regulating genes encoding molecules inhibitory for cellular responses.
Tfh cells expressed ~200 transcripts to a greater extent than memory T cells and naive T cells, whereas Tfh and memory T cells shared a group of genes that was up-regulated to a greater extent than in naive T cells. The majority of the genes, which are associated with mitochondria activities and homeostasis, did not significantly change their expression level in long-term CD4 T cells persisting in the hosts regardless of the absence or presence of cognate B cells. This observation suggests that the genetic signature shared by both Tfh and memory T cells becomes operative early in the immune response because of DC cognate interactions, but prior to cognate B-cell interactions.
On the basis of the findings reported here, we propose that CD4 memory T cells develop stepwise in response to protein antigens through sequential interaction with different types of antigen-presenting cells. The initial interaction with DCs triggers Bcl6 expression and sanctions the memory pathway, with survival and a metabolic shift, similar to CD8 memory T cells (66). Subsequently, cognate B-cell interactions regulate gene expression with the acquisition of optimal activity that establishes the memory T-cell capacity, as a consequence of down-regulation of negative regulators of T-cell responses. Thus, it seems likely that the initial metabolic shifts and homeostasis and subsequent functional development of memory T cells to support humoral memory responses are differentially regulated through interactions with different types of antigen-presenting cells. Our model is supported by a recent study demonstrating that sustained interactions between TCR and antigens promote the differentiation of CD4 memory T cells in an LCMV infection model (67). Such strategic stepwise transcriptional regulation would be essential for CD4 memory T cells to become fully committed to their precise cell fate but also would allow their plasticity to give rise to different types of memory subsets in response to external stimuli after they polarize into the memory path. Identifying the signals that direct the stepwise development of memory T cells will be important to address in the future.
There are several studies demonstrating the presence of CXCR5+ Tfh-cell memory in mice following LCMV virus infection (9, 68) and in response to protein antigens (10, 12, 13, 69) and in human peripheral blood (54, 70–72). In this context, our current studies do not support the idea that memory T cells are generated from Tfh cells undergoing development in the GC. However, the shared genetic signatures of both Tfh cells and memory T cells raise the possibility that CD4+CD62Lhigh memory T-cell precursors are derived from Tfh lineage cells early in their development.
Initial activation of antigen-specific T cells occurs in the T-cell zone through cognate interaction with DCs, and activated T cells then migrate toward the B-cell follicle after up-regulating the chemokine receptor CXCR5 (63). Tfh-cell development initiates immediately during DC priming (73), and induction of Bcl6 and CXCR5 begins within the first 48h of CD4+ T-cell priming by the second cell division in an acute virus infection (10, 62). Tfh cells finally become fate committed in the spleen within 72h of responding to an acute infection, as assessed both by transcription factor expression and cell function (68). Likewise, in response to protein antigen, a strong increase in Bcl6 expression in dividing OT-II cells was detected in the draining LN within 1–2 days post-immunization. CXCR5 was gradually up-regulated on dividing OT-II cells and expressed by a majority of the Bcl6+ population at around day 3 (65, 74). At this timepoint, Tfh cells began to enter the follicle in large numbers and underwent considerable proliferation (53, 75).
Histological studies suggest that within 1 day of exposure to antigen, responding B cells migrate to the border between the follicle and the T-cell zone. B cells pair with antigen-specific helper T cells by cognate or non-cognate interactions at 1–2 d post-immunization (75–78) and many cognate B–T interactions last much longer than non-cognate interactions between B cells and helper T cells (77). B cells may continue to survey for helper T cells, a process that could promote T-cell exchange, and long-term cognate interaction may provide sufficient time for the formation of immunological synapses between the interacting B–T interfaces (79).
Taking into account these observations, we predict that CD62Lhigh memory T-cell development starts within the CXCR5–Bcl6+ T-cell population at the T–B border, probably within 2–3 d post-immunization. Cognate B cells may deliver signals that preferentially direct memory cell precursor fates by transcriptional regulation, as observed in Fig. 8. On the other hand, the Tfh fate is decided in interfollicular regions from day 3 post-immunization, at which time a large fraction of B cells undergoes proliferation and development to GC B cells by expression of Bcl6. The Bcl6+ B cells then migrate into the follicle to form GCs by day 4 post-immunization (75, 78).
Memory T cells are clearly heterogeneous populations. Of interest, generation of CXCR5− long-term CD4 T cells, together with CXCR5+ cells, has been reported 6 weeks after adoptive response of donor T cells from TEa T-cell receptor transgenic (TCR Tg) mice (13); however, we could not detect long-lasting CXCR5+ Tfh cells of OT-II T-cell origin in adoptive hosts upon immunization with OVA and LPS as previously reported (23, 80). This does not depend on the intrinsic nature of OT-II T cells, because a small proportion of CXCR5+ Tfh cells of OT-II T-cell origin persisted for 3 weeks in dLN or spleens upon immunization with OVA in CFA (12). A similar scenario has been observed in another adoptive transfer systems utilizing polyclonal T cells after immunization with protein antigens in CFA or alum (53). As antigens in CFA or in alum persist at the injection sites much longer than OVA with LPS (81), it is conceivable that such potent antigen stimulation may prolong Tfh cell maintenance, in part owing to long-term persistence of GC reactions (24), and confer the capacity to differentiate into memory cells.
Persisting CXCR5+ Tfh cells remained after rapid contraction in the secondary recipients in the absence of cognate antigens, but a small number of cells promote the recall of Tfh cells upon potent reactivation (12, 13, 49). Thus, it seems that CXCR5+ Tfh cells can retain the effector programs to recall their lineage-specific response during the maintenance phase of memory differentiation in the absence of cognate antigen.
Finally, we show in this report that long-term Tcm cells express high levels of Gdpd3 gene transcripts and that this can distinguish them from other naive and effector T cells, including Tfh cells. Furthermore, Gdpd3 expression was induced in Tcm cells, but not Tfh cells, upon cognate GC B-cell interaction. Gdpd3 is a member of the glycerophosphodiester phosphodiesterase family, which has essential roles in the regulation of intracellular concentrations of glycerophosphodiesters (26). Whether Gdpd3 confers a metabolic shift in Tcm cells remains to be elucidated.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
This work was supported by Riken (K94-34200 to T. Takemori) and partly by a grant from the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (AI064752 to M. Shimoda).
Supplementary Material
Acknowledgements
We are grateful to Peter Burrows for critically reviewing this manuscript.
Conflict of interest statement: The authors declared no conflict of interests.
References
- 1. Crotty S. 2014. T follicular helper cell differentiation, function, and roles in disease. Immunity 41:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621. [DOI] [PubMed] [Google Scholar]
- 3. Johnston R. J., Poholek A. C., DiToro D., et al. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nurieva R. I., Chung Y., Martinez G. J., et al. 2009. Bcl6 mediates the development of T follicular helper cells. Science 325:1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yu D., Rao S., Tsai L. M., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31:457. [DOI] [PubMed] [Google Scholar]
- 6. Kaji T., Ishige A., Hikida M., et al. 2012. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 209:2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pepper M. and Jenkins M. K. 2011. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 12:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marshall H. D., Chandele A., Jung Y. W., et al. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity 35:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hale J. S., Youngblood B., Latner D. R., et al. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity 38:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pepper M. Pagán A. J. Igyártó B. Z. Taylor J. J. and Jenkins M. K. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sallusto F. Geginat J. and Lanzavecchia A. 2004. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22:745. [DOI] [PubMed] [Google Scholar]
- 12. Liu X., Yan X., Zhong B., et al. 2012. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 209:1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ise W. Inoue T. McLachlan J. B. Kometani K. Kubo M. Okada T. and Kurosaki T. 2014. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proc. Natl Acad. Sci. USA 111:11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. MacLeod M. K. David A. McKee A. S. Crawford F. Kappler J. W. and Marrack P. 2011. Memory CD4 T cells that express CXCR5 provide accelerated help to B cells. J. Immunol. 186:2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi Y. Ohta H. and Takemori T. 2001. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity 14:181 . [DOI] [PubMed] [Google Scholar]
- 16. Wu Z. J. Irizarry R. A. Gentleman R. Martinez-Murillo F. and Spencer F. 2004. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909. [Google Scholar]
- 17. Hong F. Breitling R. McEntee C. W. Wittner B. S. Nemhauser J. L. and Chory J. 2006. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22:2825. [DOI] [PubMed] [Google Scholar]
- 18. Hijikata A., Kitamura H., Kimura Y., et al. 2007. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics 23:2934. [DOI] [PubMed] [Google Scholar]
- 19. Trapnell C. Pachter L. and Salzberg S. L. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Trapnell C., Williams B. A., Pertea G., et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barnden M. J. Allison J. Heath W. R. and Carbone F. R. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34 . [DOI] [PubMed] [Google Scholar]
- 22. Thomas P. G., Brown S. A., Morris M. Y., et al. 2010. Physiological numbers of CD4+ T cells generate weak recall responses following influenza virus challenge. J. Immunol. 184:1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ichii H. Sakamoto A. Arima M. Hatano M. Kuroda Y. and Tokuhisa T. 2007. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int. Immunol. 19:427. [DOI] [PubMed] [Google Scholar]
- 24. Baumjohann D. Preite S. Reboldi A. Ronchi F. Ansel K. M. Lanzavecchia A. and Sallusto F. 2013. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity 38:596. [DOI] [PubMed] [Google Scholar]
- 25. Shulman Z. Gitlin A. D. Targ S. Jankovic M. Pasqual G. Nussenzweig M. C. and Victora G. D.. 2013. T follicular helper cell dynamics in germinal centers. Science 341:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yanaka N. 2007. Mammalian glycerophosphodiester phosphodiesterases. Biosci. Biotechnol. Biochem. 71:1811. [DOI] [PubMed] [Google Scholar]
- 27. Whitmire J. K. Asano M. S. Kaech S. M. Sarkar S. Hannum L. G. Shlomchik M. J. and Ahmed R. 2009. Requirement of B cells for generating CD4+ T cell memory. J. Immunol. 182:1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ngo V. N. Cornall R. J. and Cyster J. G. 2001. Splenic T zone development is B cell dependent. J. Exp. Med. 194:1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu H., Li X., Liu D., et al. 2013. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature 496:523. [DOI] [PubMed] [Google Scholar]
- 30. Shih T. A. Meffre E. Roederer M. and Nussenzweig M. C. 2002. Role of BCR affinity in T cell dependent antibody responses in vivo. Nat. Immunol. 3:570 . [DOI] [PubMed] [Google Scholar]
- 31. Takada K. and Jameson S. C. 2009. Naive T cell homeostasis: from awareness of space to a sense of place. Nat. Rev. Immunol. 9:823. [DOI] [PubMed] [Google Scholar]
- 32. Tan S. X. Ng Y. Burchfield J. G. Ramm G. Lambright D. G. Stockli J. and James D. E. 2012. The Rab GTPase-activating protein TBC1D4/AS160 contains an atypical phosphotyrosine-binding domain that interacts with plasma membrane phospholipids to facilitate GLUT4 trafficking in adipocytes. Mol. Cell. Biol. 32:4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobens L. L. Jr and Bouyain S. 2012. Developmental roles of tribbles protein family members. Dev. Dyn. 241:1239. [DOI] [PubMed] [Google Scholar]
- 34. Rogulski K. Li Y. Rothermund K. Pu L. Watkins S. Yi F. and Prochownik E. V. 2005. Onzin, a c-Myc-repressed target, promotes survival and transformation by modulating the Akt-Mdm2-p53 pathway. Oncogene 24:7524. [DOI] [PubMed] [Google Scholar]
- 35. Gu A. Jie Y. Sun L. Zhao S. Mingyan, E . and You Q. 2015. RhNRG-1β protects the myocardium against irradiation-induced damage via the ErbB2-ERK-SIRT1 signaling pathway. PLoS One 10:e0137337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang X. J. and Zhang D. D. 2009. Ectodermal-neural cortex 1 down-regulates Nrf2 at the translational level. PLoS One 4:e5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rockwell C. E. Zhang M. Fields P. E. and Klaassen C. D. 2012. Th2 skewing by activation of Nrf2 in CD4(+) T cells. J. Immunol. 188:1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamanouchi S., Kuwahara K., Sakata A., et al. 1998. A T cell activation antigen, Ly6C, induced on CD4+ Th1 cells mediates an inhibitory signal for secretion of IL-2 and proliferation in peripheral immune responses. Eur. J. Immunol. 28:696. [DOI] [PubMed] [Google Scholar]
- 39. Stocki P. Wang X. N. and Dickinson A. M. 2012. Inducible heat shock protein 70 reduces T cell responses and stimulatory capacity of monocyte-derived dendritic cells. J. Biol. Chem. 287:12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caserta S., Nausch N., Sawtell A., et al. 2012. Chronic infection drives expression of the inhibitory receptor CD200R, and its ligand CD200, by mouse and human CD4 T cells. PLoS One 7:e35466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang J. Lei T. Song Y. Yanes N. Qi Y. and Fu M. 2009. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J. Biol. Chem. 284:29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schichl Y. M. Resch U. Hofer-Warbinek R. and de Martin R. 2009. Tristetraprolin impairs NF-kappaB/p65 nuclear translocation. J. Biol. Chem. 284:29571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka T. Grusby M. J. and Kaisho T. 2007. PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat. Immunol. 8:584. [DOI] [PubMed] [Google Scholar]
- 44. Pan F. Sun L. Kardian D. B. Whartenby K. A. Pardoll D. M. and Liu J. O. 2007. Feedback inhibition of calcineurin and Ras by a dual inhibitory protein Carabin. Nature 445:433. [DOI] [PubMed] [Google Scholar]
- 45. Akbulut S., Reddi A. L., Aggarwal P., et al. 2010. Sprouty proteins inhibit receptor-mediated activation of phosphatidylinositol-specific phospholipase C. Mol. Biol. Cell 21:3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Betz B. C., Jordan-Williams K. L., Wang C., et al. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ise W., Kohyama M., Schraml B. U., et al. 2011. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 12:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellyard J. I. and Vinuesa C. G. 2011. A BATF-ling connection between B cells and follicular helper T cells. Nat. Immunol. 12:519. [DOI] [PubMed] [Google Scholar]
- 49. Weber J. P., Fuhrmann F., Feist R. K., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. J. Exp. Med. 212:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kroenke M. A. Eto D. Locci M. Cho M. Davidson T. Haddad E. K. and Crotty S.. 2012. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 188:3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Breitfeld D. Ohl L. Kremmer E. Ellwart J. Sallusto F. Lipp M. and Forster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 192:1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reinhardt R. L. Liang H. E. and Locksley R. M. 2009. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat. Immunol. 10:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luthje K. Kallies A. Shimohakamada Y. Belz G. T. Light A. Tarlinton D. M. and Nutt S. L. 2012. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat. Immunol. 13:491. [DOI] [PubMed] [Google Scholar]
- 54. Locci M., Havenar-Daughton C., Landais E., et al. 2013. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 39:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Delgoffe G. M., Woo S. R., Turnis M. E., et al. 2013. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature 501:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Minagawa K., Wakahashi K., Kawano H., et al. 2014. Posttranscriptional modulation of cytokine production in T cells for the regulation of excessive inflammation by TFL. J. Immunol. 192:1512. [DOI] [PubMed] [Google Scholar]
- 57. Jeong Y. H., Sekiya M., Hirata M., et al. 2010. The low-density lipoprotein receptor-related protein 10 is a negative regulator of the canonical Wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 392:495. [DOI] [PubMed] [Google Scholar]
- 58. McKinstry K. K. Strutt T. M. and Swain S. L. 2010. The potential of CD4 T-cell memory. Immunology 130:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hale J. S. and Ahmed R. 2015. Memory T follicular helper CD4 T cells. Front. Immunol. 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fazilleau N. McHeyzer-Williams L. J. Rosen H. and McHeyzer-Williams M. G. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 10:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sallusto F. Lenig D. Förster R. Lipp M. and Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708. [DOI] [PubMed] [Google Scholar]
- 62. Choi Y. S., Kageyama R., Eto D., et al. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34:932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Haynes N. M. Allen C. D. Lesley R. Ansel K. M. Killeen N. and Cyster J. G. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099. [DOI] [PubMed] [Google Scholar]
- 64. Glatman Zaretsky A. Taylor J. J. King I. L. Marshall F. A. Mohrs M. and Pearce E. J. 2009. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J. Exp. Med. 206:991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kitano M. Moriyama S. Ando Y. Hikida M. Mori Y. Kurosaki T. and Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 34:961. [DOI] [PubMed] [Google Scholar]
- 66. Pearce E. L., Walsh M. C., Cejas P. J., et al. 2009. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim C. Wilson T. Fischer K. F. and Williams M. A. 2013. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4+ memory T cells. Immunity 39:508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Choi Y. S. Yang J. A. Yusuf I. Johnston R. J. Greenbaum J. Peters B. and Crotty S. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 190:4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Weber J. P. Fuhrmann F. and Hutloff A. 2012. T-follicular helper cells survive as long-term memory cells. Eur. J. Immunol. 42:1981. [DOI] [PubMed] [Google Scholar]
- 70. Chevalier N., Jarrossay D., Ho E., et al. 2011. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J. Immunol. 186:5556. [DOI] [PubMed] [Google Scholar]
- 71. Bossaller L., Burger J., Draeger R., et al. 2006. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J. Immunol. 177:4927. [DOI] [PubMed] [Google Scholar]
- 72. Bentebibel S. E., Lopez S., Obermoser G., et al. 2013. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 5:176ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Goenka R. Barnett L. G. Silver J. S. O’Neill P. J. Hunter C. A. Cancro M. P. and Laufer T. M. 2011. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol. 187:1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Baumjohann D. Okada T. and Ansel K. M. 2011. Cutting Edge: Distinct waves of BCL6 expression during T follicular helper cell development. J. Immunol. 187:2089. [DOI] [PubMed] [Google Scholar]
- 75. Kerfoot S. M. Yaari G. Patel J. R. Johnson K. L. Gonzalez D. G. Kleinstein S. H. and Haberman A. M.. 2011. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity 34:947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Garside P. Ingulli E. Merica R. R. Johnson J. G. Noelle R. J. and Jenkins M. K. 1998. Visualization of specific B and T lymphocyte interactions in the lymph node. Science 281:96. [DOI] [PubMed] [Google Scholar]
- 77. Okada T., Miller M. J., Parker I., et al. 2005. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol 3:e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chan T. D. Gatto D. Wood K. Camidge T. Basten A. and Brink R. 2009. Antigen affinity controls rapid T-dependent antibody production by driving the expansion rather than the differentiation or extrafollicular migration of early plasmablasts. J. Immunol. 183:3139. [DOI] [PubMed] [Google Scholar]
- 79. Krummel M. F. and Cahalan M. D. 2010. The immunological synapse: a dynamic platform for local signaling. J. Clin. Immunol. 30:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Reinhardt R. L. Khoruts A. Merica R. Zell T. and Jenkins M. K. 2001. Visualizing the generation of memory CD4 T cells in the whole body. Nature 410:101. [DOI] [PubMed] [Google Scholar]
- 81. Stills H. F., Jr 2005. Adjuvants and antibody production: dispelling the myths associated with Freund’s complete and other adjuvants. ILAR J. 46:280. [DOI] [PubMed] [Google Scholar]
- 82. Suzuki R. and Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.