Abstract
A diversity reference set has been constructed for the Gossypium accessions in the US National Cotton Germplasm Collection to facilitate more extensive evaluation and utilization of accessions held in the Collection. A set of 105 mapped simple sequence repeat markers was used to study the allelic diversity of 1933 tetraploid Gossypium accessions representative of the range of diversity of the improved and wild accessions of G. hirsutum and G. barbadense. The reference set contained 410 G. barbadense accessions and 1523 G. hirsutum accessions. Observed numbers of polymorphic and private bands indicated a greater diversity in G. hirsutum as compared to G. barbadense as well as in wild-type accessions as compared to improved accessions in both species. The markers clearly differentiated the 2 species. Patterns of diversity within species were observed but not clearly delineated, with much overlap occurring between races and regions of origin for wild accessions and between historical and geographic breeding pools for cultivated accessions. Although the percentage of accessions showing introgression was higher among wild accessions than cultivars in both species, the average level of introgression within individual accessions, as indicated by species-specific bands, was much higher in wild accessions of G. hirsutum than in wild accessions of G. barbadense. The average level of introgression within individual accessions was higher in improved G. barbadense cultivars than in G. hirsutum cultivars. This molecular characterization reveals the levels and distributions of genetic diversity that will allow for better exploration and utilization of cotton genetic resources.
Key words: germplasm collection, Gossypium barbadense, Gossypium hirsutum, introgression, population structure
An understanding of the genetic diversity of cotton (Gossypium spp.) as represented in the US National Cotton Germplasm Collection is essential to develop strategies for collecting, conserving, and utilizing these germplasm resources. Of the 50 plus species of the Gossypium genus (Fryxell 1992; Wendel and Cronn 2003), the most accessible for genetic improvement efforts are the cultivated tetraploid species G. hirsutum and G. barbadense residing in the primary gene pool (Harlan and de Wet 1971). These 2 species together constitute approximately 75% of the content of the United States Department of Agriculture’s National Cotton Germplasm Collection (NCGC). Efforts to describe and classify the diversity of these species within the NCGC predate genetic analyses and began with the collection of phenotypic descriptor data using standardized descriptors (IBPGR 1980; Percival 1987; Toll 1995).
Early efforts to characterize G. hirsutum genetic diversity using isozyme and restriction fragment length polymorphism (RFLP) data revealed 2 centers of diversity for that species, one in the southern Mexico-Guatemala region and the other in the Caribbean, with variation correlated to geographical regions (Wendel et al. 1992; Brubaker and Wendel 1994). These investigations revealed that a notable genetic bottleneck occurred in the development of modern high-yielding cultivars. Further, the infraspecific division of G. hirsutum into 7 races by Hutchinson (1951), partially defined by geography, did not adequately explain observed genetic relationships. However, a recent genetic evaluation with simple sequence repeat (SSR) markers of germplasm from the CIRAD collection supported the existence of 7 races (Lacape et al. 2007).
In recent years, the majority of genetic diversity studies in cotton have focused on improved G. hirsutum cultivars, as they dominate world cotton production. These studies vary in the scope of germplasm and number of markers assayed. Tyagi et al. (2014) examined 378 upland cultivars spanning a century of cotton breeding in the United States with 120 SSR markers and identified 4 groups of germplasm that corresponded to southeastern, midsouth, southwest, and western regions of US cotton production. However, extensive admixture was observed in the cultivars. A similar study used a larger number of SSR markers (448) to examine relationships between 193 upland cultivars from 26 countries, with most cultivars originating in the United States (Fang et al. 2013). A larger number of genetic groups were identified and inferences were made based on breeding history and pedigrees rather than country of origin. Several other studies have reported various aspects of the genetic diversity of upland cotton in the NCGC, generally with a smaller number of cultivars and fewer markers (Abdalla et al. 2001; Hinze et al. 2012).
There have been fewer reported molecular analyses focusing on G. barbadense genetic diversity than there have been for G. hirsutum. One of the earliest studies used isozymes to estimate diversity and region of origin from a set of 153 G. barbadense accessions, primarily from the NCGC (Percy and Wendel 1990). This study identified geographic clusters and suggested northwestern South America as the center of diversity for the species. Four categories were proposed describing the continuum of improvement observed in G. barbadense accessions: 1) wild, 2) dooryard or commensal, 3) landraces, and 4) improved modern cultivars (Percy and Wendel 1990; Percy 2009). A subsequent study using DNA-based markers included 63 of the accessions analyzed by Percy and Wendel (1990) as well as 66 newly collected accessions from Peru, both east and west of the Andes (Westengen et al. 2005). This report further narrowed down the center of domestication and diversity of G. barbadense to the coastal region of northwestern Peru and southwestern Ecuador.
Although various genetic investigations with varying goals have been conducted on G. hirsutum and G. barbadense using accessions provided by the NCGC, there has been no comprehensive attempt to examine the diversity of these species as represented in the NCGC. The current investigation was conducted utilizing a carefully constructed reference set of germplasm representing the known diversity of the NCGC and a core set of 105 SSR markers evenly distributed across the 26 chromosomes of cotton (Yu et al. 2012). The 2256 accessions selected to create the Gossypium Diversity Reference Set (GDRS) (Hinze et al. 2015a) represent approximately 20% of the accessions held in the NCGC. A proportional strategy was used to develop the GDRS, such that the distribution of accessions within the GDRS captures the distribution of genomes, species, and intraspecific groups present within the NCGC (Brown 1989). Approximately 24.5% of the G. hirsutum accessions in the NCGC are present in the GDRS and 27.1% of G. barbadense accessions are included. The utility of the core set of markers in differentiating intra-genome variation and interspecific variation among tetraploid species has been previously reported within the broad context of all cotton genomes (Hinze et al. 2015a). However, a deeper and more comprehensive analyses of the interrelationships, and structure of genetic diversity of these commercial tetraploid species, as represented in the NCGC is warranted in view of the fact that they constitute over 98% of the world’s commercial production.
Although numerous investigations and considerable effort have gone into describing the diversity of a priori classifications, categories, or groups within G. barbadense and G. hirsutum, it has not been established whether these groups are actually useful in defining, describing, or utilizing the diversity of the germplasm collection. Our objectives were 1) to evaluate the genetic structure of G. barbadense and G. hirsutum within the NCGC, 2) to determine whether G. hirsutum races can be defined with SSR markers and are useful in classifying diversity seen in that species, 3) to re-examine the relationship between geography and genetic diversity within wild G. barbadense accessions in the collection, 4) to ascertain whether regionally and historically related breeding programs have created discernably distinctive germplasm pools useful in classifying germplasm collection diversity, and 5) to measure levels of introgression within the 2 species, its contribution to overall diversity, and its usefulness as a tool when measuring collection purity.
Materials and Methods
Germplasm
This investigation analyzed the genetic diversity of 1933 tetraploid accessions of the Gossypium Diversity Reference Set (Supplementary Table S1 online) (Hinze et al. 2015a). These accessions included 410 G. barbadense accessions and 1523 G. hirsutum accessions. Thirty-six accessions that were deemed “misclassified” in an earlier report (Hinze et al. 2015a) were removed prior to the current analysis. Improved and wild types of G. barbadense (150 and 213 accessions, respectively) and G. hirsutum (657 and 831 accessions, respectively) were compared. In this investigation, improved G. hirsutum and G. barbadense accessions have been defined as those accessions having received the greatest human manipulation (generally through intentional, systematic breeding techniques) while wild types have been defined to include truly wild accessions, dooryard cottons, and landraces (Percy and Wendel 1990). Analytical techniques have been employed that utilize a priori assignment of accessions into geographic, historical, or breeding groups based upon previous investigations, knowledge, and theory. Accessions that could not be assigned to any group were labeled “n/a”.
Improved G. barbadense accessions have been classified into 7 categories that reflect historical and regionally distinct breeding efforts that for convenience have been given geographic names:
-
1)
Egypt/Sudan: a germplasm pool developed in northeast Africa that gave rise to the Giza or “Egyptian” cottons.
-
2)
French Northern Africa: a germplasm pool, distinct from the Egyptian germplasm, originating from French colonial efforts to develop cultivars adapted to North African colonies in Morocco and Algeria.
-
3)
Sub-Saharan West Africa: germplasm from the former French colonies of Togo and the Ivory Coast.
-
4)
Pima: germplasm created by the USDA in the southwestern United States, originally derived from Egyptian ancestry, but post-1950 was derived from a hybrid pool with Egyptian, Tanguis, Sea Island, and upland contributors.
-
5)
Peru: primarily composed of Tanguis cultivars, primarily of native origin, but having some hybridization with Egyptian and Pima germplasm.
-
6)
Sea Islands: an ancestor, directly or indirectly, to all modern G. barbadense breeding germplasm pools. Originating in the Caribbean and developed in the United States “Sea Islands” off the coasts of South Carolina, Georgia, and Florida; Sea Island cottons were re-introduced into the Caribbean in the 1920’s. Individual islands of the Caribbean have developed distinctive varieties of Sea Island.
-
7)
USSR (former): primarily originating in breeding programs of Uzbekistan, with a few accessions from neighboring Turkmenistan.
Wild types of G. barbadense have been assigned a priori to regional groups based on an earlier investigation of species diversity using isozymes (Percy and Wendel 1990). These 6 regions included West of Andes, East of Andes, Caribbean Islands, Argentina-Paraguay, Central America, and Pacific Islands (no accessions from the Pacific Islands were included in the current study).
Within improved G. hirsutum germplasm, a priori categorization was made first on a global scale and then on a national scale on the basis of historically and geographically distinct breeding efforts. Globally, improved germplasm was assigned to 9 historical/geographical groups or categories, given geographic names for convenience:
-
1)
Northern Africa: includes accessions originating from countries in North and Central Africa with the equator as the southern boundary. Breeding programs in these regions were initiated by English and French colonial interests, and this influence may persist in pedigrees.
-
2)
Southern Africa: this group includes germplasm developed in South Africa, Zimbabwe, Mozambique, Madagascar, and Zambia, and may have been heavily influenced by breeding efforts of the British Empire Cotton Growers Corporation.
-
3)
Europe: mostly European countries bordering the Mediterranean, including eastern Europe, Turkey, and Syria.
-
4)
China/Southeast Asia: improved germplasm originating from China and southeast Asia; grouped together because of China’s historically strong trading and cultural influence in the area and presumptive exchange of germplasm.
-
5)
South America: improved germplasm of presumptive native origin.
-
6)
Central Asia: improved germplasm of Central Asian origin, along with the Indian subcontinent (Iran, Afghanistan, Uzbekistan, Azerbaijan, Pakistan, and India).
-
7)
Mexico/Central America: improved germplasm originating in Mexico or Central America.
-
8)
United States: improved germplasm of US origin.
-
9)
Australia: improved germplasm of Australian origin.
Within the United States, improved G. hirsutum germplasm has been assigned to 4 historical/geographical breeding categories on the basis of regional location of breeding programs, and when possible, by the regional origins of a cultivar’s parentage or pedigree. In cases where an accession could be assigned to a known regional breeding program, but upon inspection of its pedigree it was deemed to be of mixed origin (i.e. the progeny of an eastern cultivar crossed to a western cultivar, etc.), then the accession was classified as “n/a”.
-
1)
Eastern: breeding programs located in North and South Carolina, Georgia, and Alabama that were associated with development of fiber quality and with wilt disease resistance (PeeDee, Coker, Meade, Auburn, etc.).
-
2)
Midsouth: also called delta types, these breeding programs bred for the Mississippi delta and surrounding areas of Arkansas and Louisiana (Deltapine, Stoneville, Delfos, DES, Dixie King, etc.).
-
3)
Plains: programs bred for the plains environments of Texas and Oklahoma that included prominent features such as storm proof bolls, earliness, and a degree of drought tolerance (Tamcot, Rowden, Lockett, Lankart, Paymaster, etc.).
-
4)
Western: programs bred for high fiber quality cottons grown under controlled irrigation. These breeding programs were centered in California, New Mexico, and Arizona (Acala, Germains, Hopi, etc.).
Analyses of unimproved G. hirsutum germplasm centered around 7 geographical races previously described for wild types of G. hirsutum (Hutchinson 1951). These analyses included a distinct group of “mocó” cotton (G. hirsutum race marie-galante), primarily of Brazilian origin (de Menezes et al. 2010). In addition to geographic races, Hutchinson (1951) described 11 geographic regions of genetic diversity and these also have been considered in our analyses of diversity.
A primary goal of the analyses utilized in our study was to determine if the various a priori classifications based upon origin, geography, and breeding history have utility in defining and describing the diversity within G. hirsutum and G. barbadense.
SSR Genotyping
SSR data was obtained as described in a genetic diversity study of the GDRS (Hinze et al. (2015a). Briefly, DNA was extracted from a bulk of 10 root tips representing each accession. Genotyping was accomplished utilizing a core set of 105 well characterized and genetically mapped SSR markers spread across the 26 chromosomes of the tetraploid genome at a frequency of 2 markers per chromosome arm (except chromosome 5 which had 5 total markers) (Yu et al. 2012). One of the SSR markers (TMB2955) was removed from the analysis after multiple PCR failures. Due to complexity of scoring with possible amplification of duplicate loci from both A and D subgenomes of the tetraploid species, and the presence of multiple alleles due to the possible heterogeneous and/or heterozygous nature of 10 individuals in bulked DNA samples, SSRs were analyzed as dominant markers where every distinct band was scored as present or absent in each accession.
Diversity Analysis
Summary statistics used to estimate genetic diversity and differentiation within and between G. barbadense and G. hirsutum were calculated in a manner similar to that of Hinze et al. (2015a). The “Frequency…” option and “binary (diploid)” data format in GenAlEx version 6.5b3 (Peakall and Smouse 2012) along with manual calculations in an Excel 2010 spreadsheet were used to calculate descriptive statistics based on band presence/absence data for individual accessions, species, improved and wild types, and regional or race categories within improved and wild types.
Private bands included those that were restricted to a single group regardless of their frequency. Species-specific bands were defined as bands that occurred at a frequency of 50% or higher in one species and 10% or lower in the other species in G. hirsutum-G. barbadense pairwise comparisons. This method is detailed in Hinze et al. (2015a) and was used to estimate levels of introgression within accessions, categories, and races of the study.
Principal coordinate analyses (PCoA) were conducted using a matrix of genetic similarity (GS) values calculated in NTSYS (Rohlf 2000). GS values were calculated using Jaccard’s coefficient (Jaccard 1908) which is commonly applied with dominant-type data where allele frequencies cannot be calculated (Reif et al. 2005). All monomorphic bands were removed prior to calculating GS values. PCoA was first conducted on the 2 commercial tetraploid species combined, then on G. hirsutum accessions and G. barbadense accessions, independently. In addition, the wild and improved types of both species also were evaluated independently.
To examine how the difference in sample size between both species affected the diversity measures, we implemented a custom python script to randomly downsample G. hirsutum (n = 1523 accessions) to the sample size of G. barbadense (n = 410 accessions). We performed 1000 random resamplings of 410 G. hirsutum accessions, while ensuring to match as closely as possible the number of accessions per group (i.e. wild, improved, n/a) observed in G. barbadense. For each iteration, we generated a subset of downsampled G. hirsutum and calculated 5 summary statistics: the number of polymorphic bands, the number of private bands, the percentage of polymorphic bands, the percentage of private bands, and the GS among G. hirsutum accessions. Because G. barbadense had more samples in the group “n/a” (n = 47) than G. hirsutum (n = 35), all G. hirsutum accessions for this group (“n/a”) were used in each subset. The rest of the subset was composed of randomly chosen 156 improved and 219 wild G. hirsutum accessions, for a final subset size of 410 G. hirsutum. As such, the composition of the downsampled G. hirsutum subsets (35n/a, 156 improved, 219 wild) were very similar to the G. barbadense whole sample (47n/a, 150 improved, 213 wild). This allowed us to compare diversity statistics from both species at the exact same sample size (n = 410).
The number of distinct genetic pools that contributed to the extant improved accessions of G. barbadense and G. hirsutum was inferred with the software STRUCTURE version 2.3.4 (Pritchard et al. 2000). The program was run using the admixture model assuming that loci are unlinked, and with no prior regarding the population of origin or the sampling location. For each species, we ran several models assuming an increasing number of clusters K (from 1 to 8) in the population, with 20 replicates for each model. We used the method described by Evanno et al. (2005) to select the model with the uppermost hierarchical level of structure (hereafter referred to as “best model”). Accessions were a posteriori classified into distinct groups based on their estimated membership to one of the K clusters of the best model. Two different thresholds for membership were used. First, to follow the methodology used by Tyagi et al. (2014), an accession was classified as belonging to cluster K i when the genetic contribution of cluster K i to this accession was greater than 70%; otherwise the accession was deemed admixed. Second, because most accessions were admixed, we used less stringent criteria to evaluate the major genetic contribution of most accessions. An accession was mainly derived from cluster K i if its genetic background comprised at least 20% more of cluster K i than any other cluster.
Data Archiving
In fulfillment of data archiving guidelines (Baker 2013), we have deposited the primary data underlying these analyses in the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.0bn55.
Results
Descriptive Analysis of Genetic Diversity
All 104 SSR markers were polymorphic in both G. barbadense and G. hirsutum, with the exception of DPL0600, which was monomorphic in G. barbadense (Table 1). The number of bands detected by each SSR marker was used to assess the level of heterogeneity within these germplasm accessions. An average of 1.6 bands per marker was observed across all accessions with a maximum of 8 bands detected at the BNL1531 locus for both GB-1388 (G. barbadense improved type from Peru) and TX-1900 (G. hirsutum wild type from Venezuela) (Supplementary Table S1 online). An average of 3 or more alleles per accession were detected for the SSR markers BNL1531, TMB1640, BNL3545, and CIR218.
Table 1.
Summary of SSR marker diversity for the combined dataset including G. barbadense and G. hirsutum. Standard deviations are indicated in parenthesis
| G. barbadense | G. hirsutum | Overall | |
|---|---|---|---|
| Sample size | 410 | 1523 | 1933 |
| No. of polymorphic SSRs | 103 | 104 | 104 |
| No. of polymorphic bands | 827 | 1192 | 1302 |
| Percentage of polymorphic bands (%) | 63.0 | 90.8 | 99.2 |
| No. of private bands | 109 | 463 | n/a |
| Percentage of private bands (%) | 12.8 | 38.5 | n/a |
| Average no. of G. barbadense-specific bands/accession | 67.4 (6.31) | 1.7 (3.31) | n/a |
| Average no. of G. hirsutum-specific bands/accession | 2.3 (6.58) | 60.6 (13.56) | n/a |
G. hirsutum had more polymorphic bands (1192) and more private bands (463) than G. barbadense (827 and 109, respectively). A similar result was observed for the percentage of private bands. The percentage of bands that were private to G. hirsutum was almost 3 times higher than in G. barbadense (38.5% and 12.8%, respectively). To ensure that sample size was not the main driving factor explaining these differences, we downsampled G. hirsutum to 410 samples and reanalyzed the comparison of band diversity. Results show that the number of polymorphic and private bands was still higher in a G. hirsutum sample (1014.8 and 363.6, respectively) of the same size as G. barbadense (Supplementary Table S2 online). Within both species, the wild or unimproved germplasm had a greater number of polymorphic and private bands than the improved germplasm. Collectively, wild accessions of G. barbadense had 226 private bands and an 88% rate of polymorphism compared to 30 private bands and 61% polymorphism in the improved germplasm (Table 2). In G. hirsutum, wild accessions produced 416 private bands and 93% polymorphism while improved germplasm had only 36 private bands and 55% polymorphism. The average GS between accessions of G. hirsutum (0.43) was lower than the average GS value of G. barbadense (0.57) (Supplementary Table S3 online). Using the downsampling scheme to account for differences in sample size, the GS value of G. hirsutum (0.435) remained unchanged compared to the original sample size. Within both species, the average GS value of improved accessions (0.63, G. barbadense; 0.66, G. hirsutum) was higher than the GS average for wild accessions (0.52, G. barbadense; 0.37, G. hirsutum).
Table 2.
Summary of SSR marker diversity for the improved and wild types of G. barbadense and G. hirsutum. Accessions with insufficient passport information or breeding history to classify as improved or wild were grouped into the “n/a” category. Standard deviations are indicated in parenthesis
| G. barbadense | G. hirsutum | |||||||
|---|---|---|---|---|---|---|---|---|
| Improved | Wild | n/a | Overall | Improved | Wild | n/a | Overall | |
| Sample size | 150 | 213 | 47 | 410 | 657 | 831 | 35 | 1523 |
| No. of polymorphic SSRs | 103 | 102 | 103 | 103 | 104 | 104 | 102 | 104 |
| No. of polymorphic bands | 519 | 749 | 462 | 827 | 666 | 1115 | 476 | 1192 |
| Percentage of polymorphic bands (%) | 61.1 | 88.1 | 54.4 | 97.3 | 55.3 | 92.6 | 39.5 | 99.0 |
| No. of private bands | 30 | 226 | 24 | n/a | 36 | 416 | 34 | n/a |
| Percentage of private bands (%) | 5.4 | 29.0 | 4.7 | n/a | 5.2 | 36.7 | 6.8 | n/a |
| Average no. of G. barbadense-specific bands/accession | 70.2 (4.18) | 64.8 (6.44) | 70.4 (6.39) | 67.4 (6.31) | 0.7 (1.57) | 2.6 (4.03) | 1.1 (2.28) | 1.7 (3.31) |
| Average no. of G. hirsutum- specific bands/accession | 2.5 (6.62) | 1.8 (4.96) | 3.7 (11.20) | 2.3 (6.58) | 72.3 (5.00) | 50.9 (10.20) | 70.2 (13.32) | 60.6 (13.60) |
G. barbadense
Within 7 previously recognized and described historical and geographic breeding pools of improved G. barbadense, the most diverse pool based on percent band polymorphism and percent private bands were cultivars from Egypt/Sudan (53.7% and 7.4%, respectively) and Peru (48.8% and 9%, respectively) (Table 3). Sea Island cultivars were slightly less polymorphic and had a slightly lower percentage of private bands, followed by American Pima cottons and cottons of Sub-Saharan West Africa and the former USSR. The above trends among germplasm pools were generally supported by average GS values within the germplasm pools. Peru had the lowest GS value (0.58), indicative of higher diversity within the pool, while Pima had the highest GS value (0.75).
Table 3.
Summary of SSR marker diversity within improved and wild G. barbadense groups. Any accessions that have ambiguous origins and/or parentage are included in the “n/a” category. Standard deviations are indicated in parenthesis
| Improved G. barbadense groups | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Egypt/Sudan | French North Africa | Peru | Pima | Sea Islands | Sub-Saharan West Africa | USSR (former) | n/a | Overall | |
| Sample size | 35 | 6 | 15 | 42 | 22 | 6 | 7 | 17 | 150 |
| No. of polymorphic SSRs | 95 | 69 | 94 | 81 | 88 | 76 | 77 | 100 | 103 |
| No. of polymorphic bands | 298 | 108 | 271 | 202 | 222 | 141 | 129 | 322 | 519 |
| Percentage of polymorphic bands (%) | 53.7 | 19.5 | 48.8 | 36.4 | 40.0 | 25.4 | 23.2 | 58.0 | 93.5 |
| No. of private bands | 27 | 1 | 32 | 13 | 20 | 12 | 8 | 41 | n/a |
| Percentage of private bands (%) | 7.4 | 0.4 | 9.0 | 4.4 | 6.6 | 4.9 | 3.3 | 10.4 | n/a |
| Average no. of G. barbadense-specific bands/accession | 71.9 (4.84) | 73.0 (1.10) | 65.9 (4.54) | 70.1 (2.19) | 70.3 (4.12) | 68.7 (4.41) | 70.6 (3.41) | 69.9 (4.71) | 70.2 (4.18) |
| Average no. of G. hirsutum- specific bands/accession | 3.4 (9.29) | 0.7 (0.82) | 4.1 (7.10) | 0.8 (0.92) | 2.4 (3.66) | 1.0 (0.63) | 1.6 (1.40) | 5.3 (11.69) | 2.5 (6.62) |
| Wild G. barbadense groups | |||||||||
| Argentina-Paraguay | Caribbean Islands | Central America | East of Andes | West of Andes | n/a | Overall | |||
| Sample size | 20 | 37 | 23 | 29 | 87 | 17 | 213 | ||
| No. of polymorphic SSRs | 98 | 92 | 88 | 92 | 102 | 88 | 102 | ||
| No. of polymorphic bands | 387 | 365 | 242 | 351 | 653 | 264 | 749 | ||
| Percentage of polymorphic bands (%) | 49.7 | 46.9 | 31.1 | 45.1 | 83.8 | 33.9 | 96.1 | ||
| No. of private bands | 26 | 12 | 1 | 20 | 173 | 5 | n/a | ||
| Percentage of private bands (%) | 6.0 | 2.9 | 0.3 | 4.8 | 25.0 | 1.5 | n/a | ||
| Average no. of G. barbadense-specific bands/accession | 62.0 (8.10) | 67.6 (4.69) | 67.6 (3.27) | 67.1 (3.27) | 62.2 (7.27) | 67.7 (3.37) | 64.8 (6.44) | ||
| Average no. of G. hirsutum- specific bands/accession | 4.3 (9.54) | 1.3 (1.70) | 1.0 (1.26) | 0.7 (0.97) | 2.2 (6.00) | 1.2 (1.51) | 1.8 (4.96) | ||
Among previous geographically defined germplasm pools of wild G. barbadense that encompass the species’ native range (Percy and Wendel 1990), the west of Andes region was by far the most diverse with 84% polymorphic and 25% private bands. The Argentina–Paraguay region and the east of Andes region were basically equivalent in band polymorphism and private bands. Caribbean accessions, while displaying band polymorphism (47%) equivalent to the Argentina–Paraguay and the east of Andes regions, had a lower frequency of private bands. With 31% polymorphic bands and 0.3% private bands, Central American G. barbadense germplasm was the least diverse. GS coefficients of regional germplasm pools reflected trends noted in polymorphic and private bands.
G. hirsutum
For the purpose of this investigation, global cultivars were assigned to 9 regional groups assumed to have sufficiently distinct regional breeding objectives, histories, and germplasm. The United States and central Asia regions had the highest percentage of polymorphic bands (86% and 45%, respectively) and the United States cultivar region had a notably higher percentage of private bands (31.6%) than central Asia (2.4%) or any other global region (Supplementary Table S4 online). When the historical/geographical breeding regions in the United States were analyzed independently, cultivars of the mid-South region showed the most diversity (63% polymorphism and 16.7% private bands) (Table 4). The eastern, plains, and western United States regions were relatively similar in terms of diversity (39–46% polymorphism and 5.3–8.4% private bands). The Jaccard GS coefficients indicated that all 4 regions in the United States had nearly identical levels of diversity (0.65–0.66) (Supplementary Table S3 online).
Table 4.
Summary of SSR marker diversity within improved and wild G. hirsutum groups. The improved group only includes regions in the United States. Any accessions that have ambiguous origins and/or parentage are included in the “n/a” category. Standard deviations are indicated in parenthesis
| Improved US G. hirsutum groups | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eastern | Midsouth | Plains | Western | n/a | Overall | |||||
| Sample size | 69 | 89 | 73 | 49 | 92 | 372 | ||||
| No. of polymorphic SSRs | 96 | 103 | 93 | 99 | 104 | 104 | ||||
| No. of polymorphic bands | 286 | 395 | 244 | 275 | 393 | 596 | ||||
| Percentage of polymorphic bands (%) | 45.6 | 63.0 | 38.9 | 43.9 | 62.7 | 95.1 | ||||
| No. of private bands | 25 | 73 | 17 | 29 | 74 | n/a | ||||
| Percentage of private bands (%) | 7.1 | 16.7 | 5.3 | 8.4 | 16.3 | n/a | ||||
| Average no. of G. barbadense-specific bands/accession | 0.6 (0.80) | 0.4 (0.94) | 0.5 (0.75) | 0.9 (0.71) | 1.3 (3.59) | 0.7 (1.57) | ||||
| Average no. of G. hirsutum-specific bands/accession | 72.0 (4.57) | 71.8 (7.10) | 72.6 (4.27) | 72.5 (5.37) | 73.1 (3.18) | 72.3 (5.00) | ||||
| Wild G. hirsutum groups | ||||||||||
| latifolium | marie-galante | mocó | morrilli | palmeri | punctatum | richmondi | yucatanense | n/a | Overall | |
| Sample size | 82 | 60 | 31 | 14 | 10 | 17 | 9 | 4 | 604 | 831 |
| No. of polymorphic SSRs | 102 | 103 | 104 | 99 | 101 | 102 | 99 | 95 | 104 | 104 |
| No. of polymorphic bands | 648 | 684 | 549 | 389 | 388 | 445 | 329 | 269 | 1090 | 1115 |
| Percentage of polymorphic bands (%) | 57.2 | 60.4 | 48.5 | 34.3 | 34.2 | 39.3 | 29.0 | 23.7 | 96.2 | 98.4 |
| No. of private bands | 10 | 2 | 4 | 3 | 0 | 1 | 3 | 0 | 225 | n/a |
| Percentage of private bands (%) | 1.5 | 0.3 | 0.7 | 0.7 | 0.0 | 0.2 | 0.8 | 0.0 | 20.3 | n/a |
| Average no. of G. barbadense-specific bands/accession | 1.0 (1.83) | 2.7 (2.57) | 7.9 (2.85) | 1.9 (1.17) | 2.4 (2.12) | 1.8 (1.19) | 1.0 (1.22) | 3.5 (1.73) | 2.6 (4.34) | 2.6 (4.03) |
| Average no. of G. hirsutum- specific bands/accession | 58.6 (9.43) | 46.6 (8.58) | 46.8 (5.83) | 52.6 (7.01) | 52.3 (10.61) | 49.7 (9.25) | 54.1 (6.41) | 44.8 (4.57) | 50.5 (10.26) | 50.9 (10.20) |
Comparisons of the 7 geographical races described by Hutchinson (1951), as well as a Brazilian mocó cotton, were made within unimproved G. hirsutum germplasm. Mocó cotton is a distinctive arborescent shrub with a geographic distribution limited to semi-arid regions of northeastern Brazil (de Menezes et al. 2010). Of the 8 categories, latifolium (82 accessions) and marie-galante (60 accessions) are best represented in the NCGC and correspondingly, in the current dataset (Table 4). In contrast, yucatanense is only represented here by 4 accessions. Therefore, results of the comparisons within unimproved germplasm may be skewed by the unequal representation of the races within the collection. Races marie-galante and latifolium were generally similar in terms of polymorphism (60.4% and 57.2%, respectively). Latifolium had the highest percentage private bands (1.46%) among races. All races produced low percentages of private bands when compared with percentages of private bands among cultivar groups.
When these unimproved accessions were evaluated using the regional assignments outlined by Hutchinson (1951), the region comprising eastern Guatemala and El Salvador produced the highest percentage of polymorphic bands (54%) and private bands (1.5%), followed by the region of Yucatan and Campeche (43.3% polymorphism) (Supplementary Table S5 online). The extremely large set of unclassified wild accessions (i.e. “n/a”) not assigned to races or regions exhibited high levels of polymorphism and many unique alleles within undomesticated wild G. hirsutum. Those accessions that have not been classified into a race category have an astounding 225 (20%) private bands, while accessions originating outside of Mexico and Central America have 292 private bands (26.4%), indicative of much diversity waiting to be explored and utilized in those accessions with little historical or passport information.
Multivariate Analysis of Genetic Diversity
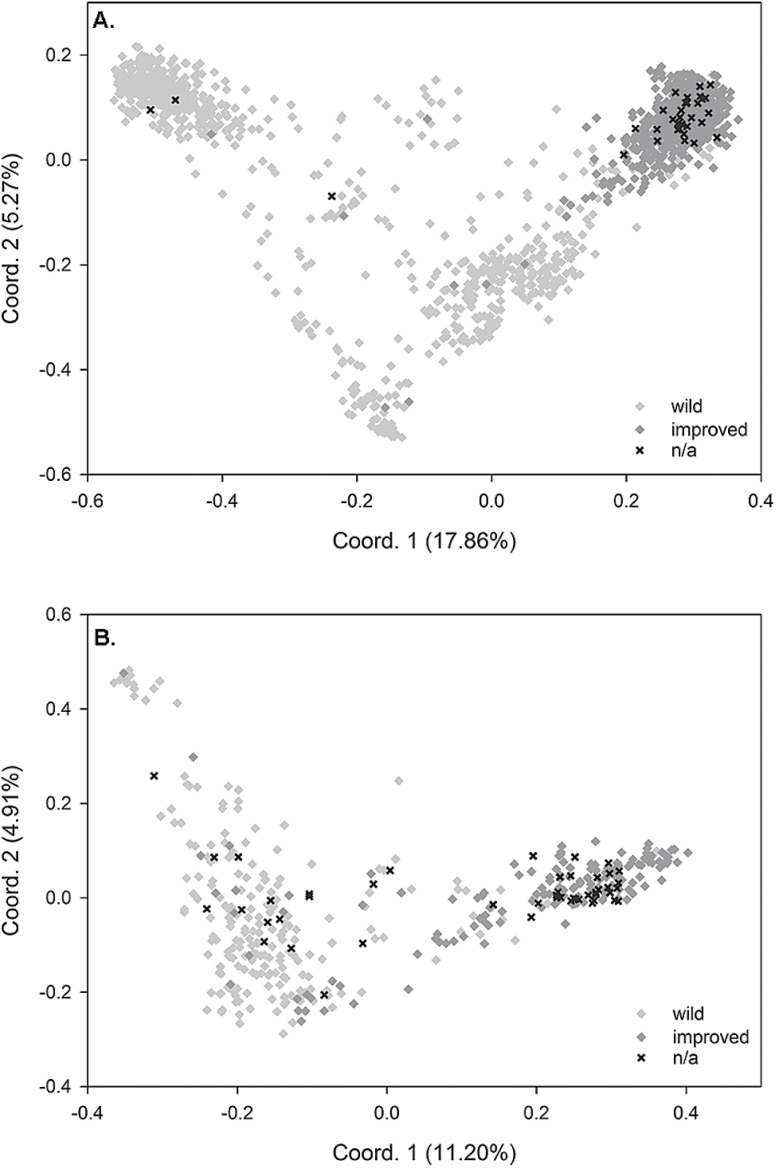
Performing a PCoA on the core set of SSR markers, we observed a distinct pattern of separation between G. barbadense and G. hirsutum accessions (Figure 1). The region between the G. hirsutum and G. barbadense clusters contained putative introgressed accessions, with introgression being supported by analyses of species-specific alleles within accessions of the 2 species. Upon independent evaluation of the G. hirsutum and G. barbadense accessions, we observed that the first principal coordinate separated wild and improved accessions into fairly distinct clusters, with some overlap occurring (Figure 2). In both species, the wild accessions tended to form broad clusters (much broader in G. hirsutum) that would indicate increased diversity relative to improved accessions. Improved accessions, especially for G. hirsutum, formed smaller, tighter groups of accessions with perceived greater similarity than wild accessions. Of interest was the fact that previously unclassified accessions of G. hirsutum overwhelmingly clustered with improved germplasm. This trend, though not as strong, seemed to be present in G. barbadense as well.
Figure 1.
Principal coordinate plot of the first 2 coordinate axes showing patterns of separation between G. hirsutum and G. barbadense that reflect 31.2% of the total variation. Thirty-six putative misclassified accessions were removed prior to analyses.
Figure 2.
Principal coordinate plots showing patterns of separation between wild and improved accessions within (A) G. hirsutum explaining 23.13% of the variation and (B) G. barbadense explaining 16.11% of the variation. Accessions which could not be classified into either category are labeled “n/a”.
G. barbadense
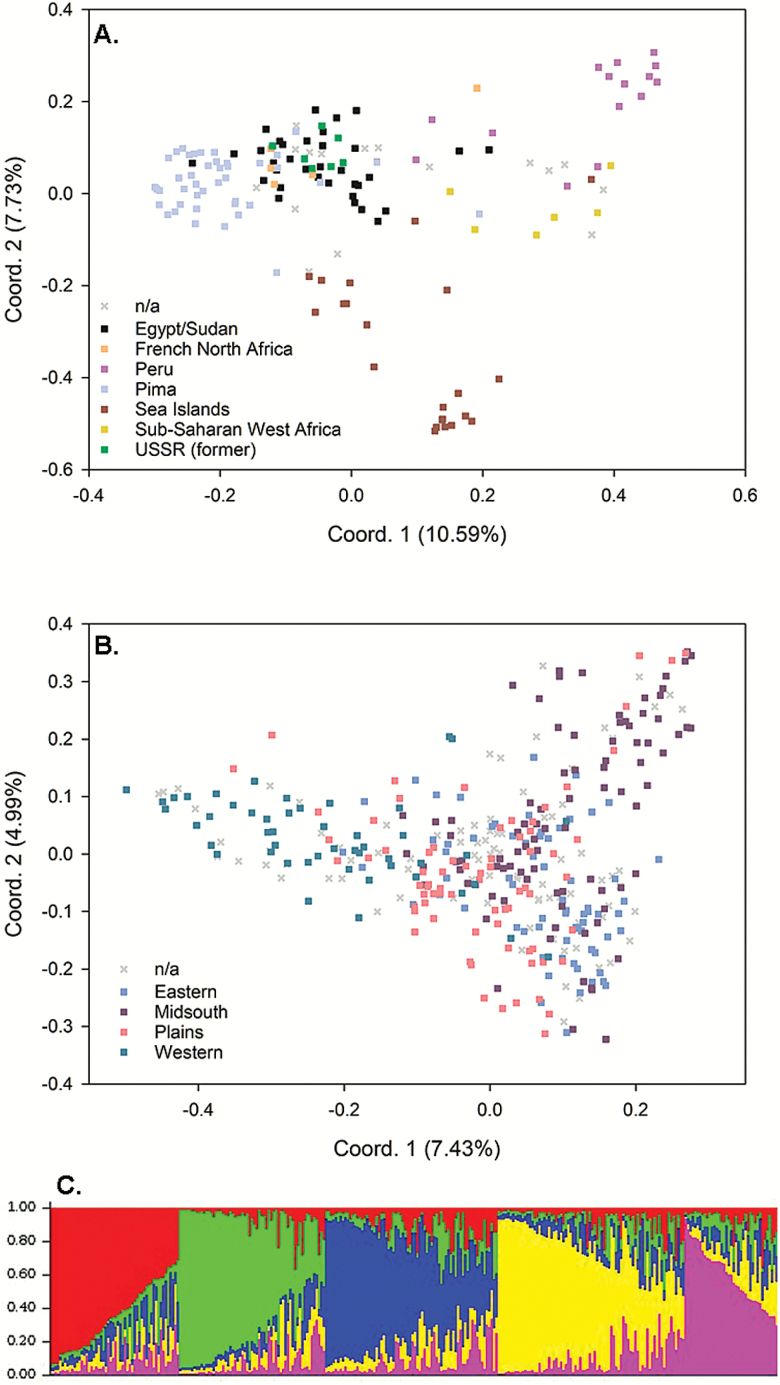
In analysis of improved G. barbadense, the first principal coordinate tended to separate Peruvian cultivars from all other cultivar groups (Figure 3A). The y-axis then further separated most accessions originating in the Sea Islands from the remaining categories. Cultivars of the Pima, Egyptian, former USSR and French North African groups tended to form a large cluster, with Pima cottons forming a slightly more distant, tighter cluster. The best STRUCTURE model (run without prior knowledge of G. barbadense breeding groups), separated the accessions into 2 significantly differentiated genetic pools (K = 2; Supplementary Figure S1 online). Accessions from Peru and from sub-Saharan West Africa had genetic backgrounds that were notably different from the remaining accessions. Forcing population priors on the model created a third significant group (K = 3 is the best model, not shown). The 3 genetic groups corresponded to Sea Island cultivars, Pima cultivars, and cultivars from Peru with the remaining accessions comprised of admixtures of those 3 genetic pools.
Figure 3.
Analysis of population structure for improved Gossypium accessions. Principal coordinate plots showing patterns of separation within (A) G. barbadense explaining 18.32% of the variation and (B) G. hirsutum from the United States explaining 12.42% of the variation. Accessions which could not be classified into a category are labelled “n/a”. (C) Results of improved US G. hirsutum genetic analysis without prior population assignment. Each accession’s genome is symbolized by a thin vertical line divided into colored segments representing the proportion of empirically defined genetic pools (K value equal to 5). Genetic pools are Western (red), Midsouth (green), Plains (blue), Eastern (yellow), and n/a (pink).
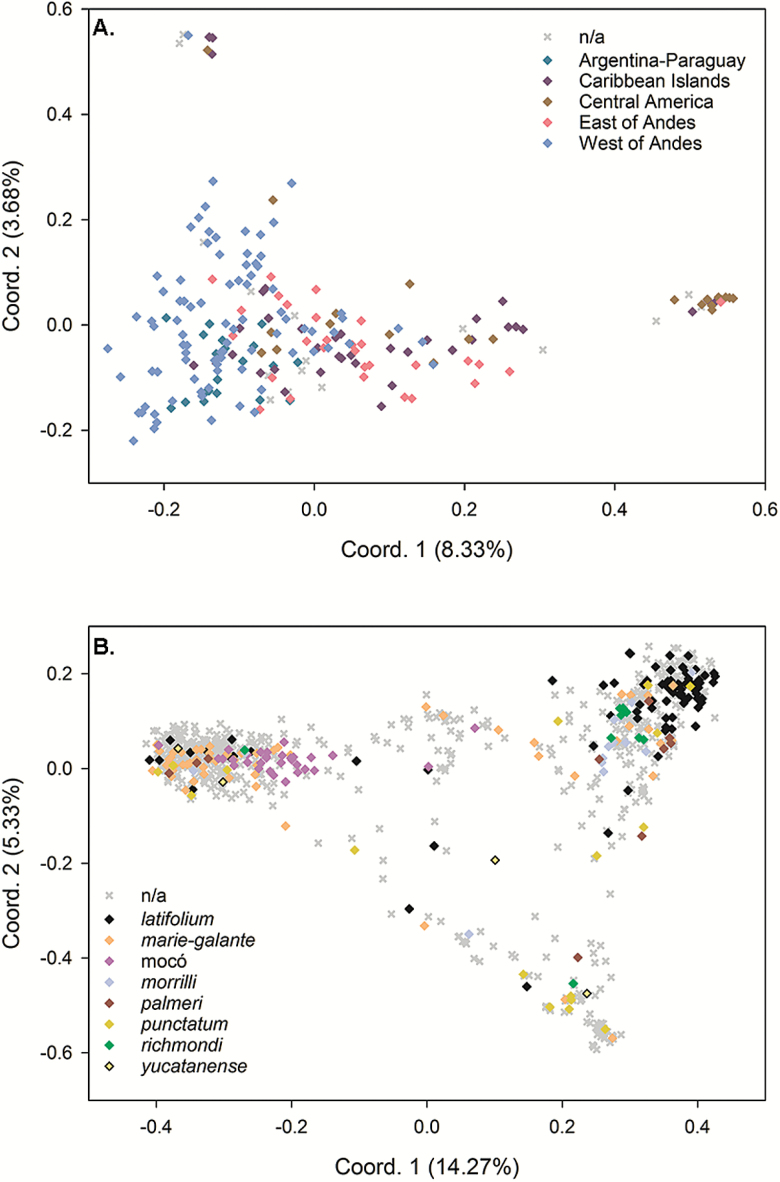
PCoA within unimproved accessions of G. barbadense produced only weak patterns that could be associated with the geographic regional groupings described by Percy and Wendel (1990). Plotting the first 2 coordinate axes from the PCoA produced a trend along the x-axis of accessions from west of the Andes merging into accessions from Argentina-Paraguay, and then merging into accessions from east of the Andes and the Caribbean Islands (Figure 4A). Approximately half of the accessions from Central America (11 out of 24 accessions) formed a distinct cluster to the right of the majority of accessions. Further investigation revealed that these accessions were collected primarily in Belize (8 accessions). Another distinct cluster of accessions was observed in the upper left quadrant and was comprised of accessions from east of the Andes, west of the Andes, the Caribbean Islands, and Central America. Rather than corresponding to a geographical region, this cluster of 7 accessions is typified by high levels of G. hirsutum-specific alleles and therefore is possibly distinct due to introgressed accessions.
Figure 4.
Principal coordinate plots showing patterns of separation between categories of wild accessions within (A) G. barbadense regions explaining 12.01% of the variation and (B) G. hirsutum races explaining 19.60% of the variation. Accessions which could not be classified into a category are labeled “n/a”.
G. hirsutum
PCoA among global G. hirsutum cultivars produced no discernable patterns of discrete clustering among cultivars that could be associated with their regional origin (Supplementary Figure S2 online). Cultivar accessions from the United States were notable in being distributed throughout a large amorphous cloud of cultivar accessions of global origins. Analysis of cultivar variation within US regions was more amenable to interpretation. Though discrete clusters were not observed, the first principal coordinate generally separated western accessions from the remaining cultivars, while the second principal coordinate tended to separate midsouth accessions from the remaining eastern and plains accessions (Figure 3B). Upon STRUCTURE analysis of G. hirsutum cultivars, the best models identified 5 unique genetic pools without prior population information (Figure 3C). Three of the pools generally corresponded to western, midsouth, and eastern regions. Accessions from the plains region had high levels of admixture, very similar to that of the “n/a” group of unknown genetic background.
PCoA of unimproved upland accessions assigned to race categories failed to form discrete clusters; rather a unique triangular pattern of dispersal was observed with large groupings of accessions in each of the 3 corners (Figure 4B). The latifolium, richmondi, and morrilli races tended to group together in the upper right quadrant of the PCoA. Race marie-galante and mocó accessions tended to form a grouping in the upper left. Many of the race punctatum accessions are found in the lower right quadrant. The race yucatanense was represented by only 4 accessions that were spread across the multivariate analysis space and were quite different. Given that the original race designations assigned to wild cottons were based as much on geographic areas as morphology, PCoA was also performed using the 11 geographic areas described by Hutchinson (1951). Accessions from Yucatan and Campeche were arranged in the lower right quadrant of the coordinate analysis, and a large group of accessions from central Guatemala, eastern Guatemala and El Salvador, and Oaxaca formed in the upper right quadrant (Supplementary Figure S3 online). Although these tendencies were observed, the clustering based on the 8 race categories was more informative.
Introgression
In the present investigation, a total of 81 bands deriving from 74 SSR markers were determined to be specific to G. barbadense (Supplementary Table S6 online) and 83 bands deriving from 72 SSR markers were determined to be G. hirsutum-specific. Overall, species-specific bands were amplified by 90 of the original core set of 105 SSR markers. Fifty-six of the markers that produced a species-specific band in one species also produced a species-specific band in the other species. Improved cultivars of both species tended to have more species-specific bands per accession than the wild accession group (Table 2; Supplementary Figure S4 online).
In the current analysis, introgression has been defined as the presence of species-specific bands of one species in a second species. Most accessions genotyped had less than 5% of the species-specific bands of the contrasting species (i.e. introgression) (Table 5). Fifteen G. barbadense and 31 G. hirsutum accessions had greater than 10% introgression (Supplementary Table S7 online). Levels of introgression varied between improved and unimproved germplasm within both species. Within G. hirsutum, 43.7% of the introgression observed was accounted for by wild accessions, whereas only 18.4% of the introgression was accounted for by improved accessions. Species wide, 36.8% of the accessions displayed no introgression. Within G. barbadense, 33.1% of the introgression observed was accounted for by wild accessions, whereas 27.1% of the introgression was accounted for by improved accessions. Species wide, 32.0% of the accessions displayed no introgression. With 68% of its accessions displaying introgression at one or more marker loci, G. barbadense had a slightly higher prevalence of introgression than did G. hirsutum, with 63.2% of its accessions displaying some level of introgression. Wild accessions accounted for more introgression in G. hirsutum than in G. barbadense.
Table 5.
Levels of introgression based on percentage of G. hirsutum species-specific bands within improved and wild types of G. barbadense accessions and percentage of G. barbadense species-specific bands within G. hirsutum improved and wild accessions. Accessions lacking information on level of cultivation are grouped as “n/a”
| Introgression (%) | G. barbadense accessions (%) | G. hirsutum accessions (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Improved | Wild | n/a | Total | Improved | Wild | n/a | Total | |
| 0 | 9.5 | 18.8 | 3.7 | 32.0 | 24.8 | 10.9 | 1.1 | 36.8 |
| 0.1–2 | 13.7 | 18.0 | 3.9 | 35.6 | 13.0 | 11.6 | 0.9 | 25.4 |
| 2.1–5 | 11.2 | 12.2 | 2.4 | 25.9 | 5.1 | 24.9 | 0.2 | 30.1 |
| 5.1–10 | 0.7 | 1.7 | 0.5 | 2.9 | 0.2 | 5.4 | 0.1 | 5.6 |
| 10.1–15 | 0.0 | 0.2 | 0.2 | 0.5 | 0.0 | 1.1 | 0.1 | 1.2 |
| 15.1–20 | 0.0 | 0.0 | 0.2 | 0.2 | 0.0 | 0.3 | 0.0 | 0.3 |
| >20 | 1.5 | 1.0 | 0.5 | 2.9 | 0.1 | 0.4 | 0.0 | 0.5 |
The distribution of introgression within the G. hirsutum and G. barbadense genomes was analyzed using the chromosome positions for markers reported in Yu et al. (2012). Introgression appeared to be fairly evenly distributed throughout the genome, with few indications of preservation of introgression in any particular chromosome. The most common distribution of introgression as observed with species-specific markers was 3 markers on each chromosome that were G. hirsutum-specific and 3 markers that were G. barbadense-specific (Supplementary Figure S5 online). Chromosome 5 was unique with 7 species-specific markers: 5 G. barbadense-specific and 2 G. hirsutum-specific markers. Chromosome 6 was unique with only 2 total species-specific markers, 1 for each species. The frequency of introgressed alleles (the number of accessions carrying a species-specific marker band on a given chromosome) suggested chromosome 17 had high levels of G. barbadense-specific bands in G. hirsutum germplasm. In addition, chromosome 14 had equally high levels of G. barbadense-specific bands in G. hirsutum germplasm, and vice versa (Supplementary Table S6 online).
Discussion
Genetic Diversity
The greater diversity within G. hirsutum than in G. barbadense, revealed by this investigation is not an artifact of sampling size (410 G. barbadense accessions vs. 1523 G. hirsutum accessions), and has been verified by a downsampling protocol. The relative greater diversity of the G. hirsutum species observed in our study agrees with previous reports (Abdalla et al. 2001; Lacape et al. 2007; Hinze et al. 2015a). The percentage of private bands in wild accessions in both species was 5–6 times that of the improved genotypes and is molecular evidence of the effects of domestication and subsequent selection. The bottleneck that occurred during domestication was caused by photoperiodism, and further narrowing of the genetic base was caused by artificial selection during the development of improved types from wild types (Meyer and Purugganan 2013). This phenomenon has been observed in many crop species, including maize (Zea mays L.) (Vigouroux et al. 2005), rice (Oryza sativa L.) (Garris et al. 2005), sunflower (Helianthus annuus L.) (Liu and Burke 2006), and soybean (Glycine spp.) (Lam et al. 2010; Li et al. 2010). However, it is also important to note that domestication can create its own diversity. This was shown in tomato (Solanum spp.) where evidence indicated that the center of diversity was the western Andes, but high diversity was also found in Central America and Europe due to heavy domestication of the edible tomato (Bauchet and Causse 2012; Koenig et al. 2013).
G. barbadense
Patterns of diversity observed within historical and geographic breeding pools of cultivated G. barbadense were in agreement with expectations based upon development history (Percy 2009). The higher percentage of polymorphic bands and private bands observed among Peruvian cottons corresponded with Peru being the center of origin and diversity of the G. barbadense species (Percy and Wendel 1990; Westengen et al. 2005) and the Peruvian Tanguis cottons being principally native in origin. Sea Island cottons, while appearing to be somewhat less diverse than Egyptian and Pima cottons, are a progenitor of both as well as of all modern cottons. Sea Island G. barbadense cottons were taken to Egypt and hybridized with Jumel’s cotton, a Peruvian-type tree cotton (native to Sub-Saharan Africa) to establish Egyptian cottons (Balls 1919). Subsequently, Egyptian cottons were imported into the American southwest to establish an American Egyptian cotton breeding program. In the 1940s, a hybrid germplasm pool was created by crossing Egyptian, Sea Island, and Tanguis germplasm, with a G. hirsutum cultivar to establish the American Pima breeding program. Egyptian cottons also were prominent in establishing breeding programs in the USSR (Uzbekistan) and northern Africa. It is not implausible to assume that some loss of allelic diversity has occurred during the establishment of successive G. barbadense improvement programs, which has been countered in Egyptian and Pima cottons by hybridization with germplasm from outside their parent pools. This review of G. barbadense improvement history is supported by our genetic structure analysis where germplasm of French North Africa, the former USSR, and Egypt generally cannot be differentiated; suggesting that they share a common genetic background and the former pools may be subsets of the latter. In contrast, accessions from the Sea Islands, western United States (Pima), Peru, and Sub-Saharan West Africa eventually resolve into unique genetic pools implying their distinct origins.
The present investigation of diversity among wild G. barbadense accessions of the NCGC supports previous phylogenetic and diversity studies conducted on smaller scales with fewer genetic markers available. The notable increase in private alleles observed in the west of Andes region relative to other regions agrees with the findings of Percy and Wendel (1990) and Westengen et al. (2005) that this region, or more specifically, the coastal regions of northwest Peru and southwest Ecuador are the center of diversity for the wild types of G. barbadense. A gradient of decreasing private bands was observed from west of the Andes to east of the Andes, the Caribbean, and Central America which supported previous reports and the postulated dispersal path of the species. GS trends of PCoA revealed a similar gradient from accessions west of the Andes, into accessions from the Caribbean Islands, grading into accessions from Central America.
G. hirsutum
Improved G. hirsutum germplasm is the preferred source of genetic diversity for use in cotton improvement programs and, therefore, much research has been conducted to evaluate the genetic diversity of this type of germplasm. While breeding programs commonly use germplasm from other breeding programs and from available collections, historically there have been pressures to breed for adaptation to local environments and adversities that may have created selective forces driving the development of distinctive germplasm pools. A prevalent question has been whether this selection has left a genetic footprint on the structure of diversity in improved germplasm that can be seen and utilized. In the present investigation, no patterns of diversity were observed among cultivated G. hirsutum, based on a global scale of historically and geographically distinct breeding efforts. However, distinctive breeding populations could be discerned on a national scale among regional breeding programs within the United States. Three populations identified by STRUCTURE analysis in the improved US accessions are consistent with previous findings (Tyagi et al. 2014). The maximum variance PCoA axis (Coord. 1) split accessions of midsouth and western breeding programs, while accessions of eastern breeding programs were only secondarily distinguished from the rest (as seen along coordinate 2 in Figure 3B here and along coordinate 3 in Figure 5 of Tyagi et al. (2014)). In addition, accessions from the midsouth had the most private bands relative to other regions in the United States. Together these results suggest that cultivars from the midsouth have unique genetic makeup.
Approximately 27% of the wild or unimproved G. hirsutum accessions in the current study have previously been assigned to races, based on a few morphological traits and geographic regions of collection (Hutchinson 1951). An objective of the current project was to determine whether these races exist on a genetic basis and whether we could classify the remaining 73% of the wild accessions into a race category. Genetic diversity, as revealed by the molecular markers of the study, could not resolve the races into distinct groups, neither when visualized in the PCoA (Figure 4B) nor did they have many private bands with which to distinguish the various categories (Table 4). Comparison of percentage of private bands in US cultivars versus private bands in G. hirsutum races reveals much lower levels of private bands among races—indicating higher band sharing among races than among cultivars. The ability to assign unclassified accessions to a race based upon SSR markers of the present study is lacking. Overall, the usefulness of races in defining and using the genetic diversity of unimproved G. hirsutum would appear to be limited.
Previous studies have documented the existence of large pooled genetic variation in the races of G. hirsutum. However, a limited number of studies have attempted to use molecular markers to distinguish among the various races, and we have not identified any studies that attempted to use molecular markers to assign the unclassified germplasm to races. Abdurakhmonov et al. (2008) evaluated 208 exotic landrace stocks from the Uzbek cotton germplasm collection, including all named races observed in the current study. The Uzbek study demonstrated that these exotic accessions had much wider diversity than the cultivars included in the same study; however, they did not detail differences among the various races. Similar research using germplasm from the cotton genetic resources conservation unit of CIRAD did specifically attempt to distinguish among the races. The 7 races of G. hirsutum along with mocó types were represented by approximately 4 accessions each from the CIRAD collection (Lacape et al. 2007). These races exhibited significantly more polymorphism than the cultivated G. hirsutum accessions, and, contrary to the current study, the SSR marker information of Lacape et al. (2007) was able to fully confirm the racial classification system of Hutchinson (1951). A previous study with isozymes (Wendel et al. 1992) showed very limited support for Hutchinson’s race classification scheme.
Introgression between G. barbadense and G. hirsutum
There is ample molecular evidence that gene introgression between G. barbadense and G. hirsutum has occurred in both species but in different intensities and in different germplasm sets (Percy and Wendel 1990; Wendel et al. 1992; Brubaker et al. 1993; Brubaker and Wendel 1994; Wang et al. 1995; Abdalla et al. 2001). These species have large native ranges that include an extensive area of sympatry in the Caribbean and Central America (Fryxell 1979; Brubaker et al. 1993) and evidence implies that introgression is happening naturally in the wild. Of the 31 wild G. hirsutum accessions with greater than 10% introgression, 21 accessions were from Brazil (4 marie-galante and 12 mocó wild types) (Supplementary Table S7 online). Mocó cottons occupy a small region of sympatry in northeastern Brazil relative to the wide geographic distribution of race marie-galante as well as being sympatric with 2 other tetraploid species, G. barbadense and G. mustelinum (Stephens 1973). The introgression identified in our study concurs with previous work that reported alleles from G. barbadense were found in mocó cotton in its native habitat in the Brazilian state of Piauí (de Menezes et al. 2010).
The frequency of occurrence of introgression was higher among improved G. barbadense accessions than among improved G. hirsutum accessions (Table 5). Likewise, the average level of introgression within individual accessions, as indicated by number of species-specific bands, was higher in improved G. barbadense cultivars than in G. hirsutum cultivars (Table 2). Frequency of occurrence of introgression was higher in wild or unimproved accessions than in cultivars in both species. However, the average level of introgression within individual accessions, as indicated by species-specific bands, was much higher in wild accessions of G. hirsutum than wild accessions of G. barbadense. These findings of non-symmetrical patterns of introgression are in accord with previous work of Brubaker et al. (1993).
The distribution of introgression was fairly uniform across the cotton genome and between the 2 species, except for an increased number of G. barbadense-specific markers observed on chromosome 5 and a reduced number of markers overall observed on chromosome 6. These differences could indicate loci that are important for species barriers or hybrid breakdown between individuals resulting from G. barbadense and G. hirsutum crosses (Jiang et al. 2000). The high number of markers could be a conferred positive selective advantage from G. barbadense relative to G. hirsutum for chromosome 5 or simply due to chromosome 5 being the genetically longest chromosome (Yu et al. 2012) with the greatest opportunity for introgression. The low number of markers could be a restriction of interspecific introgression from negative selection.
An increased frequency of introgressed bands was noted on chromosomes 14 and 17 suggesting regions of conserved introgression. Conserved introgression would be expected due to breeders’ selection for traits such as improved fiber quality and for day neutrality. Cotton breeders have historically wanted to introgress the high fiber quality genes from G. barbadense into G. hirsutum. In G. hirsutum, fiber development gene-rich islands were localized to chromosomes 5, 10, 14, and 15 (Xu et al. 2008). Photoperiod response is a crucial trait as sensitivity to photoperiod limits the use of tropical, often wild, germplasm in temperate breeding programs. Recent efforts have mapped this trait to chromosome 25 in G. barbadense (Zhu and Kuraparthy 2014). The genes responsible for photoperiod response do not appear to be connected to regions of enhanced introgression observed in the current study.
The core set of 104 SSR markers has allowed for a definitive molecular genetic separation of G. barbadense and G. hirsutum species accessions, and it has supported previous reports detailing regional groupings within improved cultivars of both species as well as the patterns of diversity reported in wild G. barbadense. However, results indicate that existing categorization schemes used in the 2 species are only partially successful in delineating the diversity of the collection. In several instances, previously defined categories could not be differentiated using SSR markers, and the usefulness of the categories in defining collection variability is doubtful. Species-specific alleles or bands have been identified that are useful in identifying and tracking introgression. Novel outlying clusters of accessions with unique combinations of alleles putatively beneficial to a cotton breeding or research program have been identified. With this information, previous publications, and information in the CottonGen database (www.cottongen.org), users of the collection will have available a molecular profile of these accessions (Hinze et al. 2015a) along with a set of descriptor data (J. Frelichowski, personal communication) and seed traits including oil and protein content (Hinze et al. 2015b) with which to make better informed decisions when identifying accessions from the Gossypium Diversity Reference Set of the National Cotton Germplasm Collection.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
International Atomic Energy Agency Coordinated Research Project: Isolation and Characterization of Genes Involved in Mutagenesis of Plants, Instituto Nacional de Tecnología Agropecuaria-International Atomic Energy Agency Research Contract No 15671: Isolation and Characterization of Genes Involved in Chloroplast Genes Mutagenesis, Agencia Nacional de Promoción Científica y Tecnológica Proyectos de Investigación Científica y Tecnológica-Fondo para la Investigación Científica y Tecnológica 2007 Nº 620: The barley chloroplast mutator as a tool to originate plastome genetic variability, and Instituto Nacional de Tecnología Agropecuaria Proyecto Específico Area Estratégica en Biotecnología 244631: Mutagenesis techniques for diversity generation on characters of agricultural and/or agro-industrial interest.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge members of the USDA-ARS cotton genetics and breeding projects for their valuable technical assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
References
- Abdalla A, Reddy O, El-Zik K, Pepper A. 2001. Genetic diversity and relationships of diploid and tetraploid cottons revealed using AFLP. Theor Appl Genet. 102:222–229. [Google Scholar]
- Abdurakhmonov IY, Kohel RJ, Yu JZ, Pepper AE, Abdullaev AA, Kushanov FN, Salakhutdinov IB, Buriev ZT, Saha S, Scheffler BE, et al. 2008. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics. 92:478–487. [DOI] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of Heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Balls WL. 1919. The Cotton Plant in Egypt. London: Macmillan. [Google Scholar]
- Bauchet G, Causse M. 2012. Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives. In: Caliskan M, editor. Genetic Diversity in Plants. InTech; p. 133–162. [Google Scholar]
- Brown AHD. 1989. Core collections: a practical approach to genetic resources management. Genome. 31:818–824. [Google Scholar]
- Brubaker CL, Koontz JA, Wendel JF. 1993. Bidirectional cytoplasmic and nuclear introgression in the New World cottons, Gossypium barbadense and G. hirsutum (Malvaceae). Am J Bot. 80:1203–1208. [Google Scholar]
- Brubaker CL, Wendel JF. 1994. Reevaluating the origin of domesticated cotton (Gossypium hirsutum; Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am J Bot. 81:1309–1326. [Google Scholar]
- de Menezes I, Barroso P, Hoffmann L, Lucena V, Giband M. 2010. Genetic diversity of mocó cotton (Gossypium hirsutum race marie-galante) from the northeast of Brazil: implications for conservation. Botany Botanique. 88:765–773. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Fang D, Hinze L, Percy R, Li P, Deng D, Thyssen G. 2013. A microsatellite-based genome-wide analysis of genetic diversity and linkage disequilibrium in Upland cotton (Gossypium hirsutum L.) cultivars from major cotton-growing countries. Euphytica. 191:391–401. [Google Scholar]
- Fryxell PA. 1979. The natural history of the cotton tribe (Malvaceae, tribe Gossypieae). College Station: (TX: ): Texas A&M University Press. [Google Scholar]
- Fryxell PA. 1992. A revised taxonomic interpretation of Gossypium L. (Malvaceae). Rheedea. 2:108–165. [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. 2005. Genetic structure and diversity in Oryza sativa L. Genetics. 169:1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ. 1971. Toward a rational classification of cultivated plants. Taxon. 20:509–517. [Google Scholar]
- Hinze L, Dever J, Percy R. 2012. Molecular variation among and within improved cultivars in the U.S. Cotton Germplasm Collection. Crop Sci. 52:222–230. [Google Scholar]
- Hinze L, Fang D, Gore M, Scheffler B, Yu J, Frelichowski J, Percy R. 2015a. Molecular characterization of the Gossypium Diversity Reference Set of the US National Cotton Germplasm Collection. Theor Appl Genet. 128:313–327. [DOI] [PubMed] [Google Scholar]
- Hinze LL, Horn PJ, Kothari N, Dever JK, Frelichowski J, Chapman KD, Percy RG. 2015b. Nondestructive measurements of cottonseed nutritional trait diversity in the US National Cotton Germplasm Collection. Crop Sci. 55:770–782. [Google Scholar]
- Hutchinson JB. 1951. Intra-specific differentiation in Gossypium hirsutum . Heredity. 5:161–193. [Google Scholar]
- IBPGR 1980. Descriptors for Cotton Species. International Board for Plant Genetic Resources Working Group; Rome, Italy. [Google Scholar]
- Jaccard P. 1908. Nouvelles recherches sur la distribution florale. Bulletin de la Société vaudoise des sciences naturelles. 44:223–270. [Google Scholar]
- Jiang CX, Chee PW, Draye X, Morrell PL, Smith CW, Paterson AH. 2000. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton). Evolution. 54:798–814. [DOI] [PubMed] [Google Scholar]
- Koenig D, Jiménez-Gómez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, Kumar R, Covington MF, Devisetty UK, Tat AV, et al. 2013. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci. 110:E2655–E2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacape J, Dessauw D, Rajab M, Noyer J, Hau B. 2007. Microsatellite diversity in tetraploid Gossypium germplasm: assembling a highly informative genotyping set of cotton SSRs. Mol Breed. 19:45–58. [Google Scholar]
- Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, et al. 2010. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 42:1053–1059. [DOI] [PubMed] [Google Scholar]
- Li YH, Li W, Zhang C, Yang L, Chang RZ, Gaut BS, Qiu LJ. 2010. Genetic diversity in domesticated soybean (Glycine max) and its wild progenitor (Glycine soja) for simple sequence repeat and single-nucleotide polymorphism loci. New Phytol. 188:242–253. [DOI] [PubMed] [Google Scholar]
- Liu A, Burke JM. 2006. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics. 173:321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Purugganan MD. 2013. Evolution of crop species: genetics of domestication and diversification. Nat Rev Genet. 14:840–852. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics. 28:2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival AE. 1987. The National Collection of Gossypium Germplasm. Southern Cooperative Series Bulletin No. 321. [Google Scholar]
- Percy RG. 2009. The worldwide gene pool of Gossypium barbadense L. and its improvement. In: Paterson A, editor. Genetics and Genomics of Cotton. Springer US; p. 53–68. [Google Scholar]
- Percy RG, Wendel JF. 1990. Allozyme evidence for the origin and diversification of Gossypium barbadense L. Theor Appl Genet. 79:529–542. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif JC, Melchinger AE, Frisch M. 2005. Genetical and mathematical properties of similarity and dissimilarity coefficients applied in plant breeding and seed bank management. Crop Sci. 45:1–7. [Google Scholar]
- Rohlf FJ. 2000. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, version 2.2. Release 2.1.
- Stephens SG. 1973. Geographical distribution of cultivated cottons relative to probably centers of domestication in the new world. In: Adrian M, editor. Genes, Enzymes and Populations. New York: Plenum Press; p. 239–254. [DOI] [PubMed] [Google Scholar]
- Toll JA. 1995. Processing of germplasm, associated material and data. In: Guarino L, Ramanatha RV, Reid R, editors. Collecting Plant Genetic Diversity: Technical Guidelines. Rome: (Italy: ): International Plant Genetic Resources Institute (IPGRI), Plant Production and Protection Division, FAO; p. 577–595. [Google Scholar]
- Tyagi P, Gore M, Bowman D, Campbell B, Udall J, Kuraparthy V. 2014. Genetic diversity and population structure in the US Upland cotton (Gossypium hirsutum L.). Theor Appl Genet. 127:283–295. [DOI] [PubMed] [Google Scholar]
- Vigouroux Y, Mitchell S, Matsuoka Y, Hamblin M, Kresovich S, Smith JS, Jaqueth J, Smith OS, Doebley J. 2005. An analysis of genetic diversity across the maize genome using microsatellites. Genetics. 169:1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Dong JM, Paterson AH. 1995. The distribution of Gossypium hirsutum chromatin in Gossypium barbadense germplasm - molecular analysis of introgressive plant breeding. Theor Appl Genet. 91:1153–1161. [DOI] [PubMed] [Google Scholar]
- Wendel J, Brubaker C, Percival A. 1992. Genetic diversity in Gossypium hirsutum and the origin of upland cotton. Am J Bot. 79:1291–1310. [Google Scholar]
- Wendel J, Cronn R. 2003. Polyploidy and the evolutionary history of cotton. Adv Agron. 78:139–186. [Google Scholar]
- Westengen O, Huaman Z, Heun M. 2005. Genetic diversity and geographic pattern in early South American cotton domestication. Theor Appl Genet. 110:392–402. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohel RJ, Song G, Cho J, Alabady M, Yu J, Koo P, Chu J, Yu S, Wilkins TA, et al. 2008. Gene-rich islands for fiber development in the cotton genome. Genomics. 92:173–183. [DOI] [PubMed] [Google Scholar]
- Yu J, Fang D, Kohel R, Ulloa M, Hinze L, Percy R, Zhang J, Chee P, Scheffler B, Jones D. 2012. Development of a core set of SSR markers for the characterization of Gossypium germplasm. Euphytica. 187:203–213. [Google Scholar]
- Zhu LL, Kuraparthy V. 2014. Molecular genetic mapping of the major effect photoperiod response locus in Pima cotton (Gossypium barbadense L.). Crop Sci. 54:2492–2498. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.