Abstract
Hawksbill turtle (Eretmochelys imbricata) populations have experienced global decline because of a history of intense commercial exploitation for shell and stuffed taxidermied whole animals, and harvest for eggs and meat. Improved understanding of genetic diversity and phylogeography is needed to aid conservation. In this study, we analyzed the most geographically comprehensive sample of hawksbill turtles from the Indo-Pacific Ocean, sequencing 766bp of the mitochondrial control region from 13 locations (plus Aldabra, n = 4) spanning over 13500 km. Our analysis of 492 samples revealed 52 haplotypes distributed in 5 divergent clades. Diversification times differed between the Indo-Pacific and Atlantic lineages and appear to be related to the sea-level changes that occurred during the Last Glacial Maximum. We found signals of demographic expansion only for turtles from the Persian Gulf region, which can be tied to a more recent colonization event. Our analyses revealed evidence of transoceanic migration, including connections between feeding grounds from the Atlantic Ocean and Indo-Pacific rookeries. Hawksbill turtles appear to have a complex pattern of phylogeography, showing a weak isolation by distance and evidence of multiple colonization events. Our novel dataset will allow mixed-stock analyses of hawksbill turtle feeding grounds in the Indo-Pacific by providing baseline data needed for conservation efforts in the region. Eight management units are proposed in our study for the Indo-Pacific region that can be incorporated in conservation plans of this critically endangered species.
Key words: marine conservation, marine turtles, mitochondrial DNA, population genetic structure
Understanding the population dynamics of marine migratory species has long presented a challenge because of the difficulties in identifying population boundaries and interrelationships. The application of genetic markers to study population genetic structure and demography has uncovered contrasting population histories and behaviors among migratory marine species. A variable degree of genetic structure within and between ocean basins has been found among many highly migratory populations, including those of sharks (Keeney and Heist 2006; Castro et al. 2007), marine mammals (Baker et al. 1994; Dalebout et al. 2005), and marine turtles (Jensen et al. 2013a).
In some marine species, populations overlap at feeding grounds, but unique breeding populations are maintained because individuals migrate to discrete natal regions for breeding (Hoffman et al. 2006; Jensen et al. 2013a). In contrast, discrete feeding-ground aggregations that are maternally determined are observed in some species in which migrations to shared breeding grounds result in gene flow among the maternal subgroups (Palsbøll et al. 1997). Our ability to define populations and determine the extent of gene flow among populations, and to relate this to migratory patterns of individuals within populations, is critical to understanding population function and for effective conservation management of what are often threatened species.
Identifying genetic lineages and estimating the times of divergence between them allows an understanding of ancestral demographic connections across ocean basins. In turn, this can reveal the influence of past changes in climate, sea level, and ocean currents on population structure (Foote et al. 2013). Insights from genetic analyses have included evidence of postglacial colonization and population expansion (O’Corry-Crowe et al. 1997; Stamatis et al. 2004; Pellerito et al. 2009) and shared vicariant events among diverse species (Gopurenko and Hughes 2002; Dethmers et al. 2006). Evidence of population expansion is often specific to particular ocean basins (Lavery et al. 1996; Duncan et al. 2006; Lukoschek et al. 2007).
Most species show global phylogeographic structure but the extent of genetic structure within or between ocean basins is not predictable and there is considerable variation in where phylogeographic splits occur (Hoffman et al. 2006; Keeney and Heist 2006; Castro et al. 2007; Díaz-Jaimes et al. 2010). Some studies have indicated isolation-by-distance effects in large marine vertebrates (Dethmers et al. 2006; Schultz et al. 2008; Mendez et al. 2010), but the processes driving these patterns across species function at different geographic scales and are influenced by dispersal capacity, natal philopatry, environmental conditions, and recent or historic physical barriers.
Genetic markers have been particularly valuable for delineating boundaries in marine turtle populations, understanding colonization history, the extent of natal philopatry, and establishing management units (MUs) for effective conservation of these vulnerable species (Jensen et al. 2013a). In general, marine turtles show natal philopatry in their selection of breeding grounds and nesting beaches, as well as geographically influenced variation in the extent to which different breeding populations overlap on feeding grounds. Within marine turtles there is considerable variation in the extent of genetic diversity, and regional or global genetic structure among, as well as within, species. Loggerhead turtles (Caretta caretta) show low genetic diversity among Indo-Pacific populations in comparison with Atlantic Ocean populations, whereas green turtles (Chelonia mydas) have similarly high levels of genetic diversity in these ocean basins (Jensen et al. 2013a). Strong natal homing within some populations has led to genetic divergence of rookeries located <200 km apart (Dethmers et al. 2006; Browne et al. 2010), in contrast with some rookeries located over 1000 km apart that do not display genetic differentiation (Dethmers et al. 2006; Dutton et al. 2007). Colonization of some rookeries appears to occur through isolated founder events, with resultant genetic bottlenecks, while other rookeries display a diversity of divergent mitochondrial haplotypes, suggesting colonization from multiple locations (Lahanas et al. 1994; Dethmers et al. 2006).
Hawksbill turtles (Eretmochelys imbricata) were once abundant in tropical and subtropical regions throughout the world. However, the species is now listed as critically endangered by the IUCN (2015) as a result of global declines in many populations across all ocean basins. These have been caused by pervasive exploitation through a large commercial trade in hawksbill shell (“tortoise shell” or “bekko”), as well as harvests for eggs, meat, and taxidermied whole animals (Witzell 1983; Milliken and Tokunaga 1987; Mortimer and Donnelly 2008; Hamilton et al. 2015). There remain concerns over domestic trade that continues in some countries (e.g., Bräutigam et al. 2006).
The decline of hawksbill turtles is of broader concern for reef ecosystems because these turtles often play an important role in controlling the growth of sponges that overgrow corals, thus influencing competition for space (Hill 1998; Bjorndal and Jackson 2003). Hawksbill turtles nest on island and mainland beaches and, like other marine turtle species, specific populations typically use a diversity of feeding grounds that often overlap with those of other populations (Diaz-Fernandez et al. 1999; Bowen et al. 2007; Blumenthal et al. 2009). Improved knowledge of population connectivity, historic gene flow, and patterns of long-distance dispersal are critical for managing their recovery (Broderick et al. 1994; Bass et al. 1996; Monzón-Argüello et al. 2011). Previous genetic studies of hawksbill turtles in the Indo-Pacific have identified eastern and western Australia populations (Broderick et al. 1994). They have also indicated a genetic connection between Seychelles and Chagos rookeries, but not with western Australia (Mortimer and Broderick 1999), and suggested genetic differentiation between 2 rookeries in Iran (Tabib et al. 2014). However, a comprehensive genetic study of the species in the Indo-Pacific is lacking.
In this study, we sequence a portion (766bp) of the mitochondrial control region in samples taken from nesting turtles at 13 rookeries in the Indian Ocean and western Pacific Ocean. We combine these data with previously published sequence data, allowing us to investigate population genetic structure and phylogeographic patterns. Using tests for genetic divergence, we identify MUs (sensu Moritz 1994) among Indo-Pacific populations to aid in the conservation of this species. This study aims to 1) quantify the genetic diversity at rookeries in the Indo-Pacific region; 2) determine the extent of genetic divergence among rookeries to define population boundaries and identify MUs; 3) determine the relationships among mitochondrial lineages globally and estimate divergence times between lineages; 4) uncover phylogeographic patterns in the Indo-Pacific region; 5) establish baseline data for future genetic studies of feeding grounds throughout the Indo-Pacific; and 6) contribute toward more effective conservation actions for hawksbill turtle populations.
Materials and Methods
Data Collection
Collections of tissues from nesting females were taken from 13 locations (plus Aldabra, n = 4), either from single rookeries or from a group of nearby rookeries in the western Pacific Ocean and Indian Ocean (Figure 1). Samples were collected between 1990 and 2010, and ranged in from 4 to 71 per site (Supplementary Table S1 online). Samples from the Aldabra group (n = 4) were excluded from the statistical analyses.
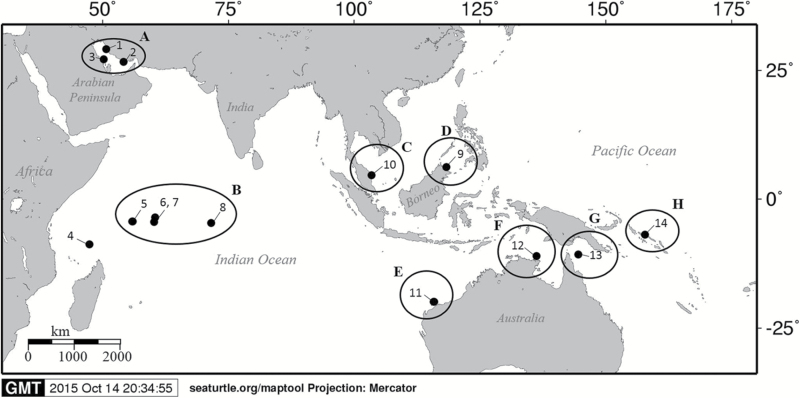
Figure 1.
Sampling locations of hawksbill turtle nesting populations in the Indo-Pacific: 1, Iran Northwest; 2, Iran Southeast; 3, Saudi Arabia; 4, Aldabra Group, Seychelles; 5, Amirantes Islands, Seychelles; 6, Platte Island, Seychelles; 7, Granitics Islands, Seychelles; 8, Chagos Archipelago; 9, East Malaysia; 10, Peninsular Malaysia; 11, Western Australia; 12, northeast Arnhem Land, Northern Territory; 13, Milman Island, north Queensland; and 14, Solomon Islands. Letters represent the 8 identified genetic stocks: A, Persian Gulf; B, western/central Indian Ocean; C, western Peninsular Malaysia; D, Sulu Sea; E, Western Australia; F, NE Arnhem Land; G, north Queensland; H, Solomon Islands.
From each individual, a small (~5mm2) piece of skin was taken from the neck region or trailing edge of the rear flipper using a sterilized scalpel and collected into either a 20% DMSO solution that was saturated with NaCl or in EtOH (≥70%). At some remote locations, tissue samples were collected from a single dead embryo or hatchling from nests that had previously hatched. When hatchlings or embryos were sampled, care was taken to restrict sampling of clutches to those laid within a 2-week period (minimum renesting interval) to avoid sampling of successive clutches from the same female. For some of the earlier sampling efforts, blood samples were taken from the dorsal cervical sinus (FitzSimmons et al. 1997). Total genomic DNA was extracted using a salting-out protocol after digestion with proteinase K (Jensen et al. 2013b). We checked this for quality and quantity by running the samples through 1.2% agarose gels and visualizing them using SyBRSafe gel stain and a UV illuminator.
The mitochondrial control region (d-loop) was amplified using the marine turtle primers LTEi9 and H950, which amplify ~800bp. This includes a smaller region (~385bp) used in previous studies (Abreu-Grobois et al. 2006). Polymerase chain reaction (PCR) volumes of 25 μL included ~40ng of genomic DNA, 1U of Taq polymerase (Astral), 200 μM of each dNTP, 1×Tris–KCl buffer, 1.5mM MgCl, 0.5 μM of each primer, and 0.5M betaine. The amplification program consisted of 5min at 94 °C, followed by 35 cycles of 30s at 94 °C, 30s at 52 °C, 30s at 72 °C, and a final extension step of 5min at 72 °C. Negative controls, in which template DNA was omitted, were included in all amplification runs. PCR products were checked for amplification quality and lack of contamination by electrophoresis in 1.2% agarose gels as described above. Before sequencing, PCR products were cleaned by precipitation using polyethylene glycol (20% PEG 8000, 2.5M NaCl). Forward and reverse sequencing reactions were performed by Macrogen, Inc. (Seoul, South Korea). Sequences were aligned using Geneious 4.5.3 or 6.1.7 (Drummond et al. 2010), using a global alignment with free end gaps, and the consensus sequences were checked manually.
Data Availability
In agreement with the data archiving rule (Baker 2013), all sequences generated in this study have been deposited in GenBank (accession numbers KT934050–KT934101—see Supplementary Table S1 online).
Population Genetics
We used the program Arlequin 3.5 (Excoffier and Lischer 2010) to calculate haplotype diversity (h), nucleotide diversity (π), the average number of nucleotide differences (k), and number of variable sites. We conducted exact tests of population differentiation and determined population pairwise F ST values (based on haplotype frequencies) to describe the genetic structure of the Indo-Pacific populations and to propose MUs (Moritz 1994) for the region. We tested for a correlation between genetic distance (F ST/[1 − FST]) and geographic distance (logarithm [ln] of distance [km]) matrices using a Mantel nonparametric permutation (n = 10000) test in Arlequin.
Analyses of molecular variance (AMOVA; Excoffier and Lischer 2010) among a hierarchical grouping of populations were used to depict regional genetic structure using Arlequin. We performed 2 different AMOVAs using a reduced alignment of 740bp to maintain compatibility with previously published data that were included in the analyses. These groupings were: 1) all sampled populations in the Indian Ocean (8 populations from our study) versus those in the Pacific Ocean (5 populations from our study); and 2) all sampled Indo-Pacific populations versus those in the Atlantic Ocean. The Atlantic population data were obtained from Diaz-Fernandez et al. (1999), Lara-Ruiz et al. (2006), Velez-Zuazo et al. (2008), Browne et al. (2010), Trujillo-Arias et al. (2014), and LeRoux et al. (2012) and included 11 populations from the Caribbean and western Atlantic Ocean. These analyses were done twice, either only considering haplotype frequencies or also accounting for Tamura-Nei distances among haplotypes.
We used a median network analysis (Bandelt et al. 1999) in the software Network 4 (http://www.fluxus-engineering.com) to depict the relationships among haplotypes (based on 766bp). Sequence divergences among haplotypes and clades were estimated using the Tamura-Nei substitution model (gamma shape parameter = 0.04) in MEGA 6 (Tamura et al. 2013).
To test for demographic expansion, we conducted neutrality tests (Tajima’s D and Fu’s Fs) using Arlequin. Using DnaSP 5 (Librado and Rozas 2009), we tested for changes in population size using the R 2 test (Ramos-Onsins and Rozas 2002).
Phylogenetic Inference of Demography and Timescale
To infer past changes in population size, sequence data from the entire Indo-Pacific population were analyzed using a Bayesian phylogenetic approach in BEAST 1.8.1 (Drummond et al. 2012). Using the Bayesian information criterion in ModelGenerator (Keane et al. 2006), we identified the TN + G model as the best-fitting model of nucleotide substitution. Owing to the intraspecific nature of the data, we did not consider models that included a parameter for the proportion of invariable sites (Jia et al. 2014). Rate variation among sites was modeled using a gamma distribution with 6 rate categories (Yang 1993). We assumed rate homogeneity among branches (strict molecular clock) and fixed the rate to 1.0. This allowed us to compare demographic models and to track relative population sizes through time, but precluded us from estimating the evolutionary timescale.
Posterior distributions of parameters were estimated using Markov chain Monte Carlo (MCMC) sampling. Samples were drawn every 104 steps over a total of 108 steps. The first 10% of samples were discarded as burn-in, and convergence to the stationary distribution was checked by inspection of MCMC traces. We compared 2 coalescent-based tree priors: constant population size and Bayesian skyline plot (BSP; Drummond et al. 2005). This comparison was done by calculating the Bayes factor, which is the ratio of marginal likelihoods between the 2 candidate models. The marginal likelihoods were calculated by stepping-stone analysis (Xie et al. 2011).We also analyzed the data using the extended BSP (Heled and Drummond 2008).
The Bayesian phylogenetic analyses were repeated for each group of most genetically divergent populations, according to pairwise F ST: Iran NW, Iran SE and Saudi Arabia, Solomon Islands, SW Indian Ocean, Western Australia, Northern Australia, Peninsular Malaysia, and East Malaysia. To decrease the error associated with the small sample sizes, we also conducted separate analyses of geographically defined groups: Australian group consisting of Western Australia and Northern Australia (n = 182), Malaysian group consisting of Peninsular Malaysia and East Malaysia (n = 48), and Persian Gulf group consisting of Iran NW and Iran SE + SA (n = 95). For each subset of samples, we identified the best-fitting model of nucleotide substitution: HKY for Iran NW, Iran SE + SA, Western Australia, Solomon Islands, East Malaysia, and the Malaysian group; HKY + G for Northern Australia, SW Indian Ocean, the Persian Gulf group, and the Australian group; and F81 for Peninsular Malaysia. Samples from the posterior were drawn every 103 MCMC steps over a total of 107 steps (except for the Australian group, for which we drew samples every 5×103 steps over a total of 5×107 steps).
To estimate the divergence times among mitochondrial lineages, we used BEAST to analyze an alignment of 440 sequences representing the Atlantic population. This data set comprised 118 random samples, representing all 23 of the hawksbill turtle haplotypes found by LeRoux et al. (2012), Lara-Ruiz et al. (2006), Diaz-Fernandez et al. (1999), and Velez-Zuazo et al. (2008), and 322 randomly selected sequences that represented all 57 of the haplotypes for the Indo-Pacific populations from Tabib et al. (2011) and from this study. The TN + G model of nucleotide substitution was selected, with rate variation among sites modeled using a gamma distribution with 6 rate categories. We used a Bayesian skyline coalescent prior and assumed a strict molecular clock. Samples from the posterior were drawn every 5×104 MCMC steps over a total of 5×108 steps.
To calibrate the molecular clock, we used a normal calibration prior with a mean of 5.63 million years (standard deviation [SD] = 1.38) for the age of the root of the tree and a truncated lognormal prior for the substitution rate, ranging from 10−6 to 0.02 substitutions/site/million years (Myr). Our age calibration is based on a molecular estimate by Duchene et al. (2012), obtained using whole mitochondrial genomes. However, its application to population-level data is likely to lead to overestimates of divergence times (Ho et al. 2005; Ho and Shapiro 2011). Therefore, the date estimates obtained in this study need to be interpreted as maximum ages.
To evaluate support for major clades in the tree, we conducted additional phylogenetic analyses using a Bayesian approach in MrBayes 3.2 (Ronquist et al. 2012) and maximum-likelihood in RAxML 7.3.1 (Stamatakis 2006). For both analyses, we used sequences from 2 species of marine turtles, C. caretta (GenBank accession number JF837824.1) and Lepidochelys olivacea (GenBank accession number AY920523.1), as outgroups. The best-fitting model of nucleotide substitution was selected using the Bayesian information criterion. In MrBayes, samples from the posterior distribution were drawn every 103 steps over a total of 2×106 steps. In the RAxML analysis, node support was evaluated using 1000 bootstrap replicates.
Results
Genetic Diversity
Mitochondrial sequence data were obtained for 492 turtles, and aligned for a 766bp region for all individuals. Fifty-two mitochondrial haplotypes were identified from the 13 Indo-Pacific sample locations (plus Aldabra, n = 4). A total of 67 polymorphic sites were observed, including 6 transversions and 61 transitions. Comparing our longer 766bp fragments with a 740bp fragment used in earlier studies (Diaz-Fernandez et al. 1999; Lara-Ruiz et al. 2006; Velez-Zuazo et al. 2008; Tabib et al. 2011; LeRoux et al. 2012; Trujillo-Arias et al. 2014), we found only one additional polymorphic site at position 1, which defines haplotype EiIP-30 (GenBank accession number KT934077). Consequently, we used the 740bp fragments in further analyses to maintain compatibility with previous data.
Eleven of the 52 haplotypes were observed at multiple rookeries, with the remainder only observed at a single rookery each (Supplementary Table S1 online). Haplotype EiIP-08 was the most common (n = 99), but it was only shared among the 3 Australian rookeries, whereas the common (n = 86) haplotype EiIP-33 was shared by 8 of 13 populations (plus Aldabra, n = 4). The other 9 shared haplotypes were found among closer geographic populations, including: EiIP-09 in Northeast Arnhem Land and Milman Island; EiIP-16 and EiIP-17 in the Amirante, Platte, and Granitics islands, and the Chagos Archipelago; and EiIP-36 in Iran and Saudi Arabia. The remaining 41 haplotypes observed at individual rookeries had frequencies that ranged from a single individual (19 haplotypes) up to 22 individuals for EiIP-53 in Peninsular Malaysia.
The number of different haplotypes observed per rookery ranged from 2 for Peninsular Malaysia to 10 for north Queensland, with an average of 3.7 haplotypes per rookery. Haplotype diversity among the 13 rookeries ranged from h = 0.087 to h = 0.782, with an overall estimate of h = 0.897 (SD = 0.007) (Table 1). Nucleotide diversities within rookeries ranged from π = 0.0002 to π = 0.0214, with an overall estimate of π = 0.0216 (SD = 0.0107). The number of nucleotide differences within populations ranged from k = 0.174 to k = 15.6, with an overall number of nucleotide differences across all samples of k = 15.7.
Table 1.
Standard and molecular diversity indices for hawksbill turtle rookeries in the Indo-Pacific based on 740bp of the mitochondrial control region
| Rookery | n | S | H | k | h | ±SD | π | ±SD |
|---|---|---|---|---|---|---|---|---|
| IranNW | 41 | 5 | 6 | 0.463 | 0.383 | 0.092 | 0.0006 | 0.0006 |
| IranSE | 41 | 32 | 8 | 2.38 | 0.727 | 0.062 | 0.0003 | 0.0020 |
| SaudArab | 13 | 2 | 3 | 0.847 | 0.500 | 0.136 | 0.0012 | 0.0009 |
| Amrt | 28 | 20 | 6 | 8.56 | 0.725 | 0.054 | 0.0118 | 0.0062 |
| Plat | 27 | 35 | 6 | 9.14 | 0.687 | 0.077 | 0.0125 | 0.0066 |
| Gran | 48 | 36 | 9 | 10.3 | 0.782 | 0.041 | 0.0141 | 0.0073 |
| Chag | 18 | 19 | 5 | 6.75 | 0.660 | 0.102 | 0.0092 | 0.0051 |
| EastMaly | 25 | 31 | 5 | 12.1 | 0.743 | 0.051 | 0.0165 | 0.0086 |
| PenMaly | 23 | 2 | 2 | 0.174 | 0.087 | 0.078 | 0.0002 | 0.0004 |
| WestAust | 47 | 6 | 4 | 1.30 | 0.373 | 0.086 | 0.0018 | 0.0013 |
| neAl | 71 | 32 | 6 | 14.9 | 0.571 | 0.030 | 0.0204 | 0.0102 |
| nQld | 64 | 39 | 10 | 15.6 | 0.725 | 0.038 | 0.0214 | 0.0107 |
| Solo | 42 | 7 | 6 | 1.39 | 0.660 | 0.045 | 0.0019 | 0.0013 |
| Overall | 488 | 67 | 52 | 15.7 | 0.897 | 0.007 | 0.0216 | 0.0107 |
Values are given for sample number (n), variable sites (S), haplotype number (H), average number of nucleotide differences (k), haplotype diversity (h), and nucleotide diversity (π). Iran NW, Iran Northwest; Iran SE, Iran Southeast; SauArab, Saudi Arabia; Aldb, Aldabra Group, Seychelles; Amrt, Amirantes Islands, Seychelles; Plat, Platte Island, Seychelles; Gran, Granitics Islands; Chag, Chagos Archipelago; East Maly, East Malaysia; Pen Maly, Peninsular Malaysia; West Aust, Rosemary and Varanus Islands, Western Australia; neAl, northeast Arnhem Land, Northern Territory; nQld, Milman Island, north Queensland; Solo, Solomon Islands.
Interpopulation Analysis
Population pairwise F ST tests indicated non-significant genetic differentiation (P > 0.05) among 9 pairs of sampled rookeries (Table 2). All other F ST values were significant (P < 0.01), and ranged from 0.084 between Iran NW and Iran SE to 0.754 between Saudi Arabia and Peninsular Malaysia. Exact tests of population differentiation showed nonsignificant (P > 0.05) genetic differentiation among 8 of the 9 pairs of rookeries identified as not differentiated in pairwise F ST tests. The exception was the north Queensland and northeast Arnhem Land rookeries, which were differentiated by this test (P = 0.005).
Table 2.
P-values of exact tests of population differentiation method (below diagonal) between Indo-Pacific hawksbill turtle rookeries and population pairwise F ST based on haplotype frequencies (above diagonal)
| IranNW | IranSE | SauArab | Amrt | Plat | Gran | Chag | East Maly | Pen Maly | West Aust | neAl | nQld | Solo | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IranNW | — | 0.084 | 0.028* | 0.4625 | 0.466 | 0.381 | 0.509 | 0.459 | 0.733 | 0.622 | 0.509 | 0.408 | 0.237 |
| IranSE | 0.0019 | — | 0.050* | 0.2741 | 0.279 | 0.221 | 0.302 | 0.266 | 0.540 | 0.456 | 0.359 | 0.257 | 0.136 |
| SauArab | 0.1109 | 0.0732 | — | 0.369 | 0.375 | 0.298 | 0.413 | 0.361 | 0.754 | 0.587 | 0.451 | 0.336 | 0.179 |
| Amrt | <0.0001 | <0.0001 | <0.0001 | — | −0.020* | −0.011* | −0.021* | 0.266 | 0.574 | 0.474 | 0.366 | 0.275 | 0.310 |
| Plat | <0.0001 | <0.0001 | <0.0001 | 0.7796 | — | −0.006* | −0.036* | 0.285 | 0.597 | 0.493 | 0.382 | 0.290 | 0.318 |
| Gran | <0.0001 | <0.0001 | <0.0001 | 0.4301 | 0.7043 | — | −0.001* | 0.236 | 0.499 | 0.421 | 0.330 | 0.246 | 0.259 |
| Chag | <0.0001 | <0.0001 | <0.0001 | 0.7204 | 0.7928 | 0.5102 | — | 0.296 | 0.652 | 0.520 | 0.396 | 0.301 | 0.340 |
| EastMaly | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | — | 0.576 | 0.471 | 0.359 | 0.267 | 0.302 |
| PenMaly | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | — | 0.732 | 0.593 | 0.511 | 0.575 |
| WestAust | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | — | 0.207 | 0.185 | 0.488 |
| neAl | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | — | 0.011* | 0.389 |
| nQld | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0045 | — | 0.292 |
| Solo | <0.0001 | <0.0001 | 0.0004 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | — |
Rookery abbreviations are defined in Table 1.
*Indicates nonsignificant FST values.
The Mantel test for isolation by distance among Indo-Pacific populations indicated a positive correlation between genetic and geographic distance (r = 0.29, P = 0.0009), although the relationship (r 2) was weak and only accounted for 8.1% of the variation in the data (Figure 2). AMOVAs revealed differences in the partitioning of genetic variation depending on the test used (Table 3). In the analysis of Indian Ocean versus Pacific Ocean, only 1–3% of the variation was partitioned among groups. Based on haplotype frequencies, most variation (64%) occurred within populations, but if molecular distances were included, similar levels of variation were distributed among populations (53%) and within (46%) populations. In the analysis of Indo-Pacific versus Atlantic Ocean, most variation (64%) was partitioned among groups when molecular distance was included in the analysis.
Figure 2.
Correlation between genetic distance (F ST/1 − F ST) and geographic distance (logarithm of distance in km) matrices for Indo-Pacific hawksbill turtles. The Mantel test identified significant isolation by distance (r = 0.29; P = 0.0009). A trendline is provided.
Table 3.
Percent variation and fixation indices for 2 AMOVA analyses of hawksbill turtle rookeries in the Indo-Pacific
| 2 groups (Indo × Pac)1 | 2 groups (IndoPac × Atl) | |||
|---|---|---|---|---|
| Tamura-Nei distances | Haplotype frequencies | Tamura-Nei distances | Haplotype frequencies | |
| % Variation | ||||
| Among groups | 1.24 | 2.85 | 64.0 | 13.8 |
| Among populations within groups | 52.9 | 33.5 | 19.8 | 37.4 |
| Within populations | 45.9 | 63.6 | 16.2 | 48.8 |
| Fixation index | ||||
| Φ SC | 0.536 | 0.345 | 0.549 | 0.434 |
| Φ ST | 0.541 | 0.364 | 0.838 | 0.512 |
| Φ CT | 0.012 | 0.020 | 0.640 | 0.138 |
The Indian Ocean group included 8 rookeries: Iran NW, Iran SE, Saudi Arabia, Amirantes, Platte Island, Granitics, Chagos, and Western Australia. The Pacific Ocean group included 5 populations: Solomon Islands, North Queensland, Arnhem Land, Peninsular Malaysia, and East Malaysia. The Atlantic Ocean group included 11 populations from 6 previous studies: Antigua (n = 72), Costa Rica (n = 60), Guadeloupe – TroisIlets, Marie Galante (n = 72), Nicaragua (n = 95), and United States Virgin Islands (n = 67) from LeRoux et al. (2012); Barbados–Leeward (n = 54). and Barbados–Windward (n = 30) from Browne et al. (2010); Brazil, nonhybrids (n = 62) from Lara-Ruiz et al. (2006); Cuba (n = 65) from Díaz-Fernádez et al. (1999), Colombia Caribe (n = 29) from Trujillo-Arias et al. (2014), and Puerto Rico (n = 109) from Velez-Zuazo et al. (2008).
Phylogenetic Relationships, Divergence Times, and Demographic History
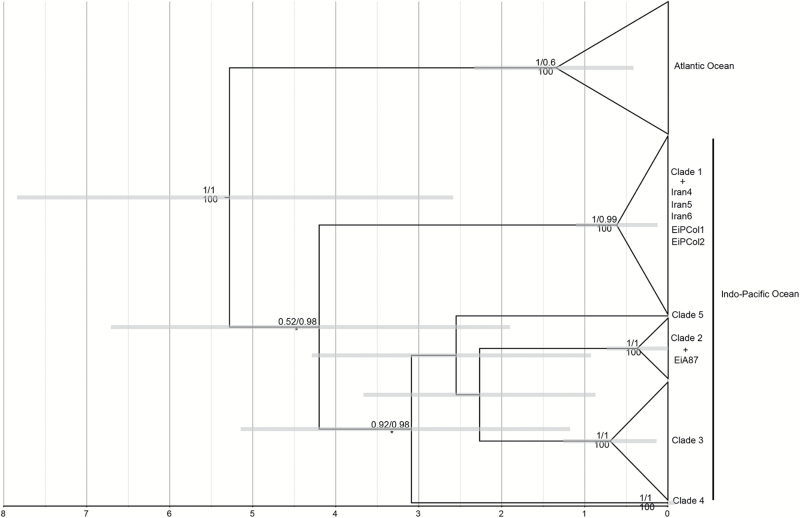
The median network grouped the 52 haplotypes (740bp) into 5 clades, 2 of which only contained a single haplotype each (Figure 3). Clades were separated by a minimum of 15 mutations. The range in sequence divergence among the clades was 0.044–0.131 (mean 0.0914) and the mean divergence within clades with more than 1 haplotype ranged from 0.004 to 0.008. The monophyly of clades 1, 2, and 3 and the western Atlantic and Caribbean clade was supported in the Bayesian analyses using BEAST and MrBayes, and in the maximum-likelihood analysis (Figure 4).
Figure 3.
Median network of 53 Indo-Pacific mitochondrial control region haplotypes of 766bp found in nesting hawksbill turtles. Red diamonds represent hypothesized median vectors.
Figure 4.
Bayesian chronogram inferred from the mitochondrial control region (740bp) of the hawksbill turtle. Numbers above branches correspond to posterior probabilities calculated in BEAST/MrBayes. Numbers below branches denote percentage bootstrap support from a maximum-likelihood analysis. Branch lengths are proportional to time, with the horizontal axis given in millions of years. Clade names correspond to those in the median network. Horizontal grey bars denote the 95% highest posterior density intervals values estimated for tree nodes. Haplotype designations for the Indo-Pacific clades are based on Table 1 (this study), Trujillo-Arias et al. (2014), Monzón-Argüelo et al. (2011), and Tabib et al. (2011). For the Atlantic Ocean clade, we used haplotypes from LeRoux et al. (2012), Lara-Ruiz et al. (2006), Díaz-Fernádez et al. (1999) and Velez-Zuazo et al. (2008).
Neutrality tests yielded concordant significant negative values of Fu’s Fs and Tajima’s D only for Iran NW and Persian Gulf group (Table 4), indicating demographic expansion. The R 2 test revealed evidence of a change in population size only for Iran NW. Bayesian inference of demographic history indicated a pattern of population expansion across the Indo-Pacific (Figure 5), but limited evidence of expansion within regions. For the combined Indo-Pacific populations, we found a log Bayes factor of 24.4 for the BSP model compared with the constant-size model (Table 4). Log Bayes factors greater than 5 indicates very strong support (Kass and Raftery 1995). In regional groupings of populations, the constant-size model received better support for the Persian Gulf group (log BF = 18.34), Malaysian group (log BF = 6.34), and Australian group (log BF = 6.40).
Table 4.
Results of neutrality tests and population-size changes in hawksbill turtle rookeries and groups of rookeries in the Indo-Pacific including Tajima’s D, Fu’s F S, R 2, and Bayes factors
| Neutrality Tests | Demographic expansion | |||||||
|---|---|---|---|---|---|---|---|---|
| N | D | P-value | Fs | P-value | R 2 | P-value | Bayes factor constant (c) vs. skyline (s) | |
| IranNW | 41 | −1.53 | 0.034 | −3.80 | 0.003 | 0.061 | 0.047 | — |
| IranSE | 41 | −2.34 | 0.000 | −0.150 | 0.503 | 0.121 | 0.626 | — |
| SauArab | 13 | 0.878 | 0.822 | 0.436 | 0.559 | 0.212 | 0.734 | — |
| Amrt | 28 | 1.78 | 0.978 | 8.03 | 0.990 | 0.204 | 0.988 | — |
| Plat | 27 | 0.118 | 0.604 | 7.35 | 0.982 | 0.140 | 0.748 | — |
| Gran | 48 | 0.818 | 0.856 | 7.56 | 0.972 | 0.142 | 0.892 | — |
| Chag | 18 | 0.720 | 0.799 | 4.90 | 0.968 | 0.167 | 0.821 | — |
| EastMaly | 25 | 1.71 | 0.962 | 11.5 | 0.998 | 0.191 | 0.978 | — |
| PenMaly | 23 | −1.52 | 0.048 | −0.153 | 0.209 | 0.204 | 0.860 | — |
| WestAust | 47 | −0.357 | 0.420 | 1.26 | 0.771 | 0.107 | 0.498 | — |
| neAl | 71 | 3.88 | 1.000 | 23.4 | 0.999 | 0.229 | 1.000 | — |
| nQld | 64 | 2.83 | 0.999 | 14.6 | 0.998 | 0.197 | 0.999 | — |
| Solo | 42 | −0.408 | 0.391 | −0.260 | 0.465 | 0.102 | 0.427 | — |
| Persian Gulf group | 95 | −2.47 | 0.001 | −4.38 | 0.037 | 0.070 | 0.230 | 18.3 (c) |
| Malaysia group | 48 | 0.288 | 0.696 | 8.23 | 0.987 | 0.121 | 0.733 | 6.34 (c) |
| Australian group | 182 | 2.71 | 0.995 | 14.4 | 0.979 | 0.167 | 0.998 | 6.4 (c) |
| Indo-Pacific | 488 | 1.55 | 0.954 | 0.763 | 0.645 | 0.115 | 0.951 | 24.4 (s) |
Rookery abbreviations are defined in Table 1.
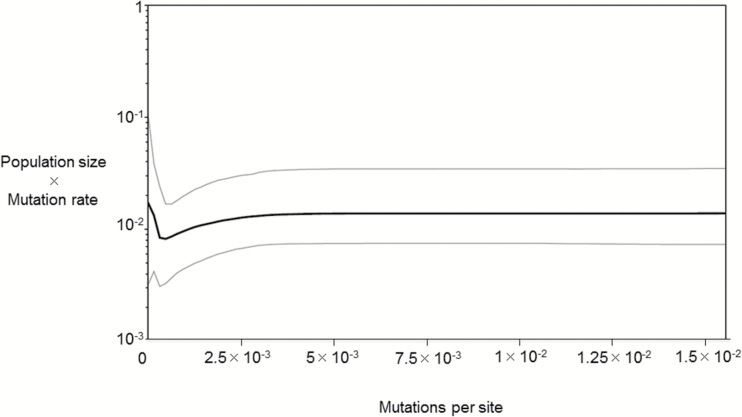
Figure 5.
Bayesian skyline plot of demographic history for the combined Indo-Pacific populations of hawksbill turtles (n = 488). The vertical axis gives the product of effective female population size and mutation rate and the horizontal axis represents genetic distance (in mutations per site).
Using the BSP model, we estimated the time to most recent common ancestor (TMRCA) for the 5 clades identified in our median network to be 4.27 Myr (95% highest posterior density interval: 1.93–6.74 Myr) (Figure 4). The divergence between clade 4 (EIIP-04) and the other clades (clades 2, 3, and 5) was estimated to have occurred 3.23 Myr ago, (95% HPD interval: 1.33–5.29 Myr) followed by the divergence of clade 5 (2.65 Myr, 95% HPD interval: 1.08–4.50) and the split between clades 2 and 3 at 2.32 Myr (95% HPD interval: 0.88–3.82) (Figure 4). The basal divergence in each Indo-Pacific clade was estimated to have occurred prior to the earliest divergence among the Atlantic lineages (1.35 Myr, 95% HPD interval: 0.44–2.36). The most diverse clades from the Indo-Pacific Ocean (clades 1 and 3) were estimated to have similar within-clade diversification times (clade 3 = 0.68 Myr, 95% HPD interval = 0.20–1.28 and clade 1 = 0.58 Myr, 95% HPD interval = 0.15–1.28), and clade 2 diversified more recently (0.34 Myr, 95% HPD interval = 0.07–0.73). All of our date estimates represent maximum ages because they have been calibrated using a rate that is likely to be lower than the intraspecific rate.
Discussion
Genetic Diversity and Structure
Hawksbill turtle rookeries in the Indo-Pacific are characterized by variable patterns of genetic diversity and structure that reflect dynamic processes of population history and gene flow. In general, genetic diversity within Indo-Pacific hawksbill rookeries is high in comparison with other ocean basins. In the populations from the Caribbean and western Atlantic Oceans the haplotype diversity (h) averaged 0.43 (overall h = 0.79) and 8 of 12 populations had relatively low diversity (h ≤ 0.50) (LeRoux et al. 2012), based on the same 740bp mitochondrial fragment used in this study. With larger sample sizes (average 65.3 samples/rookery) only 23 haplotypes were found among 717 samples (LeRoux et al. 2012). However, the greater haplotype diversity observed in the Indo-Pacific populations is not mirrored by nucleotide diversity, which is similar in the Caribbean/western Atlantic (average population π = 0.011±009; overall = 0.023). This is consistent with observations that several rookeries in the Caribbean/western Atlantic are characterized by a collection of divergent haplotypes from different clades, while other rookeries only contain haplotypes within the same clade, as is also observed in the Indo-Pacific. In comparison, rookeries in the U.S. Virgin Islands and Cuba had haplotypes from each of 3 identified haplotype clades (LeRoux et al. 2012) similar to the eastern Australia (Milman) and the Seychelles (Platte and Granitics) rookeries, whereas rookeries in Brazil (Bahia) and Barbados (Leeward) (LeRoux et al. 2012), Iran-NW, Saudi Arabia, Peninsular Malaysia, Western Australia, and the Solomon Islands all were observed to have haplotypes restricted to a single clade.
Genetic structure among Indo-Pacific rookeries indicated that most rookeries separated by 500 km in the Indo-Pacific had significant genetic divergence and can be considered as demographically distinct populations, with some exceptions. Of the 13 rookeries sampled in the Indo-Pacific, we identified 8 populations that were typically differentiated because they did not share a common haplotype and had several unique haplotypes. The only rookeries <500 km apart that were genetically differentiated were the Iran SE and Iran NW rookeries, located only 200 km apart, but these were each not differentiated from the Saudi Arabia rookery (165–375 km distant). Although rookeries in the Persian Gulf were grouped as a single population they may function more as a metapopulation, given these results and as also suggested by genetic differentiation observed by Tabib et al. (2014) between 2 rookeries located to the east of our Iran SE samples.
In Australia, rookeries in the Northern Territory and northern Queensland, located 800 km part, were not genetically differentiated. Broderick et al. (1994) also found a lack of differentiation between these rookeries using analyses of restriction fragment length polymorphisms of whole mitochondrial DNA. This lack of differentiation is interesting because it contradicts what is known about the breeding dynamics at these rookeries. The 2 rookeries are biologically distinguished by a shift in the timing of nesting, with peak nesting in summer in north Queensland (Dobbs et al. 1999) and winter/spring nesting in Arnhem Land (Limpus and Fien 2009). This substantial difference in the timing of breeding cycles would be regulated by physiological triggers (Hamann et al. 2003) and selective pressure to allow successful incubation of eggs and favorable hatchling dispersal conditions. Therefore, interbreeding between these rookeries is unlikely (Limpus and Fien 2009). We have some evidence to support this, in that the exact test indicated genetic differentiation between these rookeries. Based on this and the important biological differences between these rookeries, we consider them to function as separate populations. Therefore, we hypothesize that the lack of genetic differentiation between these 2 populations is not due to ongoing gene flow but results from a recent separation event, as suggested for other marine turtle populations (Encalada et al. 1996). One caveat to this is that there are low numbers of turtles nesting outside the peak nesting periods in both areas (Chatto and Baker 2008; Miller et al. 2008), so these turtles could represent avenues for gene flow. Research on turtles nesting outside peak nesting periods may provide insights into this question.
Comparisons of genetic diversity and structure in hawksbill populations to those of green turtle populations in the Indo-Pacific suggest regional differences in colonization histories and patterns of gene flow. Whereas hawksbill turtles in northeast Arnhem Land and the northern Great Barrier Reef (nGBR) show relatively high levels of haplotype diversity, the nGBR green turtle population have relatively low haplotype diversity (Dethmers et al. 2006), even though that population supports one of the world’s largest green turtle rookeries (Limpus et al. 2003). Populations of both species in this region contain highly divergent genetic lineages, suggestive of multiple colonization events, but colonization frequency or subsequent gene flow may have varied in the 2 species. Several island rookery sites in this region were not available for nesting during the higher sea levels of the Holocene around 6000 year ago (Limpus et al. 2003), so some variation in genetic diversity may be due to differences in the choice of nesting beaches by the 2 species and their availability through time. Beaches used by both species in Peninsular Malaysia were not available until sea level rose within the last 15000 years ago as the coastline shifted over 500 km westwards (Sathiamurthy and Voris 2006). There, lower haplotype and nucleotide diversity was observed in hawksbill turtles in comparison to green turtles, which may have been influenced by a combination of variable colonization patterns as sea level rose and anthropogenic pressures that have reduced hawksbill turtle populations to a larger extent than green turtles (Chan 2006).
The geographic scale of genetic divergence among hawksbill turtles in this study is similar to that of green turtle populations in the Indo-Pacific. Most green turtle rookeries separated by 500 km or more are genetically distinct (Dethmers et al. 2006), but some rookeries less than 200 km apart show significant genetic differentiation (Dethmers et al. 2006; Cheng et al. 2008; Nishizawa et al. 2011), and others 1000 km apart are similar (Dethmers et al. 2006). Additionally, a similar pattern of isolation by distance is shared between the 2 species, with a significant relationship found between genetic and geographic distance that explains a low proportion (6–12% for green turtles) of the pattern of genetic diversity within the Indo-Pacific (Dethmers et al. 2006). Nuclear markers will allow further testing of hypotheses of genetic flow among populations as demonstrated by microsatellite analyses that revealed genetic divergence in leatherback turtles (Dermochelys coriacea) not detected by mitochondrial DNA analysis (Dutton et al. 2013).
In comparisons of genetic structure within and among ocean basins, there was low genetic divergence between hawksbill turtles in the Indian Ocean and Pacific Ocean, moderate differentiation among Indo-Pacific populations, and high differentiation between turtles in the Indo-Pacific versus the western Atlantic/Caribbean oceans. The lack of genetic differentiation between Indian Ocean and Pacific Ocean populations occurred regardless of whether or not genetic distances among haplotypes were considered. In contrast, differentiation between Indo-Pacific and western Atlantic/Caribbean populations was much greater when genetic distances were included in the analysis. This reflects a lack of shared haplotypes observed between the nesting turtles in the western Atlantic (LeRoux et al. 2012) and those in the Indo-Pacific. Within the Indo-Pacific, most genetic variation was distributed within populations, rather than among populations, but when molecular distance was considered, about half of the genetic variability was partitioned among populations. This is consistent with the existence of some widespread haplotypes that is countered by moderate phylogeographic structure in the distribution of related haplotypes. For example, the very common EiIP-33 haplotype was found in 8 of the 13 rookeries (plus Aldabra, n = 4) sampled from the Persian Gulf (this study) and haplotype Iran 3 in Tabib et al. (2014), central Indian Ocean, north Queensland and the Solomon Islands. In contrast, 3 other common haplotypes (EiIP-08, EiIP-17, and EiIP-09) were only observed either in Australia or the central Indian Ocean.
Among western Atlantic and Caribbean populations of hawksbill turtles, although the overall level of genetic differentiation was similar to that of the Indo-Pacific population, there are some striking differences. First, the overall level of genetic differentiation changes very little if the genetic distance is considered, reflecting a low level of phylogeographic structure in the western Atlantic and Caribbean. Second, there is a high level of genetic differentiation between some neighboring populations <500 km distant (LeRoux et al. 2012). In Barbados, turtles nesting on a single beach on the windward side of the island are highly differentiated from those using nesting on beaches along the leeward side, some of which are <50 km distant (Browne et al. 2010; LeRoux et al. 2012). Differentiation on such a small scale is not expected. Tagging data from hawksbill turtles in north Queensland indicates that a substantial number of hawksbill turtles use multiple nesting beaches, either within seasons or across seasons, at least as much as 38 km distant (Miller et al. 2008). The extent to which nest site selection defines boundaries for gene flow among rookeries will depend in part upon the distribution of suitable nesting beaches between rookeries and perhaps regional behavioral differences among populations.
Phylogenetic Relationships, Phylogeography, and Demographic History
Support for the monophyly of the clade including all sequences from the Indian and Pacific Ocean was low, but the monophyly of Indo-Pacific clades 1, 2, 3, and 4 and lineages from the Atlantic Ocean was strongly supported. The lack of strong support for the Indo-Pacific clade is expected because of the absence of observed or assumed interconnecting haplotypes among the clades. Many rookeries throughout the Indo-Pacific are yet to be included in genetic analyses. These will be needed to clarify the relationships among the lineages and better understand how hawksbill turtle populations in the region have responded to dramatic changes in climate, sea level, and oceanic currents over the past 4 million years. Our divergence time estimate should be regarded as a maximum value, because it is based on the estimate of Duchene et al. (2012) and its application to a population scale is likely to lead to overestimates of divergence times (Ho et al. 2005; Ho et al. 2011).
Our date estimates suggest that Indo-Pacific lineages of the hawksbill turtle diverged during or subsequent to the early Pliocene, which was a time of dramatic changes to Indo-Pacific oceanic currents. The earth was cooling after the warm Miocene, Australia collided with the Banda arc, New Guinea was moving northwards, and the Isthmus of Panama formed (Cane and Molnar 2001; Hall 2002). Prevailing Miocene ocean currents that travelled westwards across the Pacific Ocean, north of New Guinea into the Indian Ocean were blocked by the movement of New Guinea northwards (Cane and Molnar 2001; Hall 2002). This shifted the origin of water within the Indonesian through-flow to the North Pacific, with resultant impacts upon global climate (Cane and Molnar 2001), the dispersal of marine organisms (Srinivasan and Sinha 1998), and probably the migratory life stages of hawksbill turtles. Our age estimates for the diversification within clades 1, 2, and 3 clades are broadly consistent with inter- or post-glacial expansion. In contrast, diversification of clades 4 and 5 may have occurred in the Pliocene or early Pleistocene, bearing mind that our estimates of divergence times should be regarded as maximum values.
Our Bayesian demographic analysis of the combined Indo-Pacific populations revealed a recent growth following a decrease in population size (Supplementary Figure S1 online). This is similar to the demographic pattern found for leatherback turtles (Molfetti et al. 2013) and might be associated with recoveries from bottlenecks after the Last Maximum Glacial about 18000 years ago. However, potential violations of the coalescent assumptions involved in BSP analyses suggest that our results should be interpreted cautiously (Grant 2015). For example, in our analyses of subpopulations, small sample sizes are likely to have reduced our power to resolve past changes in population size (Ho and Shapiro 2011). Our non-Bayesian analysis of demographic history shows additional evidence of expansion for the Persian Gulf region, suggesting a relatively recent colonization history, as also indicated by the star-shaped phylogeny around a central haplotype (EiIP-33). Persian Gulf nesting beaches were differentially inundated after the Last Glacial Maximum from 12000 years ago and higher sea levels of the Holocene (Lambeck 1996) would have flooded some of the low-lying islands used by hawksbill turtles, so multiple colonization events are likely.
Our genetic studies of hawksbill turtles in the Indo-Pacific have uncovered complex patterns of phylogeography and genetic structure. Most previous genetic studies of female hawksbill turtles have indicated the strong prevalence of natal homing, whereby females return to the region of their birth to nest (Bass 1999; LeRoux et al. 2012). Our study confirms that the scale at which this behavior operates is variable, and depends in part on geographic location and the influence of changing ocean currents (as also indicated for green turtles; Dethmers et al. 2006). Obviously, some degree of flexibility is needed across evolutionary time; otherwise colonization of new beaches, as needed in changing environments, would not be possible (Carr et al. 1978; Bowen et al. 1992).
Our results suggest there have been long-distance migrations to distribute the EiIP-33 haplotype found from Iran to the Solomon Islands (and also in the eastern Pacific feeding aggregation; Trujillo-Arias et al. 2014). Whether this haplotype was transported to Western Australia (or the Northern Territory) and subsequently lost from the population is unclear, but all haplotypes found in Western Australia are closely related to EiIP-33. Long-distance migrations have also transported haplotypes between Indo-Pacific and western Atlantic rookeries. The Oka24/EATL haplotype found in all nesting turtles (n = 20) in Príncipe in the eastern Atlantic (Monzón-Argüello et al. 2011) was originally observed in 2 foraging turtles in the Seychelles (Okayama et al. 1999). Based on this, Monzón-Argüello et al. (2011) hypothesized that the Príncipe population was founded by a long-distance migration event from the central Indian Ocean around the Cape of Good Hope. We find support for this hypothesis because the Oka24/EATL haplotype, which was based on a shorter d-loop sequence (481bp), corresponds to 2 haplotypes (based on 740bp) in clade 2 (EiIP-16, EiIP-76) that we found only in the central/western Indian Ocean. Additionally, the lack of genetic divergence between the Seychelles and Chagos rookeries, located 1800–2100 km apart, is based on the sharing of clade 2 haplotypes.
These examples of long-distance gene flow among island rookeries is quite different from the pattern observed in green turtles in the Indo-Pacific, where long-distance gene flow occurs when there are intermediate nesting beaches between the sampled sites (Dethmers et al. 2006). For the Granitic and Amirantes islands of Seychelles, Phillips et al. (2014) found that 8 hawksbill rookeries spanning a distance of 500 km showed considerable gene flow among the sampled sites, a relatively large effective population size (N e) estimated to exceed 1000, and no evidence of a genetic bottleneck. Molecular data suggest that the mating system of the Seychelles hawksbills turtles is conducive to maintaining a large N e, with a relatively large and widely distributed male population that promotes considerable gene flow among nesting sites across the Seychelles area (Phillips et al. 2013, 2014).
The striking lack of observed differences between the isolated Chagos and Seychelles rookeries may reflect occasional ongoing gene flow, or a relatively recent colonization event with insufficient time for genetic divergence. The Chagos Archipelago is relatively isolated from other coral reef systems in the Indian Ocean, with the nearest shallow reef system (with nesting beaches) existing in the Seychelles (Sheppard et al. 2012). Ocean currents in this region shift seasonally between flowing easterly and westerly, which has led to the Chagos Archipelago functioning as a stepping stone for marine species in both directions, and to genetic links with the Seychelles (Sheppard et al. 2012). The archipelago is characterized by low-lying coral atolls formed of Holocene reef platforms less than 3000–5000 years old (Eisenhauer et al. 1999). This suggests that nesting beaches in the area are relatively recent, and that turtles may have colonized Chagos beaches only 100–150 generations ago, based on age-to-maturity estimates of around 30 years for hawksbill turtles in the Indo-Pacific (Bell and Pike 2012).
The parallel lack of genetic divergence observed between the Northern Territory and north Queensland rookeries is also interesting because the distance spans a barrier to gene flow that emerged at various times throughout the Pleistocene resulting from the formation of the Torres Strait land bridge (Chivas et al. 2001). In green turtles (Dethmers et al. 2006), flatback turtles (Pittard 2010) and dugong (Blair et al. 2014), a major divergence among clades is associated with this Indo-Pacific barrier (Gaither and Rocha 2013), and across several species there is evidence of vicariance dated to different periods of land bridge formation and variable indications of secondary contact (Mirams et al. 2011). That the hawksbill phylogeography does not show this pattern suggests recent (<8000 years; Chivas et al. 2001) colonization across the Torres Strait.
Elucidating the processes underlying the distribution of genetic variation in marine migratory species is challenging because of the complex mix of factors that influence the species distribution and behavior. These include the dispersal capacity of migratory life stages, changing patterns of oceanic currents, sea level and surface temperatures, the potential for multiple colonization events and secondary contact and, more recently, habitat loss caused by human disturbance. Gaps in our knowledge of how marine species respond to changing environments add to the challenge of effectively protecting endangered migratory species.
MUs and Implications for Conservation
Our study of mitochondrial DNA suggests that the 13 Indo-Pacific hawksbill rookeries studied belong to 8 different genetic stocks that should be considered as independent MUs (sensu Moritz 1994) (Figure 1). Differentiation of the northeast Arnhem Land and north Queensland MUs is based on variation in the peak nesting period and the significant difference in allele frequencies observed in the exact test. Indo-Pacific hawksbill rookeries are genetically very distinct from those in the Atlantic and Caribbean. To date, the only mitochondrial haplotype (Oka24/EAT) observed for these distant ocean basins is hypothesized to have travelled to the eastern Atlantic from the central Indian Ocean around the Cape of Good Hope (Monzón-Argüello et al. 2011). Additional genetic studies of hawksbill turtles nesting and using foraging grounds around southern Africa may improve our understanding of the pattern of long-distance migrations around this region. Such events are hypothesized for green turtles, though in the opposite direction (Bourjea et al. 2007). Regardless of the dynamics of colonization, conservation of genetic diversity in hawksbill turtles requires the conservation of unique populations. Particular regions may warrant additional protection either because of the existence of unique genetic lineages or due to high levels of genetic diversity, as may be further determined using nuclear makers. The Peninsular Malaysia MU is interesting because it has an endemic haplotype (EiIP-53) at a high frequency, suggesting that this isolated rookery will be not replaced on ecological time frames once depleted (Palsbøll et al. 2007).
Identification of MUs and assessment of levels of gene flow among populations is the first step in understanding the population dynamics of hawksbill turtles in the Indo-Pacific. Importantly, because this study has included 13 rookeries scattered throughout the Indo-Pacific, it can provide the baseline for further analyses to determine the origin of animals from feeding grounds using mixed-stock analysis (Vargas et al. 2008; Velez-Zuazo et al. 2008; Dutton et al. 2013).
Understanding the spatial scale, uniqueness, and vulnerability of MUs assists recovery plans by combining relevant anthropogenic pressures to assess risk. Additional studies should be done using nuclear markers such as microsatellites or SNPs (e.g., Roden et al. 2013) to test for concordance (e.g., Dutton et al. 2013), investigate genetic differentiation across various time frames, and determine the value of assignment tests for studies of hawksbill turtles in the Indo-Pacific. Our results also suggest the origins for some haplotypes previously found in studies of foraging turtles in the Indo-Pacific (Okayama et al. 1999) and eastern Atlantic (Monzón-Argüello et al. 2010, 2011). We found that 2 of 3 haplotypes found in nesting and foraging hawksbill turtles in Príncipe (Monzón-Argüello et al. 2011) and 2 unknown haplotypes found in foraging turtles in Cape Verde (Monzón-Argüello et al. 2010) all group with the clade 2 haplotypes observed in the western/central Indian Ocean. Further research to determine the distribution of all clades in the western Indian Ocean and eastern Atlantic is needed to better understand the migratory behavior of hawksbill turtles that leads to gene flow.
International cooperation will be required to manage critically endangered hawksbill turtle populations in the Indo-Pacific. Assessment by the IUCN for the red list of threatened species indicates that all rookeries in the Indo-Pacific are declining, depleted, or lack data to indicate trends, with the exception of possibly stable populations in Oman, Qatar, and Sabah, East Malaysia (since 1985; Mortimer and Donnelly 2008), Arnavon Islands in Solomon Islands (Hamilton et al. 2015), and protected islands in the Seychelles (Mortimer JA, unpublished data). Throughout the region there are only 6 large MUs with estimates of over 1000 females nesting per year, including the Persian Gulf MU, western/central Indian Ocean MU, Indonesia, northeast Arnhem Land MU, north Queensland MU, and Western Australia MU (Mortimer and Donnelly 2008). Oman and Papua New Guinea have relatively large populations (600–800 and ~500–1000 nesting females per year, respectively; Mortimer and Donnelly 2008) that need to be assessed for genetic diversity, along with several smaller rookeries throughout the region.
Two of the identified MUs, the Persian Gulf MU and the western/central Indian Ocean MU have rookeries in multiple countries, and it is expected that most of the MUs will use foraging grounds in other countries. The north Queensland MU is known to use foraging grounds in the Solomon Islands, Papua New Guinea, and Vanuatu, whereas the Solomon Island MU uses foraging grounds in Australia and Papua New Guinea (flipper tag data; Miller et al. 1998; Limpus and Fien 2009). The Sulu Sea MU uses foraging grounds in Indonesia and the Philippines and the western Peninsular Malaysia MU uses foraging grounds in Indonesia and Singapore (reviewed by FitzSimmons and Limpus 2014). It is crucial to reverse the declines in hawksbill turtle populations throughout the Indo-Pacific, and this will be aided by a better understanding of the links between nesting and foraging populations.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/
Funding
This work was supported by CNPq - Conselho Nacional de Desenvolvimento Científico e Tecnológico (Post-Doctoral scholarship number 201968/2014-2 to S.M.V.). Sample collection in Chagos, was supported by the 1996 and 2006 Chagos Expeditions, Fauna & Flora International, a grant from UK Foreign & Commonwealth Office in 1999, and the BIOT Administration on Diego Garcia, US Naval Support Facility. Sample collection in Seychelles was funded by the GEF EMPS-J1: Turtle & Tortoise Conservation Project (1995–1998), the GEF SEYMEMP: Turtle Component (2001–2004), Seychelles Islands Foundation (SIF), Island Conservation Society (ICS), Islands Development Company (IDC), D’Arros Reseach Centre (DRC), North Island, and the Government of Seychelles. Sampling of turtles in Queensland was supported by Queensland Department of Environment and Heritage Protection with partial funding from Japan Bekko Association. The genetic analyses were funded by a grant from the Marine Turtle Conservation Fund of the U.S. Fish & Wildlife Service (Award 98210-6-G073 in 2006). Sample collection in Iran was supported by the Deputy for Natural Environment, Department of the Environment of Iran.
Supplementary Material
Acknowledgments
The authors thank Libby Howitt of Apache Energy for collecting some of the tissue samples from Varanus Island and Anna Vitenbergs for collecting some of the tissue samples from Rosemary Island. The collection of samples from Groote Eylandt occurred through the support of the Anindilyakwa Land Council and the following individuals: Keith Lambert, Gavin Enever, Phillip Mamarika, Simeon Lalara, and Russell Lalara. Eric Roest kindly collected the samples from Platte Island in 1998. Earl Possardt kindly facilitated the grant support from U.S. Fish & Wildlife Service. All samples were collected in accordance with relevant permit agencies and all international shipments were in compliance with CITES requirements. The manuscript was substantially improved by reviewer comments from Brian Bowen.
References
- Abreu-Grobois FA, Horrocks JA, Formia A, LeRoux R, Velez-Zuazo X, Dutton P, Soares S, Meylan P, Browne D. 2006. New mtDNA dloop primers which work for a variety of marine turtle species may increase the resolution capacity of mixed stock analyses. In: Frick M, Panagopoulou A, Rees AF, Williams K, editors. 26th Annual Symposium on Sea Turtle Biology and Conservation: Book of Abstracts; 2006 Apr 2–8, Island of Crete. Athens: International Sea Turtle Society; p. 179. [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Baker CS, Slade RW, Bannister JL, Abernathy RB, Weinrich MT, Lien J, Urban J, Corkeron P, Calmabokidis J, Vasquez O, et al. 1994. Hierarchical structure of mitochondrial DNA gene flow among humpback whale Megaptera novaeangliae, world-wide. Mol Ecol. 3:313–327. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Bass AL. 1999. Genetic analysis to elucidate the natural history and behaviour of hawksbill turtles (Eretmochelys imbricata) in the Wider Caribbean: a review and re-analysis. Chelonian Conserv Biol. 3:195–199. [Google Scholar]
- Bass AL, Good DA, Bjorndal KA, Richardson JI, Hillis ZM, Horrocks JA, Bowen BW. 1996. Testing models of female reproductive migratory behaviour and population structure in the Caribbean hawksbill turtle, Eretmochelys imbricata, with mtDNA sequences. Mol Ecol. 5:321–328. [PubMed] [Google Scholar]
- Bell I, Pike DA. 2012. Somatic growth rates of hawksbill turtles Eretmochelys imbricata in a northern Great Barrier Reef foraging area. Mar Ecol Prog Ser. 446:275–283. [Google Scholar]
- Bjorndal KA, Jackson JB. 2003. Roles of sea turtles in marine ecosystems: reconstructing the past. In: Lutz PL, Musick JA, Wyneken J, editors. The biology of sea turtles, Volume II. Boca Raton (FL): CRC Press; p. 259–274. [Google Scholar]
- Blair D, McMahon A, McDonald B, Tikel D, Waycott M, Marsh H. 2014. Pleistocene sea level fluctuations and the phylogeography of the dugong in Australian waters. Mar Mammal Sci. 30:104–121. [Google Scholar]
- Blumenthal JM, Abreu-Grobois FA, Austin TJ, Broderick AC, Bruford MW, Coyne MS, Ebanks-Petrie G, Formia A, Meylan PA, Meylan AB, et al. 2009. Turtle groups or turtle soup: dispersal patterns of hawksbill turtles in the Caribbean. Mol Ecol. 18:4841–4853. [DOI] [PubMed] [Google Scholar]
- Bourjea J, Lapègue S, Gagnevin L, Broderick D, Mortimer JA, Ciccione S, Roos D, Taquet C, Grizel H. 2007. Phylogeography of the green turtle, Chelonia mydas, in the Southwest Indian Ocean. Mol Ecol. 16:175–186. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Grant WS, Hillis-Starr Z, Shaver DJ, Bjorndal KA, Bolten AB, Bass AL. 2007. Mixed-stock analysis reveals the migrations of juvenile hawksbill turtles (Eretmochelys imbricata) in the Caribbean Sea. Mol Ecol. 16:49–60. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Meylan AB, Ross JP, Limpus CJ, Balazs GH, Avise JC. 1992. Global population structure and natural history of the green turtle (Chelonia mydas) in terms of matriarchal phylogeny. Evolution. 46:865–881. [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Karen LE, Eckert KL. 2006. Turning the tide: exploitation, trade and management of marine turtles in the Lesser Antilles, Central America, Colombia and Venezuela. Cambridge (UK): Traffic International. [Google Scholar]
- Broderick D, Moritz C, Miller J, Guinea M, Prince R, Limpus CJ. 1994. Genetic studies of the hawksbill turtle Eretmochelys imbricata: evidence for multiple stocks in Australian waters. Pac Conserv Biol. 1:123–131. [Google Scholar]
- Browne DC, Horrocks J, Abreu-Grobois F. 2010. Population subdivision in hawksbill turtles nesting on Barbados, West Indies, determined from mitochondrial DNA control region sequences. Conserv Genet. 11:1541–1546. [Google Scholar]
- Cane MA, Molnar P. 2001. Closing of the Indonesian seaway as a precursor to east African aridification around 3–4 million years ago. Nature. 411:157–162. [DOI] [PubMed] [Google Scholar]
- Carr A, Carr MH, Meylan AB. 1978. The ecology and migration of sea turtles, 7. The west Caribbean green turtle colony. Bull Am Mus Nat Hist. 162:1–46. [Google Scholar]
- Castro AL, Stewart BS, Wilson SG, Hueter RE, Meekan MG, Motta PJ, Bowen BW, Karl SA. 2007. Population genetic structure of Earth’s largest fish, the whale shark (Rhincodon typus). Mol Ecol. 16(24):5183–5192. [DOI] [PubMed] [Google Scholar]
- Chan E-H. 2006. Marine turtles in Malaysia: on the verge of extinction? Aquat Ecosyst Health. 9:175–184. [Google Scholar]
- Chatto R, Baker B. 2008. The distribution and status of marine turtle nesting in the Northern Territory - Technical Report 77/2008. Palmerston (NT): Parks and Wildlife Service of the NT. [Google Scholar]
- Cheng IJ, Dutton P, Chen CL, Chen HC, Chen YH, Shea JW. 2008. Comparison of the genetics and nesting ecology of two green turtle rookeries. J Zool. 276:375–384. [Google Scholar]
- Chivas AR, García A, Van Der Kaars S, Couapel MJ, Holt S, Reeves JM, Wheeler DJ, Switzer AD, Murray-Wallace CV, Banerjee D. 2001. Sea-level and environmental changes since the last interglacial in the Gulf of Carpentaria, Australia: an overview. Quat Int. 83:19–46. [Google Scholar]
- Dalebout ML, Robertson KM, Frantzis A, Engelhaupt D, Mignucci-Giannoni AA, Rosario-Delestre RJ, Baker CS. 2005. Worldwide structure of mtDNA diversity among Cuvier’s beaked whales (Ziphius cavirostris): implications for threatened populations. Mol Ecol. 14:3353–3371. [DOI] [PubMed] [Google Scholar]
- Dethmers KE, Broderick D, Moritz C, Fitzsimmons NN, Limpus CJ, Lavery S, Whiting S, Guinea M, Prince RI, Kennett R. 2006. The genetic structure of Australasian green turtles (Chelonia mydas): exploring the geographical scale of genetic exchange. Mol Ecol. 15:3931–3946. [DOI] [PubMed] [Google Scholar]
- Diaz-Fernandez R, Okayama T, Uchiyama T, Carrillo E, Espinosa G, Marquez R, Diez C, Koike H. 1999. Genetic sourcing for the hawksbill turtle, Eretmochelys imbricata, in the northern Caribbean region. Chelonian Conserv Biol. 3:296–300. [Google Scholar]
- Díaz-Jaimes P, Uribe-Alcocer M, Rocha-Olivares A, García-de-León FJ, Nortmoon P, Durand JD. 2010. Global phylogeography of the dolphinfish (Coryphaena hippurus): the influence of large effective population size and recent dispersal on the divergence of a marine pelagic cosmopolitan species. Mol Phylogenet Evol. 57:1209–1218. [DOI] [PubMed] [Google Scholar]
- Dobbs KA, Miller JD, Limpus CJ, Landry AM., Jr 1999. Hawksbill turtle, Eretmochelys imbricata, nesting at Milman Island, Northern Great Barrier Reef, Australia. Chelonian Conserv Biol. 3:344–362. [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J. 2010. Geneious v5.5 [cited 2014 August 20]. Available from: http://www.geneious.com
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 22:1185–1192. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene S, Frey A, Alfaro-Nunez A, Dutton PH, Thomas PGM, Morin PA. 2012. Marine turtle mitogenome phylogenetics and evolution. Mol Phylogenet Evol. 65:241–250. [DOI] [PubMed] [Google Scholar]
- Duncan KM, Martin AP, Bowen BW, DE Couet HG. 2006. Global phylogeography of the scalloped hammerhead shark (Sphyrna lewini). Mol Ecol. 15:2239–2251. [DOI] [PubMed] [Google Scholar]
- Dutton PH, Hitipeuw C, Zein M, Benson SR, Petro G, Pita J, Rei V, Ambio L, Bakarbessy J. 2007. Status and genetic structure of nesting populations of leatherback turtles (Dermochelys coriacea) in the Western Pacific. Chelonian Conserv Biol. 6:47–53. [Google Scholar]
- Dutton PH, Roden SE, Stewart KR, LaCasella E, Tiwari M, Formia A, Thomé JC, Livingstone SR, Eckert S, Chacon-Chaverri D, et al. 2013. Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv Genet. 14:625–636. [Google Scholar]
- Eisenhauer A, Heiss G, Sheppard C, Dullo W. 1999. Reef and island formation and Late Holocene sea-level changes in the Chagos islands. In: Sheppard CRC, Seaward MRD, editors. Ecology of the Chagos Archipelago. Otley, UK: Westbury Publishing. pp. 21–33. [Google Scholar]
- Encalada SE, Lahanas PN, Bjorndal KA, Bolten AB, Miyamoto MM, Bowen BW. 1996. Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: a mitochondrial DNA control region sequence assessment. Mol Ecol. 5:473–483. [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- FitzSimmons NN, Limpus CJ. 2014. Marine turtle genetic stocks of the Indo-Pacific: identifying boundaries and knowledge gaps. Indian Ocean Turtle Newslett. 20:2–18. [Google Scholar]
- FitzSimmons NN, Moritz C, Limpus CJ, Pope L, Prince R. 1997. Geographic structure of mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male-biased gene flow. Genetics. 147:1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, Kaschner K, Schultze SE, Garilao C, Ho SYW, Post K, Higham TF, Stokowska C, van der Es H, Embling CB, et al. 2013. Ancient DNA reveals that bowhead whale lineages survived Late Pleistocene climate change and habitat shifts. Nat Commun. 4:1677. [DOI] [PubMed]
- Gaither MR, Rocha LA. 2013. Origins of species richness in the Indo-Malay-Philippine biodiversity hotspot: evidence for the center of overlap hypothesis. J Biogeogr. 40:1638–1648. [Google Scholar]
- Gopurenko D, Hughes JM. 2002. Regional patterns of genetic structure among Australian populations of the mud crab, Scylla serrata (Crustacea: Decapoda): evidence from mitochondrial DNA. Mar Freshwater Res. 53:849–857. [Google Scholar]
- Grant WS. 2015. Problems and cautions with sequence mismatch analysis and Bayesian skyline plots to infer historical demography. J Hered. 106:333–346. [DOI] [PubMed] [Google Scholar]
- Hall R. 2002. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer based reconstruction, model and animations. J Asian Earth Sci. 20:353–431. [Google Scholar]
- Hamann M, Limpus CJ, Owens DW. 2003. Reproductive cycles of males and females. In: Lutz PL Musick JA and Wyneken J, editors. The biology of sea turtles. Vol. II. Boca Raton (FL): CRC Press; p. 135–161. [Google Scholar]
- Hamilton RJ, Bird T, Gereniu C, Pita J, Ramohia PC, Walter R, Goerlich C, Limpus C. 2015. Solomon Islands largest hawksbill turtle rookery shows signs of recovery after 150 years of excessive exploitation. PLoS One. 10:e0121435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heled J, Drummond AJ. 2008. Bayesian inference of population size history from multiple loci. BMC Evol Biol. 8:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MS. 1998. Spongivory on Caribbean reefs releases corals from competition with sponges. Oecologia. 117:143–150. [DOI] [PubMed] [Google Scholar]
- Ho SY, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. 2011. Time-dependent rates of molecular evolution. Mol Ecol. 20:3087–3101. [DOI] [PubMed] [Google Scholar]
- Ho SY, Phillips MJ, Cooper A, Drummond AJ. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 22:1561–1568. [DOI] [PubMed] [Google Scholar]
- Ho SY, Shapiro B. 2011. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour. 11:423–434. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Matson CW, Amos W, Loughlin TR, Bickham JW. 2006. Deep genetic subdivision within a continuously distributed and highly vagile marine mammal, the Steller’s sea lion (Eumetopias jubatus). Mol Ecol. 15:2821–2832. [DOI] [PubMed] [Google Scholar]
- IUCN 2015. The IUCN red list of threatened species. Version 2015.3 [cited 2015 October 21]. Available from: http://www.iucnredlist.org
- Jensen MP, FitzSimmons NN, Dutton PH. 2013a. Molecular genetics of sea turtles. In: Wyneken J, Lohmann KJ, Musik JA, editors. The biology of sea turtles. Vol. 3. Boca Raton (FL): CRC Press; p. 135–161. [Google Scholar]
- Jensen MP, Limpus CJ, Whiting SD, Guinea M, Prince RTI, Dethmers K, Adnyana IBW, Kennett K, FitzSimmons NN. 2013b. Defining olive ridley turtle management units in Australia and assessing the potential of mortality in ghost nets. Endang Species Res. 21:241–253. [Google Scholar]
- Jia F, Lo N, Ho SY. 2014. The impact of modelling rate heterogeneity among sites on phylogenetic estimates of intraspecific evolutionary rates and timescales. PLoS One. 9:e95722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass RE, Raftery AE. 1995. Bayes factors. J Am Stat Assoc. 90:773–795. [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. 2006. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 6:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney DB, Heist EJ. 2006. Worldwide phylogeography of the blacktip shark (Carcharhinus limbatus) inferred from mitochondrial DNA reveals isolation of western Atlantic populations coupled with recent Pacific dispersal. Mol Ecol. 15:3669–3679. [DOI] [PubMed] [Google Scholar]
- Lahanas PN, Miyamoto MM, Bjorndal KA, Bolten AB. 1994. Molecular evolution and population genetics of Greater Caribbean green turtles (Chelonia mydas) as inferred from mitochondrial DNA control region sequences. Genetica. 94:57–66. [DOI] [PubMed] [Google Scholar]
- Lambeck K. 1996. Shoreline reconstructions for the Persian Gulf since the last glacial maximum. Earth Planet Sci Lett. 142:43–57. [Google Scholar]
- Lara-Ruiz P, Lopez GG, Santos FR, Soares LS. 2006. Extensive hybridization in hawksbill turtles (Eretmochelys imbricata) nesting in Brazil revealed by mtDNA analyses. Conserv Genet. 7:773–781. [Google Scholar]
- Lavery S, Moritz C, Fielder D. 1996. Genetic patterns suggest exponential population growth in a declining species. Mol Biol Evol. 13:1106–1113. [Google Scholar]
- LeRoux RA, Dutton PH, Abreu-Grobois FA, Lagueux CJ, Campbell CL, Delcroix E, Chevalier J, Horrocks JA, Hillis-Starr Z, Troëng S, et al 2012. Re-examination of population structure and phylogeography of hawksbill turtles in the Wider Caribbean using longer mitochondrial DNA sequences. J Hered. 103:806–820. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Limpus CJ, Fien L. 2009. A biological review of Australian marine turtles. 3. Hawksbill turtle, Eretmochelys imbricata (Linnaeus). Brisbane (QLD): Environmental Protection Agency, The State of Queensland. [Google Scholar]
- Limpus CJ, Miller JD, Parmenter CJ, Limpus DJ. 2003. The green turtle, Chelonia mydas, population of Raine Island and the Northern Great Barrier Reef: 1843–2001. Mem Queensl Mus. 49:349–440. [Google Scholar]
- Lukoschek V, Waycott M, Marsh H. 2007. Phylogeography of the olive sea snake, Aipysurus laevis (Hydrophiinae) indicates Pleistocene range expansion around northern Australia but low contemporary gene flow. Mol Ecol. 16:3406–3422. [DOI] [PubMed] [Google Scholar]
- Mendez M, Rosenbaum HC, Subramaniam A, Yackulic C, Bordino P. 2010. Isolation by environmental distance in mobile marine species: molecular ecology of franciscana dolphins at their southern range. Mol Ecol. 19:2212–2228. [DOI] [PubMed] [Google Scholar]
- Miller JD, Dobbs KA, Limpus CJ, Mattocks N, Landry AMJ. 1998. Long-distance migrations by the hawksbill turtle, Eretmochelys imbricata, from north-eastern Australia. Wildlife Res. 25:89–95. [Google Scholar]
- Miller JD, Limpus CJ, Bell I. 2008. The nesting biology of Eretmochelys imbricata in the northern Great Barrier Reef. In: Limpus CJ, Miller JD, editors. Australian hawksbill turtle population dynamics project. Brisbane: Queensland Government Environmental Protection Agency; p. 38–80. [Google Scholar]
- Milliken T, Tokunaga H. 1987. The Japanese sea turtle trade, 1970–1986. Washington (DC): Center for Environmental Education. [Google Scholar]
- Mirams A, Treml E, Shields J, Liggins L, Riginos C. 2011. Vicariance and dispersal across an intermittent barrier: population genetic structure of marine animals across the Torres Strait land bridge. Coral Reefs. 30:937–949. [Google Scholar]
- Molfetti E, Vilaça ST, Georges JY, Plot V, Delcroix E, Le Scao R, Lavergne A, Barrioz S, dos Santos FR, de Thoisy B. 2013. Recent demographic history and present fine-scale structure in the Northwest Atlantic leatherback (Dermochelys coriacea) turtle population. PLoS One. 8:e58061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzón-Argüello C, Loureiro NS, Delgado C, Marco A, Lopes JM, Gomes MG, Abreu-Grobois FA. 2011. Príncipe island hawksbills: genetic isolation of an eastern Atlantic stock. J Exp Mar Biol Ecol. 407:345–354. [Google Scholar]
- Monzón-Argüello C, Rico C, Marco A, López P, López-Jurado LF. 2010. Genetic characterization of eastern Atlantic hawksbill turtles at a foraging group indicates major undiscovered nesting populations in the region. J Exp Mar Biol Ecol. 387:9–14. [Google Scholar]
- Moritz C. 1994. Defining ‘evolutionarily significant units’ for conservation. Trends Ecol Evol. 9:373–375. [DOI] [PubMed] [Google Scholar]
- Mortimer J, Broderick D. 1999. Population genetic structure and developmental migrations of sea turtles in the Chagos Archipelago and adjacent regions inferred from mtDNA sequence variation. In: Sheppard CRC, Seaward MRD, editors. Ecology of the Chagos Archipelago. Otley, UK: Westbury Publishing. p. 186–194. [Google Scholar]
- Mortimer J, Donnelly M. 2008. (IUCN SSC Marine Turtle Specialist Group) Eretmochelys imbricata. The IUCN red list of threatened species 2008: e.T8005A12881238 [cited 2015 October 21]. Available from: http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T8005A12881238.en
- Nishizawa H, Abe O, Okuyama J, Kobayashi M, Arai N. 2011. Population genetic structure and implications for natal philopatry of nesting green turtles Chelonia mydas in the Yaeyama Islands, Japan. Endang Species Res. 14:141–148. [Google Scholar]
- O’Corry‐Crowe G, Suydam R, Rosenberg A, Frost K, Dizon A. 1997. Phylogeography, population structure and dispersal patterns of the beluga whale Delphinapterus leucas in the western Nearctic revealed by mitochondrial DNA. Mol Ecol. 6:955–970. [Google Scholar]
- Okayama T, Díaz-Fernández R, Baba Y, Halim M, Abe O, Azeno N, Koike H. 1999. Genetic diversity of hawksbill turtle in the Indo-Pacific and the Caribbean regions. Chelonian Conserv Biol. 3:362–367. [Google Scholar]
- Palsbøll PJ, Allen J, Bérubé M, Clapham PJ, Feddersen TP, Hammond PS, Hudson RR, Jørgensen H, Katona S, Larsen AH, et al. 1997. Genetic tagging of humpback whales. Nature. 388:767–769. [DOI] [PubMed] [Google Scholar]
- Palsbøll PJ, Bérubé M, Allendorf FW. 2007. Identification of management units using population genetic data. Trends Ecol Evol. 22:11–16. [DOI] [PubMed] [Google Scholar]
- Pellerito R, Arculeo M, Bonhomme F. 2009. Recent expansion of Northeast Atlantic and Mediterranean populations of Melicertus (Penaeus) kerathurus (Crustacea: Decapoda). Fisheries Sci. 75:1089–1095. [Google Scholar]
- Phillips KP, Jorgensen TH, Jolliffe KG, Jolliffe SM, Henwood J, Richardson DS. 2013. Reconstructing paternal genotypes to infer patterns of sperm storage and sexual selection in the hawksbill turtle. Mol Ecol. 22:2301–2312. [DOI] [PubMed] [Google Scholar]
- Phillips KP, Mortimer JA, Jolliffe KG, Jorgensen TH, Richardson DS. 2014. Molecular techniques reveal cryptic life history and demographic processes of a critically endangered marine turtle. J Exper Marine Biol Ecol. 455:29–37. [Google Scholar]
- Pittard S. 2010. Genetic population structure of the flatback turtle (Natator depressus): a nuclear and mitochondrial DNA analysis [Honours Thesis]. [Canberra (NSW)]: University of Canberra; p. 104. [Google Scholar]
- Ramos-Onsins SE, Rozas J. 2002. Statistical properties of new neutrality tests against population growth. Mol Biol Evol. 19:2092–2100. [DOI] [PubMed] [Google Scholar]
- Roden SE, Morin PA, Frey A, Balazs GH, Zarate P, Cheng I-J, Dutton PH. 2013. Green turtle population structure in the Pacific: new insights from single nucleotide polymorphisms and microsatellites. Endang Species Res. 20:227–234. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathiamurthy E, Voris HK. 2006. Maps of Holocene sea level transgression and submerged lakes on the Sunda Shelf. Nat Hist J Chulalongkorn Univ Suppl. 2:1–44. [Google Scholar]
- Schultz JK, Feldheim KA, Gruber SH, Ashley MV, McGovern TM, Bowen BW. 2008. Global phylogeography and seascape genetics of the lemon sharks (genus Negaprion). Mol Ecol. 17:5336–5348. [DOI] [PubMed] [Google Scholar]
- Sheppard CR, Ateweberhan M, Bowen BW, Carr P, Chen CA, Clubbe C, Craig MT, Ebinghaus R, Eble J, Fitzsimmons N, et al. 2012. Reefs and islands of the Chagos Archipelago, Indian Ocean: why it is the world’s largest no-take marine protected area. Aquat Conserv. 22:232–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Sinha D. 1998. Early Pliocene closing of the Indonesian Seaway: evidence from north-east Indian Ocean and Tropical Pacific deep sea cores. J Asian Earth Sci. 16:29–44. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatis C, Triantafyllidis A, Moutou KA, Mamuris Z. 2004. Mitochondrial DNA variation in Northeast Atlantic and Mediterranean populations of Norway lobster, Nephrops norvegicus . Mol Ecol. 13:1377–1390. [DOI] [PubMed] [Google Scholar]
- Tabib M, Frootan F, Hesni MA. 2014. Genetic diversity and phylogeography of hawksbill turtle in the Persian Gulf. J Biodivers Environ Sci. 4:51–57. [Google Scholar]
- Tabib M, Zolgharnein H, Mohammadi M, Salari-Aliabadi MA, Qasemi A, Roshani S, Rajabi-Maham H, Frootan F. 2011. mtDNA variation of the critically endangered hawksbill turtle (Eretmochelys imbricata) nesting on Iranian islands of the Persian Gulf. Genet Mol Res. 10:1499–1503. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Arias N, Amorocho DF, López-Álvarez D, Mejía-Ladino LM. 2014. Relaciones filogeograficas de algunas colônias de alimentacion y anidacion de la tortuga carey (Eretmochelys imbricata) en el Pacifico y Caribe Colombianos. Bol Invest Mar Cost. 43:159–182. [Google Scholar]
- Vargas SM, Araújo FC, Monteiro DS, Estima SC, Almeida AP, Soares LS, Santos FR. 2008. Genetic diversity and origin of leatherback turtles (Dermochelys coriacea) from the Brazilian coast. J Hered. 99:215–220. [DOI] [PubMed] [Google Scholar]
- Velez-Zuazo X, Ramos WD, van Dam RP, Diez CE, Abreu-Grobois A, McMillan WO. 2008. Dispersal, recruitment and migratory behaviour in a hawksbill sea turtle aggregation. Mol Ecol. 17:839–853. [DOI] [PubMed] [Google Scholar]
- Witzell W. 1983. Synopsis of biological data on the hawksbill turtle, Eretmochelys imbricata (Linnaeus, 1766). Miami (FL): Food and Agriculture Organization of the Nations. [Google Scholar]
- Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. 2011. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol. 60:150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1993. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol. 10:1396–1401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In agreement with the data archiving rule (Baker 2013), all sequences generated in this study have been deposited in GenBank (accession numbers KT934050–KT934101—see Supplementary Table S1 online).