Abstract
High levels of resistance to phosphine in the rice weevil Sitophilus oryzae have been detected in Asian countries including China and Vietnam, however there is limited knowledge of the genetic mechanism of resistance in these strains. We find that the genetic basis of strong phosphine resistance is conserved between strains of S. oryzae from China, Vietnam, and Australia. Each of 4 strongly resistant strains has an identical amino acid variant in the encoded dihydrolipoamide dehydrogenase (DLD) enzyme that was previously identified as a resistance factor in Rhyzopertha dominica and Tribolium castaneum. The unique amino acid substitution, Asparagine > Threonine (N505T) of all strongly resistant S. oryzae corresponds to the position of an Asparagine > Histidine variant (N506H) that was previously reported in strongly resistant R. dominica. Progeny (F16 and F18) from 2 independent crosses showed absolute linkage of N505T to the strong resistance phenotype, indicating that if N505T was not itself the resistance variant that it resided within 1 or 2 genes of the resistance factor. Non-complementation between the strains confirmed the shared genetic basis of strong resistance, which was supported by the very similar level of resistance between the strains, with LC50 values ranging from 0.20 to 0.36mg L−1 for a 48-h exposure at 25 °C. Thus, the mechanism of high-level resistance to phosphine is strongly conserved between R. dominica, T. castaneum and S. oryzae. A fitness cost associated with strongly resistant allele was observed in segregating populations in the absence of selection.
Keywords: dihydrolipoamide dehydrogenase, fitness cost, mode of inheritance, rph2 gene
Sitophilus oryzae (Linnaeus) is a cosmopolitan insect pest of stored grain that, if left uncontrolled, will cause significant losses in both grain quality and quantity (Cotton 1920). Phosphine, hydrogen phosphide gas (PH3), is the treatment of choice in most cases against this pest as it has many advantages including acceptance by markets, low or no residue, ease of application, low cost, and the fact that it is environmentally benign (Chaudhry 2000). However, high-level resistance to this fumigant in S. oryzae has been reported from countries including India, China, Morocco, Brazil, and Australia (Rajendran 1999; Zeng 1999; Athié et al. 1998; Benhalima et al. 2004; Nguyen et al. 2015). Recently, high-level resistance to phosphine in S. oryzae has been detected in Vietnam as well. These reports are of concern as there is currently no practical replacement for phosphine that can match its range of advantages.
Phosphine resistance in S. oryzae shows 2 phenotypes, labeled weak and strong resistance, and it is essential that the genetics of these resistances is understood as a basis for developing rational strategies to manage phosphine resistance. Classical genetic analysis of S. oryzae (Li and Li 1994; Daglish et al. 2014; Nguyen et al. 2015) has indicated that weak resistance is controlled by a single incompletely recessive, autosomally inherited gene, whereas strong resistance is mediated by at least 2 incompletely recessive, autosomally inherited genes. In addition, both resistance phenotypes share 1 gene, which is responsible for weak resistance. This mode of inheritance of phosphine resistance also occurs in other insect pests of stored grain including the lesser grain borer (Rhyzopertha dominica) (Collins et al. 2002) and the red flour beetle (Tribolium castaneum) (Jagadeesan et al. 2012). Crosses between strongly resistant strains of R. dominica from Australia and India, revealed that genes contributing to strong resistance are shared between strains from 2 nations (Kaur 2012). Furthermore, crosses between field populations collected from widely separated regions in Australia, indicated that weak and strong resistance were the same in all strains (Mau et al. 2012a; Mau et al. 2012b). These data suggest that the genetic mechanism of phosphine resistance may also be similar among strains of S. oryzae from various regions of origin.
Studies based on genomic mapping and simple single nucleotide polymorphism (SNP) averaging methods confirmed that there are 2 loci associated with resistance in R. dominica and T. castaneum, namely rph1 (tc_rph1) and rph2 (tc_rph2) (Schlipalius et al. 2002; Jagadeesan et al. 2013). A critical discovery was that the second resistance gene (rph2), which is conserved in both species, encodes an essential enzyme involved in energy metabolism, dihydrolipoamide dehydrogenase (DLD). Five amino acid substitutions were detected in the dld gene in strongly resistant strains of R. dominica (P49S, P85S, G135S, K142E, N506H) and 1 mutation (G131S) in T. castaneum, all of which had been collected from Australia (Schlipalius et al. 2012). Analysis of DLD sequences of phosphine-resistant insect pests from India (Kaur et al. 2015), also showed 1 homologous variant in both R. dominica (P49S) and T. castaneum (P45S) were common across southern India.
There is currently very limited genomic information available for S. oryzae, and also limited information on the nature of phosphine resistance genes in these species. To address these limitations, the present study 1) compares the mode of inheritance of phosphine resistance in strongly resistant S. oryzae strains collected from Vietnam, China and Australia, 2) determines the resistance variant of DLD in S. oryzae and measures its linkage to the phosphine resistance trait, and 3) determines the fitness effects of a resistance allele.
Materials and Methods
Insect Strains
This study used a susceptible strain, LS2 (S-strain) collected from Brisbane, Queensland in 1965 (Daglish et al. 2002); a weakly phosphine-resistant strain, QSO335 (WR) collected from Millmerran, Queensland in 1990 (Daglish et al. 2002) and 4 strongly phosphine-resistant strains: the SRAus strain (referenced as NNSO7525) collected in 2009 from Griffith, New South Wales, Australia (Nguyen et al. 2015); the SRCN strain was derived from adults collected from Santai, Sichuan Province, China in 1998 (Daglish et al. 2002); 2 strains were collected from national reservation stations in Vietnam in 2009, 1 from Daklak (SRVN1)—a province in south Vietnam, the other strain from Vinhphuc province (SRVN2) in north Vietnam. The 2 Vietnamese strains were identified as strongly resistant to phosphine based on the FAO method (No.16) (Anon 1975) of survival at a discriminating dose of 0.25mg L−1 for 48h exposure. The S-strain was always maintained under phosphine-free condition while WR, SRAus, SRCN, SRVN1, and SRVN2 were selected at a discriminating doses of 0.04mg L−1 for WR and 0.25mg L−1 for SR for 48h over at least 3 generations to promote homozygosity of resistance genes in population. All strains were cultured on whole wheat at 25 ºC and 55% relative humidity (RH).
Fumigation Assays
Phosphine gas was generated from aluminium phosphide tablets dissolved in a 5% sulfuric acid solution and collected into a tube via a funnel. The source of phosphine was measured using a gas chromatograph fitted with a thermal conductivity detector (Perkin and Elmer, Clarus 580 model).
Adult insects (2–3 weeks old) of parental strains and their progenies were fumigated simultaneously to reduce variations in experiment (Preisler et al. 1990). We tested 150 insects per dose separated into 3 small plastic cups without food including the control (air, no PH3). Insects were exposed to phosphine at required concentrations for 48h at 25 ºC, 60% RH. After fumigation, insects were fed on whole wheat and mortality data were recorded after a 7-day recovery period. Each bioassay was conducted 3 times with 3 weeks gap between replications.
Complementation Analysis of Asian and Australian Strains
Reciprocal Crosses Between Strongly Resistant Strains
Virgin adults of the 4 strongly resistant strains (SRAus, SRVN1, SRVN2, SRCN) were sexed under a microscope according to the method of Halstead (Halstead 1963) to isolate the sexes. About 50 reciprocal, single-pair crosses were set up between each of SRCN, SRVN1, SRVN2, and SRAus. These parental pairs were placed on to fresh grain every 2 weeks to produce batches of F1 progeny. Each F1 generation (2–3-week-old adults) of each reciprocal cross was then inbred randomly to generate an F2 generation. In addition, mass crosses of 100 F1 individuals were placed on fresh grain at 2 weeks intervals. The F1, F2 progenies, and their parents were subsequently exposed at a range of concentrations of phosphine to test their response.
Statistical Analysis
Mortality occurring in the control treatment (<4% in these experiments) was corrected in treated samples using Abbott’s formula (Abbott 1925). Mortality of parental strains (P0) and their F1 progeny was then subjected to probit analysis using GenStat statistic package 16.1 version (www.vsni.co.uk/software/genstat). Homozygosity of P0 and F1 populations for response to phosphine was tested by modified chi-square (modified χ2) value (Nguyen et al. 2015) which incorporated the standard χ2 calculated according to Finney’s method (Finney 1971) with heterogeneity factor obtained from GenStat probit analysis.
The method developed by Robertson and Preisler (1992) was used to determine resistance ratios at LC50 between SRCN, SRVN, and SRAus and 95% confidence intervals (95% CI). Relative potency is a statistical parameter generated from Genstat probit analysis based on a comparison of responses to phosphine at range of doses between reciprocal F1 groups (Finney 1971). The fiducial limits (95% FL) of relative potency show non-significant difference between 2 response data of reciprocal hybrids if the lower and upper limits bound one (i.e. same potency) (Bengston et al. 1999).
The response of F1 generations to phosphine is used as a basis to identify whether the resistance gene(s) are shared between strains collected from China, Vietnam and Australian. If the response line of F1 progeny is close to or overlaps with that of the S-strain, then the gene(s) controlling resistance to PH3 in SRCN, SRVN are different from those conferring resistance in SRAus (Supplementary Figure 1). When the F1 response is close to or overlaps with the response of WR, then the 2 strains SRCN, SRVN likely share one common gene with SRAus (Supplementary Figure 1). In the case of the F1 response lying between the response lines of the 2 parental strains, then the genes contributing to strong resistance in the Asian and Australian strains are shared, but if 1 parental strain has a clearly stronger resistance, it is possible that there may be extra factors causing a higher level resistance in that strain. In this case, the response of F2 should elucidate the extra factors.
Identification and Sequencing of DLD in S. oryzae
Genetic Crosses
Virgin adults of the S-strain, WR, SRAus were sexed and set up in the following single pair crosses: ♀S-strain × ♂SRAus and ♀WR × ♂SRAus. Approximately 100 sibling F1 of each cross were then freely mated to produce an F2 and this was repeated to produce subsequent non-overlapping generations. A part of adults obtained at F6 were exposed to phosphine at 0.6mg L−1 for 48h, a dose expected to kill all susceptible and weak resistant insects (i.e. non-homozygous for strong resistance). Selected survivors were anticipated to be homozygous at the all the resistance loci contributing to strong resistance, whereas the rest of the F6 population was not exposed to phosphine and constituted an “unselected” group, which was anticipated to have mixed genotypes at the resistance loci.
RNA Extraction and Transcriptome Sequencing
Total RNA was extracted from 50mg insects (~15–20 individuals) of each parental strain and the selected and unselected F6 progeny using the Isolate RNA™ mini kit (Bioline, Australia). Live insects initially were homogenized by a rotor-stator homogenizer for 20–40s in Lysis buffer R and the subsequent steps were carried out according to the manufacturer’s protocol. Yield of RNA was assessed using a BioTek Epoch 16-spot microspot plate spectrophotometer. Total RNA was sequenced using the Illumina Hiseq RNAseq 100bp paired-end protocol.
Transcriptome Sequence Assembly
Illumina 100bp paired-end reads from the susceptible strain were assembled by CLC Genomics Workbench software version 7.0.3 (www.clcbio.com) using the de novo assembly function (minimum contig length = 200, word size = 23, bubble size = 50). The susceptible strain was used as reference to ensure variants that were unique to resistant strains were not incorporated into the reference transcriptome sequence. To ensure correct and full transcript sequence for DLD in the transcriptome, the assembled contigs were aligned against R. dominica DLD susceptible strain QRD14 from the NCBI database (accession number: JX434596) using the tBLASTx algorithm to identify homologous sequences. The matching contigs then were reassembled into a fully complete and annotated sequence coding DLD protein, named So_dld reference contig, which then replaced the homologous sequences in the S-strain reference contig list.
Linkage Analysis of So_dld to Phosphine Resistance Using SNP Variants
The reads from each parental strain and F6 selected and unselected groups were separately aligned against the partially annotated contigs of reference S-strain using the read-mapping function of CLC software (mismatch cost = 2, insertion cost = 3, deletion cost = 3, length fraction = 0.5, similarity fraction = 0.8) and SNP variants were detected via the Probabilistic Variant Detection function of CLC Genome Workbench (minimum coverage = 10, variant probability = 90%, required variant count = 2). Variant SNP frequencies (count/coverage) in So_dld relative to the susceptible strain evaluated for each of the resistant strains and F6 selected/unselected groups. If the SNP frequency was >95%, the strain was classed as homozygous, but heterozygous at an allele frequency of 50–80% (Jagadeesan et al. 2013).
Comparison of So_dld Sequences of All Strains
cDNA Synthesis and Sample Preparation for Sequencing
We confirmed and compared the So_dld sequences of all the Australian strains and strongly resistant strains from Vietnam and China using Sanger sequencing. Total RNA of 15 adult insects of SRVN1, SRVN2, SRCN was extracted using the Isolate RNA mini kit (Bioline, Australia). Next, we used 1000ng of purified RNA from the 6 research strains to convert to cDNA using the Invitrogen SuperScriptTM III cDNA synthesis kit. All reverse transcription steps were conducted following the manufacturer’s protocol. The DLD coding sequences (1.8kb) of all strains were PCR amplified from cDNA in a 20-µL reaction volume including 2 µL (~100ng) cDNA template, 1 µL forward primer 10nM (5′-AGTTGTCATTCCGTCGTCCT-3′), 1 µL reverse primer 10nM (5′-GCGAAGGAATTAAGCACATT-3′), 10 µL master mix of EmeraldAmp GT PCR master mix kit. The PCR reaction conditions were: denaturation at 95 °C for 3min; then 40 cycles of 95 °C for 20s, 60 °C for 30s, 72 °C for 2min; final extension at 72 °C for 5min. PCR products were subsequently purified and sequenced.
Fine-Scale Linkage Analysis
Single-Pair Genetic Crosses and Genomic DNA Extraction
A virgin female of S-strain or WR was each crossed to a virgin male of SRAus to produce an F1 population, a virgin single pair of the F1 cohort was mated to each other to produce an F2 generation. Progeny from F2 was allowed to mate freely to produce F3 and subsequent non-overlapping generations. Approximately 10 000 individual offspring of each of the crosses were selected at either the F16 for cross WR × SRAus cross or F18 of the S-strain × SRAus cross using the discriminating dose of 0.6mg L−1 for F16 and 0.5mg L−1 for F18. Individual survivors of each selection were preserved at −20 °C until DNA was extracted for subsequent genotyping at the So_dld locus. Briefly, genomic DNA was extracted from each individual of parents, hybrid F1s and survivors of F16 (WR × SRAus) and F18 (S-strain × SRAus) using a modified HotsHot DNA extraction method described by Montero-Pau et al. (2008). One insect per extraction was lysed in 75 µL Alkaline lysis buffer (25mM NaOH and 0.2mM EDTA) (pH = 12) at 95 °C for 30min, cooled down at 4 °C for 10min; then solution was neutralized by 75 µL of 40mM Tris–HCl (pH = 5).
Visualization of the Resistance Allele in the dld Gene
Primers were designed that amplified a specific 297bp region which contained a unique SNP coding for an amino acid variant, discussed below. We designed a cleaved amplified polymorphic sequence (CAPS) assay that utilized a restriction enzyme (RE) site that recognized the unique variant SNP. The assay cleaved the resistant allele but did not digest sensitive alleles at that specific site.
Individuals were genotyped by PCR amplification of the So_dld marker fragment in a reaction that consisted of 10 µL PCR master mix (EmeraldAmp), 1 µL of 10nM forward (5′-AGGAGTACGGCGCATCAT-3′) and 1 µL of 10nM reverse (5′-CGATAACAAAAAAGGGGCG-3′) primers, 2 µL of 1:10 dilution of DNA template (HotsHot), 6 µL dH2O. The reaction conditions were: denaturation for 3min at 95 ºC followed by 40 cycles of 95 °C 20s, 55 °C 30s, 72 °C 1min, with a final extension of 5min at 72 °C. For each individual, all PCR products were subsequently digested with addition of 10 µL restriction enzyme mixture including 3 µL 10× NE buffer, 0.5 µL HphI enzyme (BioLabs), 6.5 µL dH2O and incubated at 37 °C for 10h, inactivated at 65 °C for 20min then cool down at 4 °C for 10min. The HphI enzyme cuts the So_dld marker fragment of the strongly resistant strain into 2 smaller fragments of 100 and 197bp but does not cut fragments from the S-strain or WR strain. Digested samples (5 µL) were visualized by agarose gel electrophoresis loaded on (2% agarose in 1× TAE, 110V, 60min).
If there was any recombination between So_dld marker and its target resistant gene being detected on selected population, map distance was calculated by formula (Schlipalius et al. 2002):
Fitness Analysis of the So_dld Resistance Allele
The effect of the So_rph2 resistance allele on reproductive fitness was monitored as change in allele frequency over generations in the absence of phosphine selection. About 92–96 individuals each of F2, F10, F16, or F18 mapping populations that had been propagated in the absence of phosphine were genotyped for the presence or absence of the N505T variant by the CAPS procedure described above. We firstly calculated the frequency of each allele at each generation that we analyzed and compared the change in allele frequency relative to the F2 generation. We then used Chi-square analysis to determine whether the genotype ratios at each generation deviated from Hardy–Weinberg equilibrium. Significant changes in the allele frequencies across generations and genotype frequencies within populations were used to identify selective advantage or disadvantage associated with the So_rph2 resistance variant.
Data Archiving
According to the data archiving guidelines (Baker 2013), we have deposited the nucleotide sequences of dld mRNA of S. oryzae in Genbank, accessions KT893322–KT893327.
Results
Complementation Analysis of Asian and Australian Strains
SRVN1 × SRAus Cross
Modified χ2 analysis showed that both parental resistant strains were homogeneous in their response to phosphine (P = 0.587 for SRVN1 and P = 0.694 for SRAus) (Table 1). The resistance ratio at the LC50 between SRAus and SRVN1 was 1.02, indicating that the level of resistance to phosphine of these strains was the same. The response of the F1 (♀SRVN1 × ♂SRAus) relative to that of the reciprocal cross F1 (♀SRAus × ♂SRVN1) was 0.913 (0.767–1.082). Because the fiducial limits bounded 1.0, the response to phosphine of the reciprocal F1 hybrids was found to be not significantly different. This confirmed that the genes controlling strong resistance in SRVN1 and SRAus were autosomally inherited, allowing data from the F1 hybrids to be pooled.
Table 1.
Probit analysis of the international strains and the Australian strongly resistant strain with their F1 progenies exposed to phosphine for 48 h
| Strain or cross | n (number) | Slope ± SE | LC50 (mg L−1) (95% FL) | LC99.9 (mg L−1) | df | Modified χ2 | P | Rf (95%CI) at LC50 |
|---|---|---|---|---|---|---|---|---|
| Cross of SRVN1 × SRAus | ||||||||
| SRVN1 | 2827 | 2.69±0.22 | 0.286 (0.254–0.318) | 4.024 | 5 | 3.746 | 0.587 | — |
| F1A | 1589 | 3.06±0.38 | 0.272 (0.223–0.314) | 2.781 | 4 | 3.342 | 0.502 | 0.95 (0.80–1.14) |
| F1B | 1664 | 3.02±0.36 | 0.297 (0.248–0.340) | 3.123 | 4 | 3.629 | 0.459 | 1.04 (0.87–1.23) |
| F1 pooled | 3253 | 3.03±0.26 | 0.284 (0.254–0.312) | 2.972 | 4 | 7.032 | 0.134 | 0.99 (0.86–1.15) |
| SRAus | 3007 | 4.08±0.30 | 0.292 (0.268–0.317) | 1.672 | 5 | 3.036 | 0.694 | 1.02 (0.90–1.17) |
| Cross of SRVN2 × SRAus | ||||||||
| SRVN2 | 2706 | 3.49±0.22 | 0.198 (0.182–0.215) | 1.522 | 5 | 9.400 | 0.094 | — |
| F1A | 1794 | 3.04±0.29 | 0.217 (0.188–0.243) | 2.256 | 4 | 6.598 | 0.159 | 1.09 (0.95–1.25) |
| F1B | 1802 | 2.98±0.34 | 0.215 (0.179–0.247) | 2.339 | 4 | 5.189 | 0.268 | 1.08 (0.92–1.27) |
| F1 pooled | 3596 | 3.01±0.21 | 0.216 (0.196–0.234) | 2.297 | 4 | 9.167 | 0.057 | 1.09 (0.97–1.22) |
| SRAus | 3007 | 4.08±0.30 | 0.292 (0.268–0.317) | 1.672 | 5 | 3.036 | 0.694 | 1.47 (1.32–1.65) |
| Cross of SRCN × SRAus | ||||||||
| SRAus | 3007 | 4.08±0.30 | 0.292 (0.268–0.317) | 1.672 | 5 | 3.036 | 0.694 | — |
| F1A | 2407 | 3.97±0.39 | 0.299 (0.260–0.338) | 1.795 | 6 | 9.565 | 0.144 | 1.02 (0.89–1.18) |
| F1B | 2412 | 4.10±0.32 | 0.333 (0.299–0.368) | 1.89 | 6 | 7.632 | 0.266 | 1.14 (1.00–1.29) |
| F1 pooled | 4819 | 4.02±0.25 | 0.316 (0.291–0.341) | 1.853 | 6 | 16.273 | 0.012* | 1.08 (0.97–1.21) |
| SRCN | 3851 | 3.67±0.26 | 0.361 (0.327–0.395) | 2.506 | 7 | 10.562 | 0.159 | 1.23 (1.10–1.39) |
F1A is progenies of cross SRVN/SRCN (♀) × SRAus (♂); F1B is progenies of cross SRAus (♀) × SRVN /SRCN (♂). df, degree of freedom; n, number insects tested; SE, standard error, 95% FL, 95% fiducial limits; LC50, lethal concentration at 50% mortality; LC99.9, lethal concentration at 99.9% mortality; Rf, resistance factor; 95% CI, 95% confident interval.
*P < 0.05.
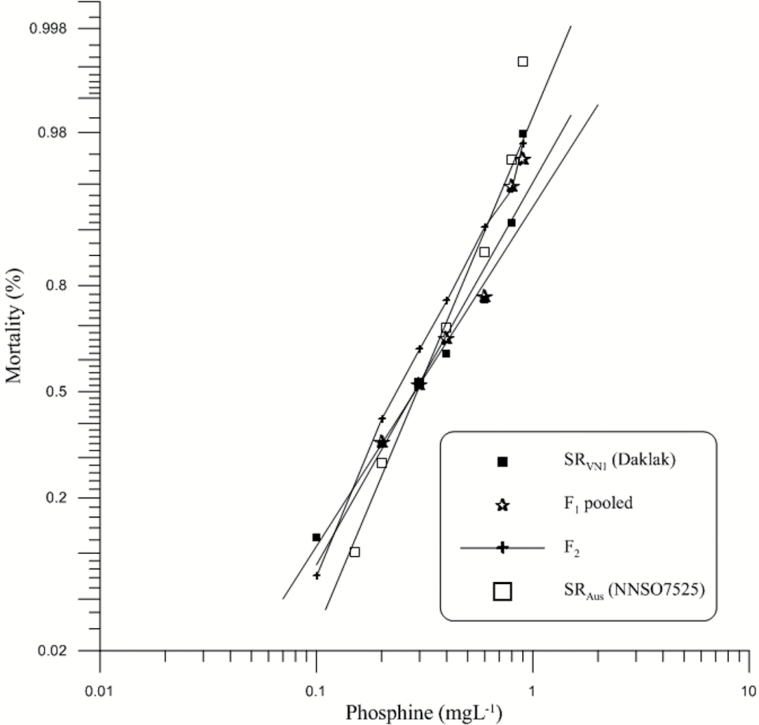
Although the mortality response lines of the 2 parental strains SRVN1 and SRAus intersected, the probit response of the F1 generation always lay between the parent lines (Figure 1) indicating that genes contributing to resistance to phosphine in SRAus were the same in SRVN1. We tested the response of the F2 progeny at a range of PH3 doses to detect the possible presence of additional gene(s) causing strong resistance that was not shared between 2 parental strains. However, there were no differences observed in the response of the F2, supporting our hypothesis that the phosphine resistance is conferred by the same genes in SRAus and SRVN1.
Figure 1.
Mortality response of parental (P0) strains (SRAus, SRVN1) as well as their F1 and F2 generations after exposure to phosphine for 48h without food at 25 °C, 60% RH. The expected response lines of the P0 and F1 were drawn using a probit linear model y = αx + β in which α and β are the slope and intercept, respectively. x is the log-transformed dose. y is the percent mortality. The observed mortality of parents and the F1, F2 generations are also displayed.
SRVN2 × SRAus Cross
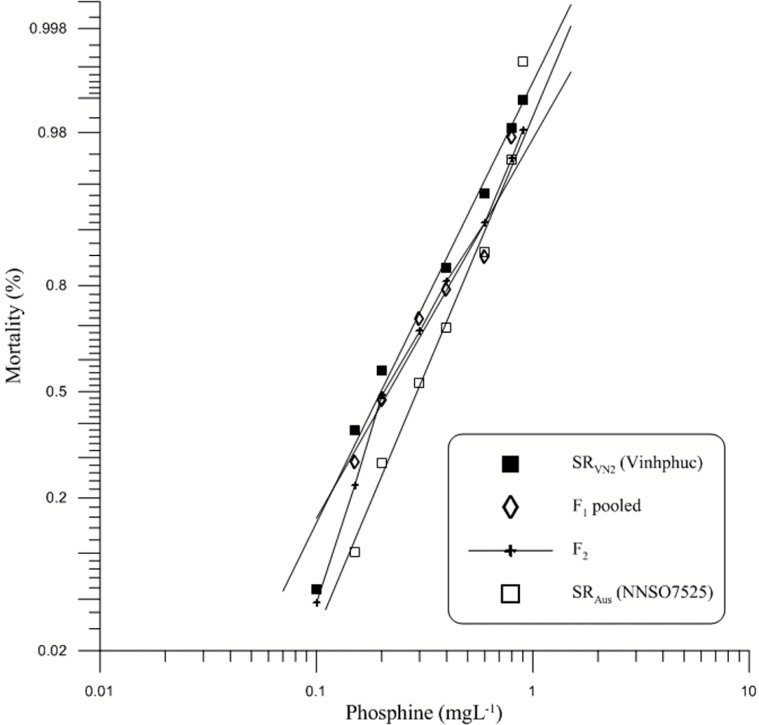
The mortality response of the second strongly resistant strain from Vietnam, SRVN2, fitted closely to the probit model (Figure 2) showing homogeneity of response to PH3 (P = 0.094) (Table 1). The resistance to phosphine of SRVN2 was slightly less than that of SRAus (Rf = 1.47), and their LC50s were distinguishable statistically as the number 1 was not included in the 95% CIs (Table 1). Relative potency (95% FL) when comparing data of reciprocal F1s was 0.995 (0.849–1.167), therefore responses of F1 hybrids were statistically identical showing the absence of a maternal effect on the trait. This allowed data of F1A and F1B to be combined in future analysis.
Figure 2.
Mortality response of parental (P0) strains (SRAus, SRVN2) as well as their F1 and F2 generations after exposure to phosphine for 48h without food at 25 ºC, 60% RH. The expected response lines of the P0 and F1 were drawn using a probit linear model y = αx + β in which α and β are the slope and intercept, respectively. x is the log-transformed dose. y is the percent mortality. The observed mortality of parents and the F1, F2 generations are also displayed.
In these crosses, the observed response of the F1 was lower than that of the parental strains at high doses (from 0.6mg L−1) while responses at low and moderate doses were in between those of the parents (Figure 2). The simplest explanation is that both parental strains share the same major resistance factors and that the higher level of resistance in the hybrid is the result of an allelic interaction. The response of the F2 almost overlapped the response of the F1 (Figure 2), showing both parental strains to share the same genes responsible for strong resistance to phosphine.
SRCN × SRAus Cross
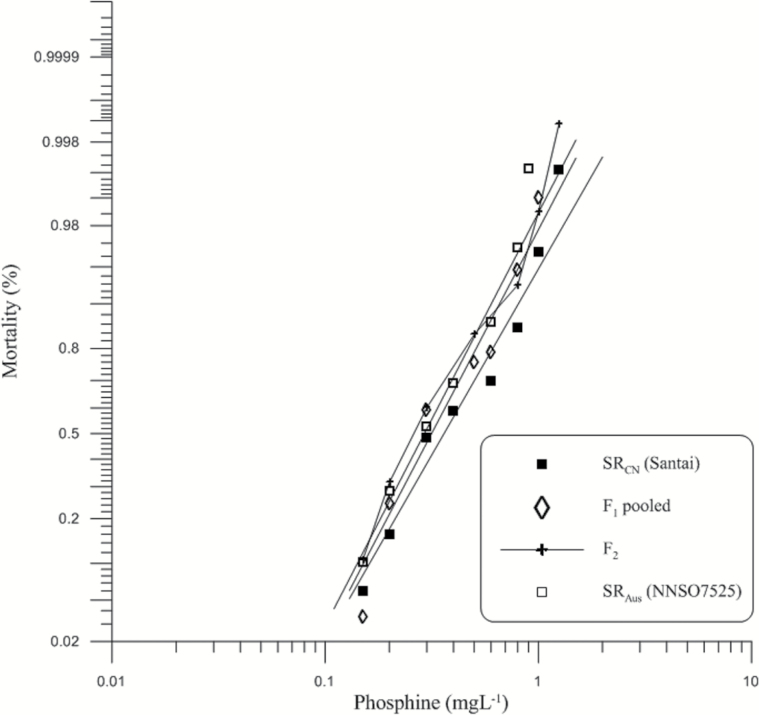
The response of SRCN to phosphine was homogeneous (P = 0.159) (Table 1) with a somewhat higher resistance, 1.23 times than that of the SRAus. The LC50 values of these 2 strains were significantly different (Table 1). Relative potency (RP) analysis showed no significant difference between responses of F1 progenies of SRCN × SRAus and SRAus × SRCN (RP [95% FL] = 1.109 [0.955–1.289]). Therefore, genes conferring strong resistance to PH3 in parental strains were not sex linked and response data of reciprocal F1 hybrids could be merged.
The response of the F1 progeny lay entirely between those of parental strains but closer to that of SRAus (Figure 3), indicating that the genes responsible for strong resistance in SRAus were shared with SRCN. The response curve of the F2 generation (Figure 3) confirmed the absence of any additional genes for resistance to PH3 in SRCN.
Figure 3.
Mortality response of parental (P0) strains (SRAus, SRCN) as well as their F1 and F2 generations after exposure to phosphine for 48h without food at 25 °C, 60% RH. The expected response lines of the P0 and F1 were drawn using a probit linear model y =αx + β in which α and β are the slope and intercept, respectively. x is the log-transformed dose. y is the percent mortality. The observed mortality of parents and the F1, F2 generations are also displayed.
Reference Transcriptome Assembly of So_dld
Assembly of the transcriptome from the S-strain resulted in 28,439 contigs with a mean length of 1116bp and with the total length of all contigs just greater than 31Mb (Supplementary Table 1). We used the DLD mRNA sequence of R. dominica (accession: JX434596) to find homologous sequences in the susceptible S. oryzae transcriptome contig list by tBLASTx and found 14 contig fragments matching DLD. These contigs were then reassembled to create one single sequence of 1956bp that covered the entire coding region of So_dld. The encoded DLD protein of S. oryzae was predicted to be 507 amino acids in length (accession number: KT893322).
Unique SNP Detection
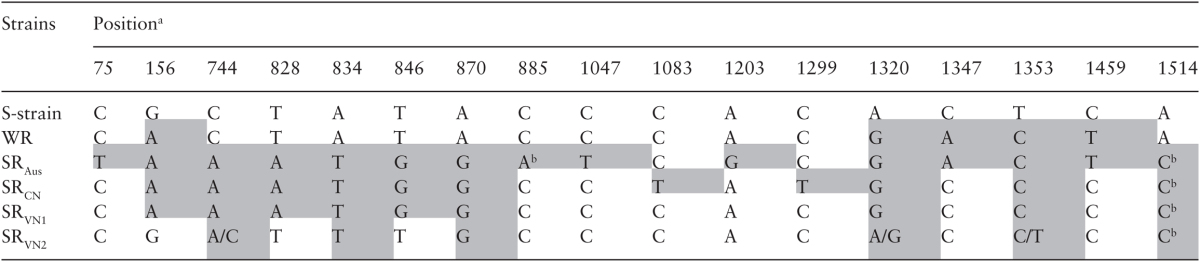
Alignment of the So_dld cDNA sequences of all S. oryzae strains used in this study (accession number: from KT893323 to KT893327) showed many SNPs along the DLD coding region of resistance strains compared to the susceptible strain, but almost all these SNPs were synonymous changes and did not alter the translated amino acid sequence of DLD (Table 2). However, this comparison showed mutations of S. oryzae strains from different geographic regions arising independently from each other.
Table 2.
SNPs in the DLD coding sequence of resistant strains compared to the susceptible reference strain of S. oryzae
SNPs are highlighted in grey.
aPositions are numbered from the start of the open reading frame of the DLD coding sequence.
bSNP causes amino acid change discussed in text.
We aligned the translations of DLD of all resistance strains to that of the S-strain reference sequence and identified an Asparagine >Threonine amino acid substitution, N505T, shared between all the strongly resistant strains (Figure 4). This variant was homologous to a previously identified resistance variant in the dld gene of R. dominica QNRD345 (N506H) (Schlipalius et al. 2012). This variant occurs near the carboxyl terminus of the DLD polypeptide (Figure 4), but close to the active site disulfide (Schlipalius et al. 2012). A cleaved amplified polymorphic sequence (CAPS) diagnostic marker was developed for this unique SNP to facilitate detailed analysis of linkage and fitness effects. We also detected another polymorphism that caused an amino acid variant in SRAus (S295R) that was not observed in resistant Asian strains (Figure 4). Alignment against the human 3-dimensional structure (1ZMC) (Brautigam et al. 2005) showed that this variant occurred at an outer loop of the DLD protein which was not close to the disulfide active site, FAD/NAD+ binding sites or dimerisation interface areas and therefore was considered far less likely to be related to resistance.
Figure 4.
Alignment of a portion of DLD protein of all S. oryzae strains and susceptible R. dominica strain. The putative resistance variant occurs at position 506 of the R. dominica sequence (505 in S. oryzae). All insects that have no DLD resistance variant have an N amino acid at this position, whereas strongly resistant individuals all have a T at the corresponding position. In contrast, a variant at position 295 in S. oryzae is unlikely to be associated with resistance.
Linkage Analysis
Investigating preliminary indication of whether So_dld was linked to phosphine resistance, was obtained by testing for lack of heterozygosity of the gene in the strongly resistant parental strains and among strongly phosphine resistant progeny relative to susceptible or weakly resistant strains and unselected progeny. In fact, strongly resistant individuals were invariably homozygous at the So_dld locus, whereas, susceptible, weakly resistant or unselected insects were heterozygous, with no individual allele present at a frequency greater than 64%.
Much stronger linkage data was obtained by analyzing strongly resistant individuals from segregating progenies established from single-pair intercrosses of WR × SRAus and S-strain × SRAus. Genotyping of 95 F16 (WR × SRAus) and 52 F18 (S × SRAus) strongly resistant survivors using the CAPS assay showed that 100% of the survivors of both crosses were homozygous resistant for the N505T variant at DLD, defining the resistance locus as an interval of 0.022 cM. As the size in centiMorgans of the genetic linkage map of S. oryzae is not known, we used the average of the map sizes of R.dominica and T.castaneum 390 and 570cM, respectively (Beeman and Brown 1999; Schlipalius et al. 2002) to provide us with an estimate of the map size of S. oryzae (480 cM). If we use the number genes identified in T. castaneum (16 404 genes) (Richards et al. 2008) as an estimate of the number of genes in beetles in general, each map unit (1 cM) of the rice weevil genome would contain roughly 16 404/480 34 genes. The strong resistance locus of S. oryzae has been mapped to an interval within 0.022 cM of the dld locus, an interval that would contain an estimated (34 genes/cM × 0022 cM x 2), 1–2 genes.
Fitness Analysis
We analyzed the segregating single-pair intercrosses in the absence of phosphine selection by applying the CAPS assay to evaluate the persistence of the resistant So_rph2 alleles for up to 16 generations. The frequency of the resistance allele (R) consistently decreased from the F2 to F10 generation (P = 0.021) and from the F2 to the F16 generation (P = 8.59E−09) of progeny from the WR × SRAus cross (Table 3). This was most apparent in the decline in the proportion of homozygous resistant individuals from 23.16 to 4.17% over 14 generations. Likewise, there was also a decrease in the frequency of the resistance allele over 16 generations among the progeny of an S-strain × SRAus cross. The decrease was only significant at the F18 generation (P < 0.001) (Table 3). Thus, there is apparently a fitness cost associated with the resistance allele responsible for strong resistance to phosphine in S. oryzae. Interestingly, the genotype ratios at each generation of each cross indicated that the populations were in Hardy–Weinberg equilibrium (P > 0.05) (Table 3). Thus, the selective pressure against the strong resistance allele is weak at each generation but is clearly apparent when the analysis encompasses many generations. Selection against the resistance allele was stronger in populations in which every individual was homozygous for resistance at the weak resistance locus than in populations that began with an equal proportion of susceptibility and resistance alleles at the weak resistance locus.
Table 3.
Fitness cost, in the absence of phosphine, associated with the So_rph2 resistance allele in the progeny of 2 crosses, S-strain × SRAus and WR × SRAus
| Generations | No. tested insects | No. of each genotype | Allelic frequency | P (df = 1) | ||||
|---|---|---|---|---|---|---|---|---|
| S/S | S/R | R/R | p (S) | q (R) | Allelea | Genotypeb | ||
| WR × SRAus cross | ||||||||
| F2 | 95 | 28 | 46 | 22 | 0.54 | 0.47 | — | 0.701 |
| F10 | 96 | 36 | 46 | 14 | 0.61 | 0.39 | 0.021* | 0.911 |
| F16 | 96 | 50 | 42 | 4 | 0.74 | 0.26 | 8.59E−09*** | 0.183 |
| S-strain × SRAus cross | ||||||||
| F2 | 92 | 17 | 49 | 26 | 0.45 | 0.55 | — | 0.469 |
| F10 | 96 | 22 | 52 | 22 | 0.50 | 0.50 | 0.173 | 0.414 |
| F18 | 96 | 33 | 47 | 16 | 0.59 | 0.41 | 0.00013*** | 0.915 |
df, degrees of freedom.
a P value for change in allele frequency was calculated as deviation of the number of copies of each allele in the F10, F16, or F18 generations relative to the allele number in the F2 generation, df = number of alleles − 1.
b P value to evaluate Hardy–Weinberg equilibrium was calculated as deviation of the observed genotype frequency from that predicted if the genotypes were in Hardy–Weinberg equilibrium (p2 S/S: 2pq S/R: q2 R/R), df = number of genotypes − number of alleles.
Significance (*P < 0.05), Significance (**P < 0.01), Significance (***P < 0.001).
Discussion
Understanding the genetic basis of phosphine resistance in insect pests between nations is vital for informing resistance management strategies globally. Our study focused on identification of the genetic mechanism of phosphine resistance in S. oryzae in China and Vietnam based on a previously genetically characterized Australian strain. The strong resistance to phosphine of the Australian S. oryzae strain (Nguyen et al. 2015) was controlled by at least 2 major genes, with one of the genes responsible for the previously characterized weak resistance phenotype (Daglish et al. 2014). Genetic crosses between the Australian and Asian strains revealed that the same genes conferred strong resistance in all 4 resistant strains, and no additional major genes were detected in any of them. Our results are consistent with a complementation study between Indian and Australian strains in R. dominica (Kaur, 2012). Although the LC50 of the Indian R. dominica strain was about 3 times greater than that of the strongly resistant Australian reference, there were 2 major resistance genes that were common to all strongly resistant strains of that species. This conservation of resistance genes (rph1, rph2) in R. dominica had previously been observed in strains from many different regions in Australia (Mau et al. 2012a; Mau et al. 2012b). Overall, classical genetic studies published to date, including our present study, suggest common mechanisms related to inheritance of phosphine resistance in all phosphine resistant stored product pests. Our subsequently molecular results that identified the gene contributing to the strong resistance phenotype of S. oryzae support this conclusion.
Phosphine resistance has been found to be conferred by 2 genes that act in synergy in both R. dominica (Schlipalius et al., 2001; Schlipalius et al. 2002; Collins et al. 2002; Schlipalius et al. 2008; Schlipalius et al. 2012; Mau et al. 2012a; Mau et al. 2012b; Kaur et al. 2012) and T. castaneum (Schlipalius et al. 2012; Jagadeesan et al. 2013). The identity of the first gene (rph1/tc_rph1) that is responsible for the weak resistance phenotype is currently unknown, however the second gene (rph2/tc_rph2) that results in high level resistance in combination with rph1/tc_rph1 was found to be the metabolic enzyme DLD in both species. Since this enzyme is very highly conserved in eukaryotes (Schlipalius et al. 2012; Kaur et al. 2015), we anticipated that at least one of the genes contributing to strong resistance in S. oryzae was also DLD. Firstly, we analyzed transcriptome structure of one susceptible strain to phosphine of S. oryzae and determined total length of transcripts being ~32Mb. From our initial sequencing of the S. oryzae transcriptome, we assembled and validated a full-length mRNA transcript of So_dld from the reference strain (S-strain). We then analyzed the progeny of a segregating genetic intercross between susceptible and strongly resistant strains to determine whether the resistance phenotype segregated with the So_dld allele of the strongly resistant strain. We also repeated the analysis with a weakly by strongly resistant intercross. RNA sequence of the dld gene from progeny that survived exposure to a high dose of phosphine clearly demonstrated inheritance of the DLD enzyme variant from the strongly resistant parent in the strongly resistant progeny. In contrast, there was no strong bias toward the DLD variant from the strongly resistant parent in siblings of the same crosses that had not been exposed to phosphine. Thus, the dld gene is clearly linked to strong resistance to phosphine in S. oryzae.
We found that the DLD sequences of the strongly resistant strains collected from Vietnam, China, and Australia, all carried the exact same missense mutation in the gene that resulted in a N505T amino acid variant in the encoded protein. A resistance variant is also found at the corresponding position of the DLD enzyme of strongly phosphine resistant R. dominica, N506H (Schlipalius et al. 2012). Although all 4 strongly resistant strains of S. oryzae shared the common N505T variant, other polymorphisms in the DNA sequence of the dld gene demonstrated that the resistance allele of each strain arose independently, as would have been expected due to the geographically diverse sites of origin. The finding of only one resistance variant in S. oryzae is quite unique, as multiple variants of DLD have been found to cause resistance in R. dominica (Schlipalius et al. 2012). In addition, the (P49S) variant of R. dominica and the corresponding (P45S) variant of T. castaneum is very common in strains of these species collected from India, Turkey, USA, and Australia (Schlipalius et al. 2012; Kaur et al. 2015; Koçak et al. 2015; Chen et al. 2015), but is not in any S. oryzae strains in the present study.
The DLD enzyme has 4 characterized and functionally distinct domains: a binding site for the FAD cofactor, a binding site for the NAD+ substrate, an active site disulfide and a domain required for homodimer formation. When 2 monomers dimerise they form a pore which leads from the surface of the protein to the active site disulfide (Brautigam et al. 2005). The N505T amino acid polymorphism found in S. oryzae is located at carboxyl terminus of the DLD polypeptide but in the quaternary structure it contributes to the pore near the active site disulfide. All phosphine resistance variants of DLD reported in insect pests including S. oryzae have been amino acid substitutions and nearly all of them are located in the pore or near the active site of the enzyme. Results with S. oryzae support the hypothesis (Schlipalius et al. 2012) that altered function of the active site disulfide bond is key to resistance and may be the target site of phosphine.
In order to confirm the strong linkage of So_dld to phosphine resistance, we genotyped the progeny of 2 single-pair intercrosses between either a susceptible or weakly resistant strain and a strongly resistant strain, one at the F16 generation and the other at the F18 generation. 100% of 95 or 52 phosphine resistant progeny were homozygous for the N505T variant amino acid which placed it within a locus spanning 1 or 2 genes (0.022 cM). Based on this tight linkage and the similarity between the candidate resistance variant in S. oryzae and those of other species, we concluded that strong resistance to phosphine involves variants in the dld gene in S. oryzae, which has now been observed in 3 pest insect species.
We observed a fitness cost associated with the resistance variant in a segregating population in the absence of phosphine selection over more than 15 generations. This was most strongly observed as a significant loss of the resistance allele across the generations. The decrease in the frequency of the resistance allele in progeny from the WR × SRAus intercross was observed more consistently than the decrease observed in progeny from the S-strain × SRAus intercross. A previous study based on the resistance phenotype of populations derived from an intercross between the weakly resistant and susceptible strains also used in the current study detected no apparent fitness cost of the weak allele over 7 generations without selection pressure (Daglish et al. 2014). Jagadeesan et al. (2012, 2013) employed both phenotype and genotype approaches to analyze fitness cost in resistant insects of T. castaneum and found a strong fitness cost associated with tc_rph2 but selective advantage for tc_rph1. In contrast, a fitness cost was not observed for a resistance allele of rph2 in the absence of phosphine exposure in R. dominica (Collins et al. 2001; Schlipalius et al. 2008). Although they did not evaluate directly mortality of resistant individuals over generations, other studies tested respiration, developmental, and population growth rates to analyze fitness of resistant phenotypes compared with susceptible strains in R. dominica, T. castaneum, and Oryzaephilus surinamensis (Pimentel et al. 2007; Sousa et al. 2009). These studies indicated that a higher level of resistance to phosphine was accompanied by lower rates of development and respiration. This is consistent with resistance being associated with DLD, a key enzyme of energy metabolism, which suggests that resistance would likely be associated with a strong fitness cost. The presence of fitness costs associated with resistance dictates that the frequency of the resistance allele will decrease in a mixed population in the absence of selection. Based on this principle, management techniques such as rotation of chemicals (Tabashnik 1990) may be effective at managing resistance. Effective resistance management strategies will benefit from regular monitoring. Our present study has contributed towards the development of molecular markers for strong resistance to phosphine in S. oryzae so that reliable assessments of resistance can be performed rapidly on insects collected from field populations.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
Australia-India Strategic Research Fund Grand Challenge Grant (GCF010006 awarded to P.J.C. and P.R.E.); the MB phase-out project in Vietnam was funded by World Bank (GEF-TF055863 awarded to T.M.D.); an AusAid Postgraduate Scholarship (http://aid.dfat.gov.au/australia-awards/pages/studyin.aspx awarded to TTN).
Supplementary Material
Acknowledgments
We thank Dr Rajeswaran Jagadeesan for useful comments on the manuscript and Dr Raman Kaur for her advice regarding statistical analysis. We acknowledge Andrew Tuck for assistance with molecular analysis as well as Hervoika Pavic and Linda Bond for their assistance with selection and maintenance of insects.
References
- Abbott WS. 1925. A method of computing the effectiveness of an insecticide. J Econ Entomol. 18:265–267. [Google Scholar]
- Anon 1975. Recommended methods for detedtion and management of resistance of agricultural pests to pesticide. Tentative method for adults of some major pest species of stored cereals, with methyl bromide and phosphine. FAO Method No.16. FAO Plant Prot Bull. 23:12–26. [Google Scholar]
- Athié I, Gomes RAR, Bolonhezi S, Valentini SRT, De Castro MFPM. 1998. Effects of carbon dioxide and phosphine mixtures on resistant populations of stored-grain insects. J Stored Prod Res. 34:27–32. [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Beeman RW, Brown SJ. 1999. RAPD-based genetic linkage maps of Tribolium castaneum . Genetics. 153:333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengston M, Collins PJ, Daglish GJ, Hallman VL, Kopittke RM, Pavic H. 1999. Inheritance of phosphine resistance in Tribolium castaneum (Coleoptera: Tenebrionidae). J Econ Entomol. 92:17–20. [Google Scholar]
- Benhalima H, Chaudhry M, Mills K, Price N. 2004. Phosphine resistance in stored-product insects collected from various grain storage facilities in Morocco. J Stored Prod Res. 40:241–249. [Google Scholar]
- Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. 2005. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J Mol Biol. 350:543–552. [DOI] [PubMed] [Google Scholar]
- Chaudhry MQ. 2000. Phosphine resistance. Pesticide Outlook. 11:88–91. [Google Scholar]
- Chen Z, Schlipalius D, Opit G, Subramanyam B, Phillips TW. 2015. Diagnostic molecular markers for phosphine resistance in US populations of Tribolium castaneum and Rhyzopertha dominica . PLoS One. 10:e0121343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins P, Daglish G, Nayak M, Ebert P, Schlipalius D, Chen W, Pavic H, Lambkin TM, Kopittke R, Bridgeman B. 2001. Combating resistance to phosphine in Australia. In: Donahaye EJ, Navarro S, Leesch JG, editors. International conference on controlled atmoshphere and fumigation in stored products, Fresno, CA, p. 593–607. [Google Scholar]
- Collins PJ, Daglish GJ, Bengston M, Lambkin TM, Pavic H. 2002. Genetics of resistance to phosphine in Rhyzopertha dominica (Coleoptera: Bostrichidae). J Econ Entomol. 95:862–869. [DOI] [PubMed] [Google Scholar]
- Cotton R. 1920. Rice Weevil (Calandra) Silophilus oryzae . J Agric Res. 20:409–422. [Google Scholar]
- Daglish GJ, Collins PJ, Pavic H, KOPITTKE RA. 2002. Effects of time and concentration on mortality of phosphine-resistant Sitophilus oryzae (L) fumigated with phosphine. Pest Manag Sci. 58:1015–1021. [DOI] [PubMed] [Google Scholar]
- Daglish GJ, Nayak MK, Pavic H. 2014. Phosphine resistance in Sitophilus oryzae (L.) from eastern Australia: Inheritance, fitness and prevalence. J Stored Prod Res. 59:237–244. [Google Scholar]
- Finney DJ. 1971. Probit analysis, 3rd edn Cambridge, UK: University Printing House. [Google Scholar]
- Halstead DGH. 1963. External sex differences in stored-products Coleoptera. Bull Entomol Res. 54:119–134. [Google Scholar]
- Jagadeesan R, Collins PJ, Daglish GJ, Ebert PR, Schlipalius DI. 2012. Phosphine resistance in the Rust Red Flour Beetle, Tribolium castaneum (Coleoptera: Tenebrionidae): inheritance, gene interactions and fitness costs. Plos One. 7:e31582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesan R, Fotheringham A, Ebert PR, Schlipalius DI. 2013. Rapid genome wide mapping of phosphine resistance loci by a simple regional averaging analysis in the red flour beetle, Tribolium castaneum . BMC Genomics. 14:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R. 2012. Molecular genetics and ecology of phosphine resistance in Lesser Grain Borer, Rhyzopertha dominica (F.) (Coleoptera: Bostrichidae). PhD thesis, University of Queensland, St Lucia, Australia. [Google Scholar]
- Kaur R, Schlipalius DI, Collins PJ, Swain AJ, Ebert PR. 2012. Inheritance and relative dominance, expressed as toxicity response and delayed development, of phosphine resistance in immature stages of Rhyzopertha dominica(F.) (Coleoptera: Bostrichidae). J Stored Prod Res. 51:74–80. [Google Scholar]
- Kaur R, Subbarayalu M, Jagadeesan R, Daglish GJ, Nayak MK, Naik HR, Ramasamy S, Subramanian C, Ebert PR, Schlipalius DI. 2015. Phosphine resistance in India is characterised by a dihydrolipoamide dehydrogenase variant that is otherwise unobserved in eukaryotes. Heredity. 115:188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koçak E, Schlipalius D, Kaur R, Tuck A, Ebert P, Collins P, Yılmaz A. 2015. Determining phosphine resistance in rust red flour beetle, Tribolium castaneum (Herbst.) (Coleoptera: Tenebrionidae) populations from Turkey. Turk J Entomol. 39:129–136. [Google Scholar]
- Li Y, Li W. 1994. Inheritance of phosphine resistance in Sitophilus oryzae (L.) (Coleoptera, Curculionidae). In Highley E Wright E. Banks HJ and Champ BR, editors. Stored Products Protection: Proceeding of the 6th International Working Conference on Stored-Product Protection, 1994. Apr 17–23, Canberra, Australia: CAB International, Wallingford, UK, 113–115. [Google Scholar]
- Mau YS, Collins PJ, Daglish GJ, Nayak MK, Ebert PR. 2012. a. The rph2 gene is responsible for high level resistance to phosphine in independent field strains of Rhyzopertha dominica . Plos One. 7:e34027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mau YS, Collins PJ, Daglish GJ, Nayak MK, Pavic H, Ebert PR. 2012. b. The rph1 Gene Is a Common Contributor to the Evolution of Phosphine Resistance in Independent Field Isolates of Rhyzopertha dominica . Plos One. 7:e31541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Pau J, Gómez A, MUÑOZ J. 2008. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnol Oceanogr Methods. 6: 218–222. [Google Scholar]
- Nguyen TT, Collins PJ, Ebert PR. 2015. Inheritance and characterization of strong resistance to phosphine in Sitophilus oryzae (L.). PloS One. 10:e0124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel MA, Faroni LR, Totola MR, Guedes RN. 2007. Phosphine resistance, respiration rate and fitness consequences in stored-product insects. Pest Manag Sci. 63:876–81. [DOI] [PubMed] [Google Scholar]
- Preisler HK, Hoy MA, Robertson JL. 1990. Statistical analysis of modes of inheritance for pesticide resistance. J Econ Entomol. 83:1649–1655. [Google Scholar]
- Rajendran S. 1999. Phosphine resistance in stored insect pests in India In: Jin ZL, Liang Y, Tan X, Guan L, editors. Proceeding of the seventh International Working Conference on Stored-product Protection, Oct 14–19. Beijing, China: Sichuan Publishing House of Science and Technology, Chengdu, China, p. 635–641. [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ. 2008. The genome of the model beetle and pest Tribolium castaneum . Nature 452:949–955. [DOI] [PubMed] [Google Scholar]
- Robertson JL, Preisler HK. 1992. Pesticide bioassays with arthropods. Boca Raton: CRC Press. [Google Scholar]
- Schlipalius D, Waldron J, Carroll B, Collins P, Ebert P. 2001. A DNA fingerprinting procedure for ultra high‐throughput genetic analysis of insects. Insect Mol Biol. 10:579–585. [DOI] [PubMed] [Google Scholar]
- Schlipalius DI, Chen W, Collins P, Nguyen T, Reilly P, Ebert P. 2008. Gene interactions constrain the course of evolution of phosphine resistance in the lesser grain borer, Rhyzopertha dominica . Heredity 100:506–516. [DOI] [PubMed] [Google Scholar]
- Schlipalius DI, Cheng Q, Reilly PEB, Collins PJ, Ebert PR. 2002. Genetic linkage analysis of the lesser grain borer Rhyzopertha dominica identifies two loci that confer high-level resistance to the fumigant phosphine. Genetics. 161:773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlipalius DI, Valmas N, Tuck AG, Jagadeesan R, Ma L, Kaur R, Goldinger A, Anderson C, Kuang J, Zuryn S. 2012. A core metabolic enzyme mediates resistance to phosphine gas. Science. 338:807–810. [DOI] [PubMed] [Google Scholar]
- Sousa A, Faroni LDA, Pimentel M, Guedes R. 2009. Developmental and population growth rates of phosphine-resistant and-susceptible populations of stored-product insect pests. J Stored Prod Res. 45:241–246. [Google Scholar]
- Tabashnik BE. 1990. Modeling and evaluation of resistance management tactics. In: Roush RT, Tabashnik BE, editors. Pesticide resistance in arthropods. Springer: p 153–182. [Google Scholar]
- Zeng L. 1999. Development and countermeasures of phosphine resistance in stored grain insects in Guangdong of China. In: Jin ZLQ, Liang Y, Tan X, Guan L, editors. Proceedings of the 7th International Working Conference on Stored-product Protection, Oct 14–19 Beijing, China: Sichuan Publishing House of Science and Technology, Chengdu, China, p. 642–647. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.