Abstract
The renewable source of highly reduced carbon provided by plant triacylglycerols (TAGs) fills an ever increasing demand for food, biodiesel, and industrial chemicals. Each of these uses requires different compositions of fatty acid proportions in seed oils. Identifying the genes responsible for variation in seed oil composition in nature provides targets for bioengineering fatty acid proportions optimized for various industrial and nutrition goals. Here, we characterized the seed oil composition of 391 world-wide, wild accessions of Arabidopsis thaliana, and performed a genome-wide association study (GWAS) of the 9 major fatty acids in the seed oil and 4 composite measures of the fatty acids. Four to 19 regions of interest were associated with the seed oil composition traits. Thirty-four of the genes in these regions are involved in lipid metabolism or transport, with 14 specific to fatty acid synthesis or breakdown. Eight of the genes encode transcription factors. We have identified genes significantly associated with variation in fatty acid proportions that can be used as a resource across the Brassicaceae. Two-thirds of the regions identified contain candidate genes that have never been implicated in lipid metabolism and represent potential new targets for bioengineering.
Key words: Arabidopsis thaliana, fatty acids, genome-wide association mapping, lipid metabolism, seed oil composition, triacylglycerol
Introduction
The triacylglycerols (TAGs) found in oilseeds provide a renewable source of highly reduced carbon for human consumption, industrial petrochemical replacement, and biodiesel (Thelen and Ohlrogge 2002; Vanhercke et al. 2013; and refs therein). Currently, 160 million metric tonnes of plant oils are being produced globally, with 80% for food stuffs and 20% for industrial uses (Mielke 2012). With a growing population, dwindling petroleum supplies, and the low carbon dioxide impact of biofuels, demand for plant oils will only increase. Many uses of seed oils require different optimal fatty acid compositions—the relative proportions of the fatty acids found in the seed oils. For example, for biodiesel, an ideal balance of high oxidative stability, and improved cold tolerance results from high proportions of monounsaturated fatty acids, especially 18:1 (Durrett et al. 2008). In canola, reduced levels of erucic acid and linolenic acid are desirable for human consumption (Yang et al. 2012). To manipulate fatty acid composition efficiently for differing purposes, we must understand the interplay between the biosynthesis of fatty acids, their incorporation into TAGs, and the regulatory networks that control these processes.
While many of the enzymes involved in the synthesis and modification of fatty acids and their subsequent incorporation into TAGs have been well studied and are conserved between most of the major oilseed species (e.g., Brassica napus, A. thaliana, castor bean and soybean) (Sharma and Chauhan 2012), comparatively little is known about the structural and regulatory differences in these genes that produce such extensive variation in seed oil composition. The processes involved in fatty acid synthesis and incorporation into TAGs have been shown to be far more complex than a simple linear biochemical pathway (Thelen and Ohlrogge 2002; Chapman and Ohlrogge 2012). Due to the identification of homologs of the lipid genes between many of the oilseed species, a more detailed understanding of how seed oil composition variation is produced within a single species in nature is likely to be useful for disentangling this process in other species.
Due to the availability of many tools for A. thaliana, it has become the reference system for the study of lipid metabolism. The value of studying lipid metabolism in A. thaliana is of additional interest because its fatty acid biosynthesis and regulation are similar to that found in B. napus (Yang et al. 2012).
Recent work on seed oil synthesis and accumulation in A. thaliana has concentrated on elucidating the regulation of this process. The availability of seed-specific genome-wide expression data sets has led to several publications devoted to identifying coexpression networks involved in seed filling (Wang et al. 2007; Peng and Weselake 2011). To date, only a few transcription factors that specifically control seed storage accumulation have been identified and validated (Baud et al. 2007; Wang et al. 2007; Mu et al. 2008).
Three quantitative trait loci (QTL) mapping studies have attempted to uncover the genetic basis of variation in seed oil composition in A. thaliana (Hobbs et al. 2004; O’Neill et al. 2012; Sanyal and Linder 2012). Sixty-one genomic regions were identified, each being a large region covering dozens to hundreds of genes. In addition, while QTL mapping provides more power than many other mapping methods to detect true associations, it is limited to variation segregating between the parents. Among the QTL studies to date, only 10 pairs of accessions have been examined. Therefore, genes important for determining seed oil composition could have been missed.
These shortcomings of QTL mapping are largely overcome in GWAS. Because GWAS uses large numbers of natural accessions, more of the genetic variation found in nature is represented and a much larger set of historical recombination events are reflected, allowing a larger number of genomic regions associated with the trait of interest to be identified. In addition, when performed with large numbers of markers, associations can be resolved to the genetic level in most instances. GWAS is now possible in species with a high degree of population structure, like A. thaliana, due to new statistical tests that control for relatedness (Kang et al. 2008). On the other hand, GWAS suffers from a high number of false positives due to the large numbers of markers tested for association with trait variation.
Our goal was to identify the genes responsible for variation in seed oil composition in A. thaliana. Using 391 accessions from across the geographic distribution of A. thaliana, we performed a GWAS on single nucleotide polymorphisms (SNPs) for the relative proportion of each of the 9 major seed oil fatty acids and 4 biologically relevant composite traits. Our results could be used to develop strategies for modifying seed oil composition for consumption or industrial purposes.
Materials and Methods
Accessions and Seed Production
Three hundred ninety-one accessions (Supplementary Table S1) from across the geographic range of A. thaliana were chosen to capture as much of the phenotypic variation in seed oil composition as possible. Seeds were obtained from the Arabidopsis Biological Resource Center (ABRC) and the Juenger lab at the University of Texas at Austin.
To minimize environmental maternal effects, we produced seeds under common garden conditions. Surface-sterilized seeds were soaked and stratified for 6 days at 4 °C before planting to break dormancy. Following stratification, plants were grown from November 2010 to March 2011 in a glasshouse with 16h of supplemental lighting from high pressure sodium, high-intensity discharge lights. Glasshouse temperatures were recorded every 4min with an automatic data logger. Temperature ranged from a minimum of 6.1 °C at night to a maximum of 28.9 °C during the day with a mean of 18.1 °C across all data points. Three replicates of each accession were planted in separate pots in a completely randomized design for a total of 1341 pots in 77 trays. An average of 5 seeds were planted in each pot and randomly thinned to 1 plant after germination and establishment. To decrease environmental variation, especially due to edge effects, pots were placed in every other tray slot such that every pot was equidistant. Twice per week the trays were subirrigated, fertilized with Dyna-Gro 7-9-5 plant food (Dyna-Gro Nutrition Solutions, Richmond, CA), and tray locations within the growing area were randomized. Once a plant bolted, it was isolated using the Arasystem (Betatech, Gent, BE). Seeds were harvested from each plant after the siliques matured and browned. Seeds were stored in coin envelopes at room temperature for 2–9 months before seed oil compositions were determined.
Seed Oil Composition Analysis
Fatty acid compositions of the accessions were determined using gas chromatography of fatty acid methyl esters (FAMEs). For each of the 3 replicate plants per accession, 2 extractions were performed and averaged on a per plant basis. Extraction procedures were modified from (Zheljazkov et al. 2008): about thirty randomly chosen seeds per plant were crushed with a glass pestle in a 2-mL glass autosampler vial and mixed with 150 µL of extraction buffer (75% hexane, 20% chloroform, and 5% 0.5M sodium methoxide in methanol). A minimum of 5min elapsed before the first sample was injected into the chromatograph. Samples were analyzed using an HP 5890A gas chromatograph with a robotic autoloader and a DB-23 capillary column (Agilent Technologies, Santa Clara, CA). An initial oven temperature of 180 °C was maintained for 5.5min and then raised 15 °C per min to a final temperature of 240 °C for 0.5min for a total run time of 10min. Two injections were performed for every tenth sample to assess the repeatability of the results. In only 5 double injections did the measured fatty acid proportions differ by more than 1%, with a maximum difference of 1.75%. Because of this high precision, only one of each of the double injections was chosen at random to be used for the analyses. FAME peaks were visualized using a flame ionization detector and were identified by comparison to FAME standards 189-4, 189-5, 189-12, and 189-19 (Supelco, Bellefonte, PA). The standards were run before each set of seed oil extractions to calibrate retention times. Peaks were integrated using Agilent Chemstation software revision A.04.02 (Agilent Technologies, Inc., Santa Clara, CA).
A total of 13 fatty acid traits were generated for each sample: the relative proportions of the 9 principle fatty acids in A. thaliana seed oil (16:0, 18:0, 18:1, 18:2, 18:3, 20:0, 20:1, 20:2, and 22:1; where the first number corresponds to the number of carbon atoms in the chain and the second, the number of double bonds), the sum of the proportions of the 3 fatty acids produced in the plastid (Plastid) (16:0, 18:0, and 18:1), the total proportion of saturated fatty acids (Sat) (16:0, 18:0, and 20:0), the total proportion of polyunsaturated fatty acids (PUFA) (18:2, 18:3, and 20:2), and the total proportion of very long chain fatty acids (VLCFA) (20:0, 20:1, 20:2, and 22:1). The 4 composite traits were chosen for their potential importance in oil quality or genetic control of oil quality. Plastid fatty acids may be subject to maternal effects due to their location of synthesis. The other 3 traits (Sat, PUFA, VLCFA) are of interest for industrial and food applications (Thelen and Ohlrogge 2002; Vanhercke et al. 2013).
The broad-sense heritability of each untransformed trait was determined using Proc mixed in SAS™ software (SAS Institute, Inc., Cary, NC). The class variable was accession and was considered a random variable. The calculations were also performed with tray as a random variable in the model and with box-cox transformed data.
Because the fatty acid biosynthetic pathway involves a complex network of reactions in which some of the fatty acids are precursors for the production of others, it is important to consider the correlations between their proportions in the seed oil. Pairwise Pearson correlations between the 9 fatty acids were performed with the cor.test function in R (R Core Team 2014).
Association Mapping
About 214 051 SNPs for A. thaliana were downloaded from http://walnut.usc.edu/2010/SNPs (Horton et al. 2012) (v3.06). Of these, we removed 13,748 minor alleles having frequencies of 5% or less (Kang et al. 2008; Atwell et al. 2010).
Three hundred ninety-one accessions were used to detect associations between SNP genotypes and variation in each of the 13 traits. Since using multiple measurements per accession can increase the power to detect associations (Kang et al. 2008), all 3 biological replicates were used in the GWAS analyses. We used the EMMA.reml.t function of the R implementation efficient mixed-model association (EMMA) v. 1.1.2 for Linux. EMMA identifies associations between SNPs and trait variation while controlling for genetic relatedness using a pairwise genetic similarity matrix (Kang et al. 2008). An identity matrix was included to identify biological replicates. The SNP genotype is modeled as a fixed effect and the genetic similarity matrix as a random effect. To meet the assumptions of EMMA, the traits were first subjected to box-cox transformations (Box and Cox 1964) to approximate normal distributions.
Because association mapping using such a large dataset is computationally expensive and EMMA models each SNP independently, the dataset was divided into 1020 sets of 210 SNPs each and run in parallel on the Lonestar cluster of the Texas Advanced Computing Center at the University of Texas at Austin.
Identification of A Posteriori Candidate Genes
Analysis of the EMMA results was limited to the 100 SNPs of lowest P value for each trait. Because linkage disequilibrium in A. thaliana decays within 10kb on average (Kim et al. 2007), all genes located between 10kb upstream and downstream of each top 100 SNP were considered associated with that SNP to generate a list of possible a posteriori candidate genes. A scoring system modified from Verslues et al. (2014) was used to rank the gene lists taking account of both the number of top 100 SNPs associated with each gene and the P value of the SNPs. Each gene was given 10 points for association with each top 10 SNP, 5 points for a top 20 SNP, and 1 point for a top 100 SNP. These values were chosen arbitrarily in an attempt to prioritize genes for future validation efforts. Strings of adjacent genes associated with traits are likely due to linkage disequilibrium, therefore regions of interest were defined rather than examining single genes. Genes with at least a score of 3 formed the starting point of a region, which then extended to all adjacent genes with a minimum score of 1. Gene functions and GO terms were obtained from The Arabidopsis Information Resource (TAIR; www.arabidopsis.org), The Arabidopsis Book (Li-Beisson et al. 2013) and http://aralip.plantbiology.msu.edu/.
Data Archiving
In accordance with data archiving guidelines (Baker 2013), accession collection information, significant SNPs and the genes linked to them are available as online Supporting Information. The following files were submitted to Dryad: Supplementary tables 1–3 and 2 csv files: 1. Fatty acid proportions for 391 Arabidopsis thaliana accessions. 2. Box-Cox transformed fatty acid proportions for 391 Arabidopsis thaliana accessions.
Results
Natural Variation in Fatty Acid Compositions
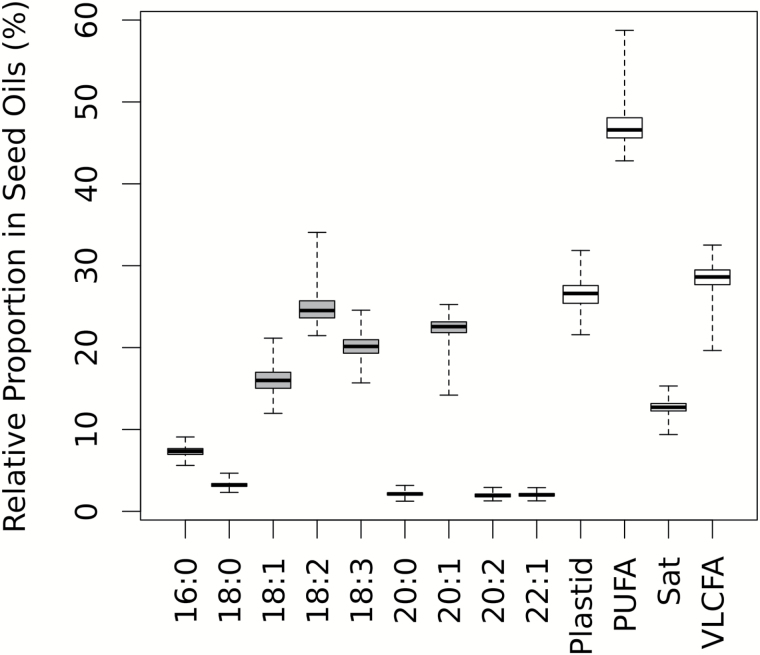
Among the 391 accessions, the 9 major fatty acids found in A. thaliana seed oil displayed substantial variation in their relative proportions (Figure 1). Four of the unsaturated fatty acids (18:1, 18:2, 18:3, and 20:1) comprise an average of 83.4% of the seed oil, while the remaining 5 fatty acids are each less than 8%. 18:2 was the most abundant fatty acid in every accession (24.8% ± 1.7%, mean ± SD), and of the saturated fatty acids, 16:0 had the highest mean proportion (7.3% ± 0.55). In general, the fatty acids with higher mean proportions also had higher variances (Table 1). Coefficients of variation (CV) were calculated to see if the amount of variation in fatty acid proportions generally scaled with the means. In general, the least abundant fatty acids had the highest CV values and the most abundant had lower CV values indicating higher relative variation in the less abundant fatty acids. The highest CVs were found for 20:2 and 22:1 (CV = 14.2% and 12.0%, respectively) and the lowest for 18:2 (CV = 6.9%) and 18:3 (CV = 6.3%).
Figure 1.
Relative proportions of the 9 major fatty acids and 4 composite traits in Arabidopsis thaliana seeds using accession averages (N = 391).
Table 1.
Summary statistics of variation and heritability for fatty acid percentages in 391 accessions of Arabidopsis thaliana.
| Trait | Mean | H 2a | Variance | SDb | CV (%)c |
|---|---|---|---|---|---|
| 16:0 | 7.31 | 0.880 | 0.302 | 0.550 | 7.52 |
| 18:0 | 3.22 | 0.871 | 0.0836 | 0.289 | 8.98 |
| 18:1 | 16.0 | 0.841 | 2.25 | 1.50 | 9.36 |
| 18:2 | 24.8 | 0.940 | 2.89 | 1.70 | 6.85 |
| 18:3 | 20.1 | 0.809 | 1.60 | 1.27 | 6.28 |
| 20:0 | 2.12 | 0.829 | 0.0479 | 0.219 | 10.3 |
| 20:1 | 22.4 | 0.946 | 1.75 | 1.32 | 5.91 |
| 20:2 | 1.96 | 0.886 | 0.0772 | 0.278 | 14.2 |
| 22:1 | 2.02 | 0.846 | 0.0586 | 0.242 | 12.0 |
| Plastid | 26.6 | 0.849 | 2.63 | 1.62 | 6.10 |
| PUFA | 46.9 | 0.931 | 3.59 | 1.89 | 4.04 |
| Sat | 12.7 | 0.892 | 0.658 | 0.811 | 6.41 |
| VLCFA | 28.5 | 0.931 | 2.77 | 1.66 | 5.84 |
aBroad-sense heritability.
bstandard deviation.
ccoefficient of variation.
Heritabilities
The broad-sense heritability of each untransformed trait was high, varying from 0.81 to 0.95 (Table 1), with the heritabilities of 2 of the commercially valuable composite traits, VLCFA and PUFA particularly high at 0.93. Models were also run with tray as a random variable and using the box-cox transformed data. These modifications changed the heritabilities by no more than ±0.02 from the values reported.
Correlations Between Fatty Acids
Thirty of the 36 fatty acid pairs were significantly correlated (P < 0.05) and 28 of the 30 were highly correlated (P < 0.01) (Table 2). Approximately half of the relationships between pairs of significantly correlated fatty acids were positive (13/30) and ranged from 0.127 to 0.562 with an average value of 0.334. The strongest positive correlation was between 20:1 and 22:1. This was a surprising result as a negative correlation was expected because 20:1 is the substrate for the production of 22:1. All saturated fatty acid pairs were positively correlated as were all but 1 pair (20:1/20:2) of the VLCFA. The significant negative correlations were slightly stronger on average (−0.364, range: −0.113 to −0.766) than the significant positive correlations.
Table 2.
Pearson correlations (r) between fatty acid pairs using the accession averages (N = 391)
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 20:2 | |
|---|---|---|---|---|---|---|---|---|
| 18:0 | 0.341*** | |||||||
| 18:1 | −0.163** | 0.173*** | ||||||
| 18:2 | −0.366*** | −0.276*** | −0.023 | |||||
| 18:3 | −0.031 | −0.174*** | −0.439*** | −0.335*** | ||||
| 20:0 | 0.217*** | 0.502*** | −0.382*** | −0.392*** | −0.026 | |||
| 20:1 | 0.219*** | 0.025 | −0.318*** | −0.693*** | −0.047 | 0.484*** | ||
| 20:2 | −0.193*** | −0.373*** | −0.766*** | 0.346*** | 0.193*** | 0.127* | 0.074 | |
| 22:1 | −0.113* | −0.358*** | −0.619*** | −0.222*** | 0.187*** | 0.468*** | 0.562*** | 0.529*** |
*P < 0.05, **P < 0.01, ***P < 0.001, significant pairs in bold.
Of the 8 fatty acid pairs with direct substrate/product relationships (expected negative correlations), 2 pairs (18:1/20:1 and 18:2/18:3) were negatively correlated and 5 were positive (16:0/18:0, 18:0/18:1, 18:0/20:0, 18:2/20:2, and 20:1/22:1). One was non-significant (18:1/18:2). Interestingly, the strongest correlation (r = −0.766) was between the proportions of 18:1 and 20:2, which are not directly connected in the biochemical pathway, whereas 18:1 and its immediate product, 18:2, were not significantly correlated. In addition, the substrate-product pair, 18:2/20:2, was positively correlated (r = 0.375).
Regions of Interest Identified by GWAS
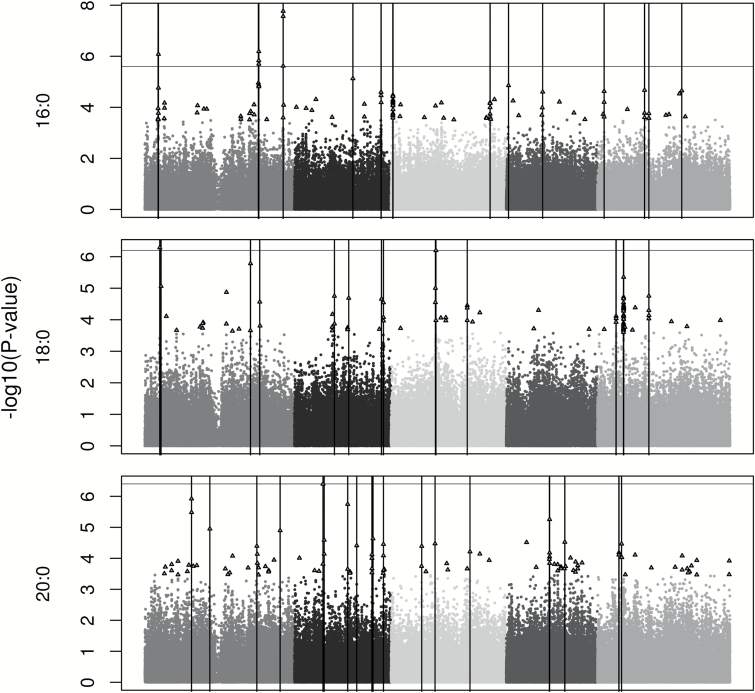
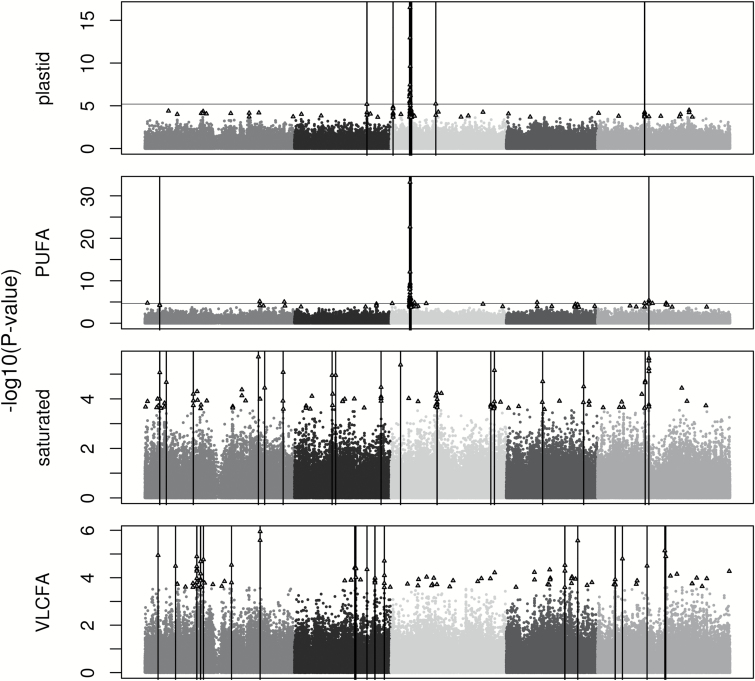
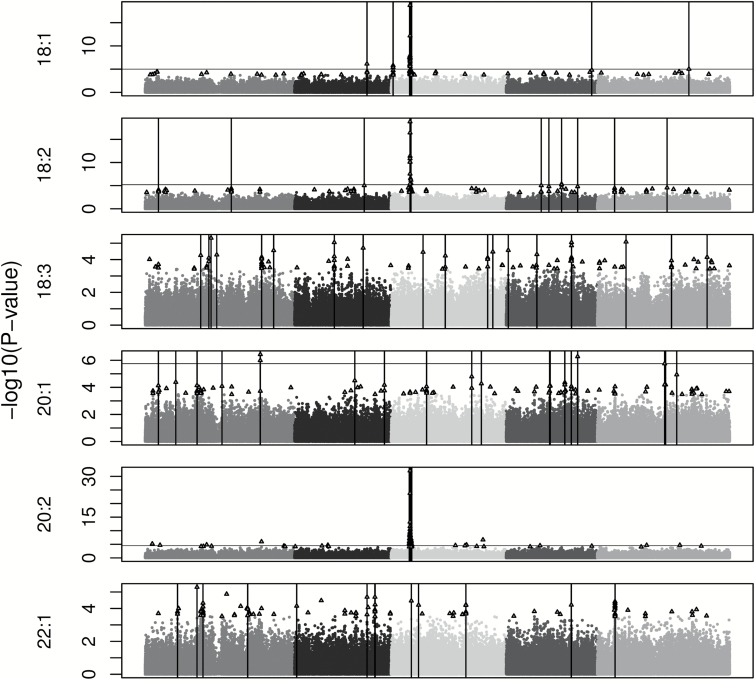
SNP-trait associations were assessed with the mixed model EMMA while controlling for population structure. Across the 13 traits, P values of the top 100 SNPs ranged from 5.8×10−34 to 3.8×10−4 (Supplementary Table S2). The top 100 SNPs were associated with 4 to 19 clusters of genes per trait (Figures 2–4), with the clusters encompassing 9.8 genes on average (range = 1–66 genes; Supplementary Table S3). The extent of each cluster of interest (0.47–209.8kb) was likely driven by linkage disequilibrium with only 1 or 2 causal genes.
Figure 2.
Genome-wide association mapping results for the 3 saturated fatty acids in Arabidopsis thaliana using the efficient mixed model association package in R. The 5 chromosomes are represented by different shades of gray with the x axis as physical position. Triangles depict the top 100 SNPs for each trait. Vertical bars represent the physical positions of the regions of interest. A genome-wide significance level of FDR < 0.1 is indicated by a horizontal line.
Figure 4.
Genome-wide association mapping results for the 4 composite traits in Arabidopsis thaliana using the efficient mixed model association package in R. The 5 chromosomes are represented by different shades of gray with the x axis as physical position. Triangles depict the top 100 SNPs for each trait. Vertical bars represent the physical positions of the regions of interest. A genome-wide significance level of FDR < 0.1 is indicated by a horizontal line. Traits lacking a horizontal line do not have significant SNPs at FDR < 0.1.
Genes by Category
Approximately one-third (52 of 166) of the regions of interest contained lipid and/or transcription factor genes. Ten regions had both lipid and transcription factor genes, while 38 regions had only lipid genes and 4 only a transcription factor gene. The remaining 114 regions did not contain genes with an obvious function in lipid metabolism or regulation within 10kb (Supplementary Table S3).
Lipid-Related Enzymes
While 34 a posteriori candidates (Supplementary Table S3) encode products with functions known to be involved in lipid synthesis, degradation, or transport, the most promising are those directly involved with fatty acids or TAGs (14 genes). The most striking signal for these genes, and for the entire genome, was a cluster of 15 highly significant SNPs (FDR < 0.0001) found on chromosome 3. Five of the 15 were found within FAD2, 9 were within 10kb, and all 15 were found within 103kb. They were strongly associated with 5 traits (18:1, 18:2, 20:2, PUFA, and Plastid) (Figures 3 and 4). FAD2 encodes a fatty acid desaturase, which catalyzes the desaturation of 18:1 to 18:2.
Figure 3.
Genome-wide association mapping results for the 6 unsaturated fatty acids in Arabidopsis thaliana using the efficient mixed model association package in R. The 5 chromosomes are represented by different shades of gray with the x axis as physical position. Triangles depict the top 100 SNPs for each trait. Vertical bars represent the physical positions of the regions of interest. A genome-wide significance level of FDR < 0.1 is indicated by a horizontal line. Traits lacking a horizontal line do not have significant SNPs at FDR < 0.1.
Remaining Genes
Eight of the a posteriori candidate genes encode transcription factors, half of which are MYB types (Supplementary Table S3). None of these genes have been previously implicated in the regulation of lipid metabolism. All of the transcription factor genes were associated with multiple traits. One hundred fourteen regions did not contain a posteriori candidate genes with a clear relationship to lipid synthesis, transport or regulation.
Discussion
The high heritabilities of the fatty acid traits (H 2 > 0.8 in all cases) and the extensive natural variation in the fatty acid proportions suggest selective breeding of many oil compositions would be possible in A. thaliana. Nonetheless, there will be limitations on traditional breeding imposed by trait means and variances and possibly by the highly significant correlations between fatty acid pairs. On the other hand, in some cases, these correlations may facilitate breeding of desired compositions. For example, the saturated fatty acids are positively correlated with one another, and it is often desirable to decrease total saturated fatty acids as a unit. The unexpected patterns of correlations we found between fatty acid pairs, such as positive or non-significant correlations between substrates and their products, support the idea of non-linearity of TAG production and the complexity of acyl fluxes (Thelen and Ohlrogge 2002; Vanhercke et al. 2013). While A. thaliana is not a target for industrial or nutritional oil production, if the high heritabilities and variances found within the species are representative of closely related commercial oil species, such as B. napus then selective breeding may be possible for some desired compositions based upon information from Arabidopsis. The least abundant fatty acids (18:0, 20:0, 20:2, and 22:1), while not capable of being bred to high proportions, can be manipulated to a greater relative extent than indicated by their low means, as indicated by their above average CV percentages.
Because the genes involved in fatty acid synthesis and TAG accumulation are conserved across many plant species, the genes associated with seed oil composition in A. thaliana provide likely candidates for QTL mapping and genetic engineering in non-model oilseed species (Sharma and Chauhan 2012). For example, the functions of FAD2 and FAD3, discovered and validated in A. thaliana, were used to identify the gene copies of FAD2 and FAD3 in B. napus responsible for variation in the proportions of 18:1 and 18:3 (Yang et al. 2012). This strategy could be applied to genes from our study to manipulate fatty acid proportions in other oilseed species. Recent advances in artificial microRNA technologies for seed-specific post-transcriptional gene silencing have been demonstrated to be highly effective in both model organisms and crop species (Sablok et al. 2011; Belide et al. 2012).
Lipid Metabolism Genes
While the 34 lipid metabolism genes associated with seed oil composition variation in this study represent good targets for industrial manipulation of fatty acid proportions, 14 of these genes encode products with functions directly related to fatty acid synthesis and metabolism. Six of the 14 known fatty acid genes were previously associated with the same traits identified here in earlier QTL mapping studies (Hobbs et al. 2004; O’Neill et al. 2012; Sanyal and Linder 2012). The associations identified in our study are in accordance with known effects on specific fatty acid proportions through previous molecular work for 6 of our fatty acid specific genes (FAD2, DES1, DES3, KASII, FATA1, FATB; Sussman et al. 2000; Buhr et al. 2002; Salas and Ohlrogge 2002; Bonaventure et al. 2003; Kachroo et al. 2007; Pidkowich et al. 2007; Belide et al. 2012; Head et al. 2012; Moreno-Pérez et al. 2012). The remaining 8 fatty acid synthesis and metabolism genes (KASIII, KAR, DGAT1, LPAAT5, LPCAT2, SDRB, beta-PDH, and ACP5) are known to contribute to fatty acid production or breakdown but have not been shown to modify fatty acid proportions and, therefore, represent new putative targets for engineering specific oil compositions. A description of these 14 genes, their associations, previous work, and suggestions for engineering are discussed below.
Genes Previously Engineered to Modify Seed Oil Composition
The most highly significant SNPs linked to FAD2 were 9–26 orders of magnitude more significant than any other SNPs and encompassed 5 traits (18:1, 18:2, 20:2, Plastid, and PUFA). Previous QTL mapping studies found QTL overlapping with this region for 18:1 (Hobbs et al. 2004; Sanyal and Linder 2012), 18:2 and 20:2 (O’Neill et al. 2012; Sanyal and Linder 2012), and Plastid (Sanyal and Linder 2012). FAD2 encodes the enzyme that desaturates 18:1 to 18:2, which are the substrates for multiple downstream products (18:1 for 20:1, and 18:2 for 18:3 and 20:2). Molecular studies have confirmed the associations we found for FAD2 in A. thaliana (Belide et al. 2012) and showed the same function for FAD2 in soybean (Buhr et al. 2002). With such a pivotal role in the fatty acid biosynthetic pathway, FAD2 is an excellent target for manipulation of oil composition and has been used to bioengineer plants with modified compositions (Buhr et al. 2002; Belide et al. 2012). In A. thaliana and soybean, silencing of FAD2 causes an increase in oleic acid proportions and a decrease in PUFA.
KASII, which catalyzes the elongation of 16:0 to 18:0, was significantly associated with variation in the proportions of 16:0 and total saturated fatty acids. Sanyal and Linder (2012) also identified a QTL for 16:0 in this region. RNAi silencing of KASII in A. thaliana (Pidkowich et al. 2007) and a soybean KASII mutant (Head et al. 2012) have both shown an increase in the proportion of 16:0 in seeds. Two of the 7 isoforms (Kachroo et al. 2007) of an additional enzyme, S-ACP-DES (DES1 and DES3), which desaturates 18:0 to 18:1 were associated with variation in 18:0 here and in a previous QTL mapping study (Sanyal and Linder 2012). Since oil high in 18:1 and low in saturated fatty acids is a desirable composition for biodiesel production (Durrett et al. 2008), engineering of this composition may be possible through up-regulation of KASII in combination with up-regulation of S-ACP-DES.
FATA1 encodes a thioesterase responsible for export of fatty acids from the plastid and was associated with variation in 18:0. The substrate preferences of 2 of the 3 thioesterases, FATA1 and FATB, have been characterized (Salas and Ohlrogge 2002). Both FATA1 and FATB can hydrolyze 18:0 acyl-ACPs for export from the plastid, but they do not have their highest activity with 18:0. Nonetheless, engineering low levels of 18:0 in TAGs by silencing FATA1 may not be successful because reduced FATA activity has drastic effects on seed storage accumulation. Reduced thioesterase activity in a double FATA T-DNA mutant caused significant alterations in the proportions of most of the fatty acids in the seeds as well as reduced seed oil accumulation, which the authors suggest was due to a build-up of acyl-ACPs in the plastid causing a negative feedback loop that shut down fatty acid production (Moreno-Pérez et al. 2012).
FATB, an acyl-acyl carrier protein thioesterase which hydrolyzes acyl-ACP to produce free fatty acids and was associated with 16:0, 18:2, and 20:1. FATB was also associated with 16:0 in an earlier QTL mapping study (O’Neill et al. 2012). FATB has a substrate preference for 16:0-ACP (Salas and Ohlrogge 2002) but will also hydrolyze 18:1-ACP (24% less activity) and 18:0-ACP (64% less activity). Because of this substrate preference for 16:0, down-regulation of FATB or protein variants of FATB could decrease the amount of 16:0 exported to the ER to be incorporated into seed oils. As expected, a T-DNA insertion mutation in FATB (CS6525) (Sussman et al. 2000) produced a 56% reduction of 16:0 in seeds as compared to wild-type (Bonaventure et al. 2003). Additionally, the insertion line had decreased 18:0 (31%), 18:1 (8%), and 18:3 (13%) and increased 18:2 (20%) and VLCFA (12%) (Bonaventure et al. 2003).
Lipid Genes Newly Implicated in Seed Oil Composition Variation
Three genes encoding enzymes involved in the initial steps of fatty acid synthesis were found in regions associated with fatty acid proportions. KASIII and KAR were linked to SNPs associated with variation in 16:0, and encode essential enzymes of the fatty acid synthesis (FAS) complex. In a 2011 QTL mapping study, O’Neill et al. identified a QTL for 16:0 that colocated with KASIII. The beta-subunit of the plastid pyruvate dehydrogenase complex, β-PDH was associated with proportions of 20:1 and VLCFA. While manipulation of these enzymes may alter total seed oil content, it is unclear from current knowledge of their functions and positions in the pathway how they might change oil composition.
Extensive research has been conducted on modifying acyltransferases through biotechnology to create designer seed oil compositions (reviewed in Snyder et al. 2009). Two of the 3 Kennedy pathway acyltransferases were associated with seed oil composition variation. LPAAT was associated with 18:3 and catalyzes the second acylation of the glycerol backbone but its substrate specificity is unknown in A. thaliana. The final acylation of the glycerol backbone to form a TAG is catalyzed by DGAT1 (associated with Sat). Although AtDGAT1 displays substrate specificity for 18:3, mutant lines for this gene have altered proportions of most fatty acids in Arabidopsis seeds (Katavic et al. 1995, Routaboul et al. 1999). B. napus DGAT1 enzymes have to been shown to have a 4–7-fold specificity for 16:0 over 18:1 but are sensitive to available relative concentrations of substrates (Aznar-Moreno et al. 2015). In addition to the Kennedy pathway enzymes, the acyltransferase LPCAT2 (associated with 18:0 in this study) was recently found to participate in acyl editing by incorporating nascent fatty acids (mostly 18:1) into the membrane lipid phosphatidylcholine, PC (Bates et al. 2012). These fatty acids can then be further desaturated and subsequently transferred to the sn-3 position of a diacylglycerol to generate a TAG (Xu et al. 2012). A double knockout of lpcat1/lpcat2 altered seed oil composition of PC, including an increase of 18:0, suggesting the LPCATs may be responsible for the removal of 18:0 from the PC during the acyl editing cycle (Bates et al. 2012).
Fatty acid accumulation in developing seeds is controlled through the coordinated effort of fatty acid synthesis and fatty acid beta-oxidation (Eccleston and Ohlrogge 1998). SDRB, which encodes a dienoyl-CoA reductase involved in fatty acid beta-oxidation, was associated with proportions of 18:1, 18:2, 20:2, plastid and PUFA. SDRB may act by preferentially hydrolyzing TAGs with these fatty acids. Previous work has shown a decrease in lipid content in seeds during the final stages of maturation (Baud and Lepiniec 2009), which is the seed development stage of highest expression for SDRB (Belmonte et al. 2013). If this mechanism of preferential degradation of TAGs with specific fatty acids can be demonstrated in natural accessions, it may be possible to engineer a decrease in these fatty acid proportions in TAGs through upregulation of this gene.
Acyl carrier proteins (ACP) carry nascent acyl chains during fatty acid synthesis. ACP5 is plastid-localized (TAIR) and was associated with 16:0 and Plastid. There has not been any research connecting ACPs to changes in fatty acid proportions in seeds, so this gene may be a less promising candidate for engineering desired compositions.
Transcription Factors
None of the 8 transcription factor genes linked to SNPs associated with fatty acid proportions in this study have been shown to regulate seed oil composition. Interestingly, Peng and Weselake (2011) found that promoter motifs of fatty acid synthesis genes were enriched for MYB factors and 4 of the 8 genes identified here encode MYB transcription factors (MYB67, MYB10, MYB85, and AT3G12730). To test the functional relationships of these 8 regulatory genes to seed oil composition, they should be silenced and their effect on seed oil composition characterized. Our study did not identify any of the transcription factors previously implicated in the regulation of fatty acid production and seed oil accumulation [FUS3 (Wang et al. 2007), LEC2 and WRI1 (Baud et al. 2007), and LEC1 and ABI3 (Mu et al. 2008)]. Trait associations with these genes may have been missed for a variety of reasons (i.e., correcting for population structure, insufficient marker coverage, rare alleles, etc) [Reviewed in Myles et al. 2009].
Remaining Regions of Interest
While the remaining 114 regions of interest identified by this study do not contain genes that encode products with a clear functional relationships to lipid metabolism, they are significantly associated with variation in fatty acid proportions. Therefore, the set of genes affecting oil composition in A. thaliana, and possibly many other oilseed species, may be much larger than previously recognized. Of the genes with maximum points per region per trait, 13% have unknown functions and 5% encoded kinases which could play a regulatory role in fatty acid and TAG synthesis. If some of these genes are shown to affect oil composition, they would be new targets for engineering seed oil composition and producing novel oils for consumption and industry.
Conclusions
The genes associated with natural variation in our study both confirm what is currently known about how seed oil composition is regulated and provide new candidates for bioengineering. While our work indicates some food uses of oils might be best produced through traditional breeding, most industrial applications will probably require genetic engineering to achieve high levels of the desired TAGs or fatty acids. Most industrial applications of plant lipids require high purity of the fatty acid of interest (sometimes in excess of 90%), and post-plant purification steps are only cost-effective when the desired fatty acids are at high levels. (Thelen and Ohlrogge 2002; Vanhercke et al. 2013).
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
National Science Foundation doctoral dissertation improvement grant (DEB-1011609) to [SEB, CRL].
Conflict of interest
The authors declare that they have no conflict of interest.
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.4p7gk
Supplementary Material
Acknowledgments
Thank you to T. Juenger for providing accession seed. We are grateful for analysis advice from G. Morrison, J. Lasky and T. Nakov. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing high performance computing resources that have contributed to the research results reported within this paper. URL: http://www.tacc.utexas.edu.
References
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 465:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar-Moreno J, Denolf P, Van Audenhove K, et al. 2015. Type 1 diacylglycerol acyltransferases of Brassica napus preferentially incorporate oleic acid into triacylglycerol. J Exp Bot. 66:6497–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C. 2012. Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol. 160:1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B. 2007. WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50:825–838. [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. 2009. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem. 47:448–455. [DOI] [PubMed] [Google Scholar]
- Belide S, Petrie JR, Shrestha P, Singh SP. 2012. Modification of Seed Oil Composition in Arabidopsis by Artificial microRNA-Mediated Gene Silencing. Front Plant Sci. 3:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. 2013. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci U S A. 110:E435–E444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Salas JJ, Pollard MR, Ohlrogge JB. 2003. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell. 15:1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box G, Cox D. 1964. An analysis of transformations. J Roy Statis Soc B, 26, 211–252. [Google Scholar]
- Buhr T, Sato S, Ebrahim F, Xing A, Zhou Y, Mathiesen M, Schweiger B, Kinney A, Staswick P. 2002. Ribozyme termination of RNA transcripts down-regulate seed fatty acid genes in transgenic soybean. Plant J. 30:155–163. [DOI] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB. 2012. Compartmentation of triacylglycerol accumulation in plants. J Biol Chem. 287:2288–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett TP, Benning C, Ohlrogge J. 2008. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54:593–607. [DOI] [PubMed] [Google Scholar]
- Eccleston VS, Ohlrogge JB. 1998. Expression of lauroyl-acyl carrier protein thioesterase in brassica napus seeds induces pathways for both fatty acid oxidation and biosynthesis and implies a set point for triacylglycerol accumulation. Plant Cell. 10:613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head K, Galos T, Fang Y, Hudson K. 2012. Mutations in the soybean 3-ketoacyl-ACP synthase gene are correlated with high levels of seed palmitic acid. Mol Breed. 30:1519–1523. [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ. 2004. Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol. 136:3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton MW, Hancock AM, Huang YS, Toomajian C, Atwell S, Auton A, Muliyati NW, Platt A, Sperone FG, Vilhjálmsson BJ, et al. 2012. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 44:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A, Shanklin J, Whittle E, Lapchyk L, Hildebrand D, Kachroo P. 2007. The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol Biol. 63:257–271. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics. 178:1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, Mackenzie SL, Covello PS, Kunst L. 1995. Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiol. 108:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. 2007. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 39:1151–1155. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F., et al. 2007. Acyl-lipid metabolism. The Arabidopsis Book, 1–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke T. (ed.). 2012. Oil World Annual. Hamburg, Germany: ISTA Mielke GmbH. [Google Scholar]
- Moreno-Pérez AJ, Venegas-Calerón M, Vaistij FE, Salas JJ, Larson TR, Garcés R, Graham IA, Martínez-Force E. 2012. Reduced expression of FatA thioesterases in Arabidopsis affects the oil content and fatty acid composition of the seeds. Planta. 235:629–639. [DOI] [PubMed] [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. 2008. Leafy cotyledon1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148:1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. 2009. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 21:2194–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill CM, Morgan C, Hattori C, Brennan M, Rosas U, Tschoep H, Deng PX, Baker D, Wells R, Bancroft I. 2012. Towards the genetic architecture of seed lipid biosynthesis and accumulation in Arabidopsis thaliana. Heredity (Edinb). 108:115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng FY, Weselake RJ. 2011. Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics. 12:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidkowich MS, Nguyen HT, Heilmann I, Ischebeck T, Shanklin J. 2007. Modulating seed beta-ketoacyl-acyl carrier protein synthase II level converts the composition of a temperate seed oil to that of a palm-like tropical oil. Proc Natl Acad Sci USA. 104:4742–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. . 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: ISBN 3-900051-07-0 http://www.R-project.org/. [Google Scholar]
- Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L. 1999. The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem. 37:831–840. [DOI] [PubMed] [Google Scholar]
- Sablok G, Pérez-Quintero AL, Hassan M, Tatarinova TV, López C. 2011. Artificial microRNAs (amiRNAs) engineering - On how microRNA-based silencing methods have affected current plant silencing research. Biochem Biophys Res Commun. 406:315–319. [DOI] [PubMed] [Google Scholar]
- Salas JJ, Ohlrogge JB. 2002. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch Biochem Biophys. 403:25–34. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Randal Linder C. 2012. Quantitative trait loci involved in regulating seed oil composition in Arabidopsis thaliana and their evolutionary implications. Theor Appl Genet. 124:723–738. [DOI] [PubMed] [Google Scholar]
- Sharma A, Chauhan RS. 2012. In silico identification and comparative genomics of candidate genes involved in biosynthesis and accumulation of seed oil in plants. Comp Funct Genomics. 2012:914843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ. 2009. Acyltransferase action in the modification of seed oil biosynthesis. N Biotechnol. 26:11–16. [DOI] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S. 2000. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124:1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Ohlrogge JB. 2002. Metabolic engineering of fatty acid biosynthesis in plants. Metab Eng. 4:12–21. [DOI] [PubMed] [Google Scholar]
- Vanhercke T, Wood CC, Stymne S, Singh SP, Green AG. 2013. Metabolic engineering of plant oils and waxes for use as industrial feedstocks. Plant Biotechnol J. 11:197–210. [DOI] [PubMed] [Google Scholar]
- Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN. 2014. Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol. 164:144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Guo J, Lambert KN, Lin Y. 2007. Developmental control of Arabidopsis seed oil biosynthesis. Planta. 226:773–783. [DOI] [PubMed] [Google Scholar]
- Xu J, Carlsson AS, Francis T, Zhang M, Hoffman T, Giblin ME, Taylor DC. 2012. Triacylglycerol synthesis by PDAT1 in the absence of DGAT1 activity is dependent on re-acylation of LPC by LPCAT2. BMC Plant Biol. 12:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Fan C, Guo Z, Qin J, Wu J, Li Q, Fu T, Zhou Y. 2012. Identification of FAD2 and FAD3 genes in Brassica napus genome and development of allele-specific markers for high oleic and low linolenic acid contents. Theor Appl Genet. 125:715–729. [DOI] [PubMed] [Google Scholar]
- Zheljazkov VD, Vick Ba, Ebelhar MW, Buehring N, Baldwin BS, Astatkie T, Miller JF. 2008. Yield, oil content, and composition of sunflower grown at multiple locations in Mississippi. Agronomy J, 100, 635–642. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.4p7gk