Abstract
Honey bees are exposed to many damaging pathogens and parasites. The most devastating is Varroa destructor, which mainly affects the brood. A promising approach for preventing its spread is to breed Varroa-resistant honey bees. One trait that has been shown to provide significant resistance against the Varroa mite is hygienic behavior, which is a behavioral response of honeybee workers to brood diseases in general. Here, we report the use of an Affymetrix 44K SNP array to analyze SNPs associated with detection and uncapping of Varroa-parasitized brood by individual worker bees (Apis mellifera). For this study, 22 000 individually labeled bees were video-monitored and a sample of 122 cases and 122 controls was collected and analyzed to determine the dependence/independence of SNP genotypes from hygienic and nonhygienic behavior on a genome-wide scale. After false-discovery rate correction of the P values, 6 SNP markers had highly significant associations with the trait investigated (α < 0.01). Inspection of the genomic regions around these SNPs led to the discovery of putative candidate genes.
Keywords: Candidate genes, hygienic behavior, SNP array, Varroa destructor, Varroa resistance.
The reproduction of Varroa destructor, an ectoparasitic mite of the honeybee (Apis mellifera), occurs only in the bees’ capped brood cells, and the damage caused to colonies is mainly a consequence of the infestation of pupae. Inside the capped brood cells, the mites puncture the hosts’ integument and suck out the hemolymph. This weakens the pupae and shortens their lifespan (Schneider and Drescher 1987). Several viral diseases are also transmitted by Varroa (Ball 1985; Allen et al. 1986; Ball and Allen 1988; Allen and Ball 1996; Martin 1998; Nordström et al. 1999; Bakonyi et al. 2002; Chen et al. 2004). Varroa poses a serious threat to the A. mellifera beekeeping industry; indeed, Varroa infestation is generally considered to be the most serious problem affecting beekeeping worldwide.
Various strategies are used to combat Varroa. The first is to keep the mite population within tolerable limits using acaricides. However, their effectiveness depends upon weather conditions and the timing of application. Residues of these substances also accumulate in bee products (Wallner 1999), and mites have developed resistance to many of them (Milani 1999). Further disadvantages of these chemical treatments include the high costs and intensive labor requirements.
The second strategy is to breed Varroa-resistant honeybees. In this case, the hygienic behavior of honeybees is of particular interest (Spivak 1996). This is defined as the honeybee workers’ ability to detect and remove pupae infected with brood diseases before the causative organisms reach the infectious stage, thereby limiting the spread of infection. There is evidence that hygienic behavior to combat Varroa destructor is triggered by odors that originate from infected hosts (Martin et al. 2002; Spivak et al. 2003; Navajas et al. 2008; Schöning et al. 2012).
It has long been known that hygienic behavior confers resistance to American Foulbrood (Woodrow and Holst 1942) and chalkbrood (Gilliam et al. 1983). More recently, it has been demonstrated that hygienic bees detect and remove brood infested with Varroa destructor (Boecking and Drescher 1992; Spivak 1996; Thakur et al. 1997; Harbo and Harris 2005; Harbo and Harris 2009). This interrupts the reproductive cycle of the mite (Rath and Drescher 1990; Fries et al. 1994).
The heritability of hygienic behavior was estimated as ~0.2 by Boecking et al. (2000) and as ~0.6 by Harbo and Harris (1999) and Lapidge et al. (2002). Heritability estimation in honeybees is methodically difficult due to their reproductive peculiarities, namely polyandry and haplodiploidy (Bienefeld and Pirchner 1990). Heritability studies of hygienic behavior concur that the trait is heritable to some degree, suggesting that there is some potential for promoting this trait through selective bee breeding.
Unfortunately, conventional breeding methods are only partially applicable to this aim for 2 reasons: this behavior is observed only in workers (only queens and drones are fertile), and it occurs at a very low frequency which is difficult to measure. A promising way to address this problem is to exploit the honeybee genome sequence (Honeybee Genome Sequencing Consortium 2006) using molecular genetic methods. The preferred strategy is to genotype a high number of genetic markers in linkage or association studies in order to identify genomic regions implicated in hygienic behavior and ultimately discover the causative genes. The first steps have already been taken to unravel the molecular genetics of hygienic behavior. Some quantitative trait loci (QTL) studies exist with moderate marker numbers; however, no previous studies have employed a large scale SNP assay, nor have QTL or candidate genes been confirmed by independent studies.
In his classic study of behavioral genetics, Rothenbuhler (1964) proposed that the 2 components of hygienic behavior, uncapping and removal, are under separate genetic control and that each component is controlled by a single, unlinked Mendelian locus. However, this 2-locus model is clearly an over-simplification. Moritz (1988) reevaluated Rothenbuhler’s data and found that they were more suggestive of 3 loci than 2, whereas Lapidge et al. (2002) proposed a quantitative pattern of inheritance for the trait that involved 7 loci. Recently, Oxley et al. (2010) identified 6 QTLs; however, it is not possible to compare the 2 previously mentioned studies because they used different genetic maps. Tsuruda et al. (2012) found one major QTL on chromosome 9 for hygienic behavior against Varroa using a small-scale SNP-Chip (1340 informative SNPs). Oxley et al. also reported a QTL for the trait on chromosome 9, but in a different region. Nonetheless, all authors agree that a strong genetic component is involved in the control of hygienic behavior and that the variation in this trait is controlled by a small number of loci. These loci affect a bee’s sensitivity to the stimulus and set a specific individual threshold governing the likelihood of a worker engaging in hygienic behavior. Therefore, a genetic predisposition to the heightened detection of abnormal brood odors may facilitate the expression of hygienic behavior in a colony (Spivak et al. 2003). Although hygienic behavior may also be influenced by environmental factors to some extent (Thompson 1964; Momot and Rothenbuhler 1971), the elucidation of gene variants that control this behavior and their propagation by breeding techniques appear to be a promising approach in the fight against Varroa destructor.
The basis for genetic mapping of QTL, the step that precedes the actual identification of gene variants, is genomic recombination, as the number of crossovers between 2 points on a chromosome correlates with their physical distance. Honey bee genetic maps have revealed a higher rate of recombination than any reported for a higher eukaryote (Hunt and Page 1995; Solignac et al. 2004). This high recombination rate is very useful for QTL mapping. It results in a higher resolution of physical chromosome distance, reducing the effort required to identify which gene (or genes) influences a trait (Rinderer et al. 2010). Large scale SNP arrays are an appropriate tool for identifying QTL in the honey bee genome because they contain so many markers that there is a high likelihood of identifying one (or several) that are situated within the actual sequence of the trait-influencing gene. We may require such markers in order to prevent the rapid erosion of linkage disequilibrium due to the high recombination rate of the honey bee genome (Rinderer et al. 2010).
Generally, hygienic behavior against diseased brood consists of the detection of the diseased (or otherwise handicapped) brood cells, their uncapping, and the removal of the diseased pupae/larvae. In our long-term video observation studies of hygienic behavior towards Varroa parasitized brood cells, we have found the removal to be the least specific component; about 25% of the observed worker bees are involved in this behavior. The most specific behavior is the detection of the Varroa parasitized brood cell through the cell caps. In an unselected population, less than 1% of the bees show this behavior. An extremely left-skewed distribution appears, with many families whose members do not show the behavior at all and a very small number of families in which up to 2% of the members show the behavior. If a bee initiates uncapping, she is usually supported by up to 10 other bees until the cell is completely uncapped. Some of these bees continue and begin removing impaired brood. Bees involved only in uncapping, without initiating this behavior, can be assumed to be less specific within this complex cooperation, as they are less able to detect the stimulus of a Varroa parasitized pupa through the closed cap. Consequently, the whole hygienic process depends mainly upon the bees who first detect the parasitized brood and initially uncap the cells. This allows the other (presumably less sensitive) bees to more clearly recognize the stimulus of the parasitized brood through the partially opened cell cap and complete the hygienic removal of the handicapped brood. We strongly assume that the bees which initiate the uncapping (detecting the infestation) are the most important ones for Varroa resistance breeding. Consequently, it is of major interest to know the genes involved in this special component of the resistance mechanism. Our specific bioassay allowed us to monitor the detection and uncapping of Varroa parasitized brood cells (DUVB) by individual bees. This DUVB behavior bioassay is assumed to be much more Varroa specific and sensitive than the freeze- or pin-killed brood assays (strong and unspecific stimulus) used in other studies.
Materials and Methods
Defense Behavior Bioassays and Phenotyping
We phenotyped single worker bees to assay their defensive behavior against Varroa destructor was using a special behavior bioassay developed at the Institute for Bee Research Hohen Neuendorf, Germany (LIB). During one replicate of the DUVB behavior bioassay, 2000 worker bees were marked individually after hatching by gluing numbered opalith tags on their thoraces (Figure 1). The marked bees were transferred to a caged experimental comb (Bienefeld et al. 2016). These combs were derived from unrelated, disease- and Varroa-free colonies. The observation area in each of these combs contained approximately 170 brood cells (ca 10×10cm). Next, 45 of these cells were artificially infested with 1 Varroa mite each by cutting a slot into the cap, inserting a mite using a small paint brush, and then resealing the cell. About 73–77 cells were left untreated and 45 cells were opened and resealed without inserting a mite to serve as controls for the effect of cap manipulation.
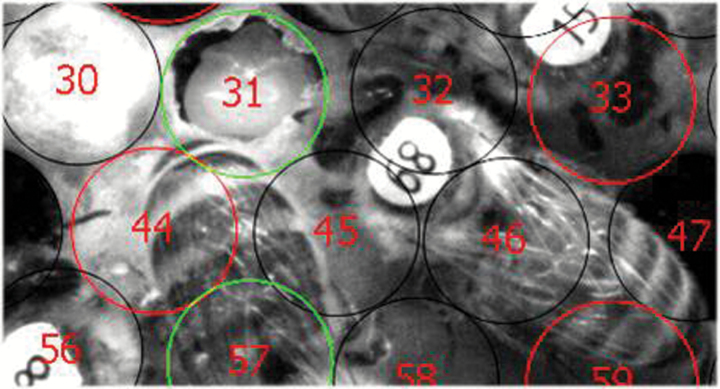
Figure 1.
Photo from the video of hygienic behavior towards Varroa-infested brood cells. Software-assisted each cell was marked differently according to its infestation status (Varroa infested, non-Varroa infested). Cell 31, pictured here, has already been completely uncapped so that the pupa is visible. The bee to the right of the cell, marked number 68, has opened this Varroa-parasitized cell.
The caged experimental combs were integrated into the hives of colonies to provide the experimental bees with adequate warmth and a natural environment. The experiments were conducted in 11 replicates between 2009 (5 replicates) and 2010 (6 replicates). An infrared sensitive camera (Panasonic WV-NP1004 megapixel color network IP) was installed in front of the comb and activities were recorded in the observation area of the comb for 7 days. Infrared LEDs (OSA Opto-Light GmbH, Germany, Type: OIS 330 880) were used to provide illumination rather than visible light. As bees do not respond to this part of the light spectrum (880±10nm), they were not disturbed during the long observation periods. The recordings were inspected by 2 independent observers and scanned to record DUVB behaviors. At the end of each replicate test, i.e. after video recording on day 8, the honey bees were killed by being brushed from the brood comb into a basin containing ethanol (96%). These bees were stored in the ethanol until DNA extraction.
Crossing Scheme and Test Animals
The honey bees (Apis mellifera carnica) used in this experiment were obtained from a special crossing scheme conducted to assemble most of the alleles for Varroa resistance segregation in German carnica bee populations and to promote the development of Varroa-resistant bees, the frequency of which is very rare (<1%) in naturally occurring honeybee populations. The crossing scheme was conducted as follows: 10 queens from a line bred specifically for the DUVB at LIB since 1997 (Bienefeld et al. 2001) were mated with drones from line shown to be extremely non-hygienic line. Ten F1 queens from these crosses, i.e. 1 per original queen, were each artificially inseminated using the same pooled sperm from ca. 250 drones. These drones were derived from various sources and were either unrelated or only marginally related to each other and the queens. During 2009 and 2010, the resulting worker offspring of these queens was analyzed to evaluate their DUVB behavior using the bioassay described above. To compensate for age effects (Thakur et al. 1998) on DUVB, freshly emerged worker bees (0–12h) were labeled individually using opalith tags and subjected to behavior bioassays at the age of 4 days. During the first 4 days, the bees tested within the same replicate were allowed to become accustomed to each other. They were kept together in a caged comb that was integrated into a colony to provide warmth and a natural environment. After this, the bees exhibited calm and normal behavior upon inspection with the infrared camera.
During the 11 replicates of the behavior bioassay, 22 000 worker bees were phenotyped. The 122 top performing DUVB bees in this population were selected for genotyping. These bees were the best 122 in terms of the number of DUVB actions observed, meaning that they began to uncap at least one Varroa-infested cell and assisted in at least one other uncapping event. Furthermore, the number of uncappings and assistive actions directed against Varroa-infested cells by these individuals was at least double the number of actions they directed toward control cells.
In the control sample, an equal number of workers was selected in which each individual was descended from the same queen, but none of the workers displayed hygienic behavior.
DNA Extraction
The honeybees used for DNA extraction were stored in ethanol (96%) after the behavior bioassays.
Two methods were used for DNA extraction: automated extraction with a QIAsymphony SP Workstation (Qiagen, Hilden, Germany) using DNA tissue according to a low content protocol, and manual extraction using the Gentra Puregene DNA kit protocol (Qiagen, Gentra Puregene Handbook, DNA Purification from Mouse Tail Tissue). A modification of the Puregene protocol was used in which whole bees were crushed in 300 µL cell lysis solution (Puregene) before adding 30 µL of proteinase K. The mixture was incubated on a heat block at 55 °C with an agitation of 500rpm overnight. The manufacturer’s handbook instructions were followed the next day.
The DNA quality and quantity were assessed using 0.8% agarose gel electrophoresis and a Picogreen quantification linked to a plate fluorometer (DTX 880 plate reader, Beckman Coulter, CA, USA).
Automated extraction yielded an appropriate DNA quality and quantity for Affymetrix SNP array genotyping, so this was used to isolate genomic DNA from the samples.
Genetic Tools and Statistical Analysis
SNP genotyping was performed using an Affymetrix 44K SNP array, which was developed in cooperation with AROS Applied Biotechnology AS. For detailed information about the development of this assay, please refer to the corresponding publication by Spötter et al. (2012). The BRLMM-P algorithm (http://media.affymetrix.com/support/technical/whitepapers/brlmmp_whitepaper.pdf) was used for genotype calling. Information related to the SNPs used in this array is deposited in the Dryad repository (DRYAD entry doi:10.5061/dryad.8635cs4h). This array can be obtained from AROS Applied Biotechnology AS, Aarhus, Denmark.
The SNP array contained 32 632 SNPs that yielded nondistorted and reliable genotyping signals. Of these, 6625 SNPs were monomorphic, leaving 26 007 SNPs to be included in the statistical analyses. In fulfillment of data archiving guidelines (Baker 2013), the data underlying the actual analyses have been deposited in the Dryad repository.
As described in the Materials section, the test animals were members of 1 of 12 families. Therefore, within each family, linkage disequilibrium between a QTL and a linked SNP in the neighborhood is to be expected. The F1 queens were not genotyped; most of those queens died during the second year of the experiment and were immediately disposed of by the workers. All queens were inseminated using a sperm pool. As the family size was relatively small, a usual linkage analysis or a combination of linkage and linkage disequilibrium analyses was not feasible. We therefore used the following approach to test the dependence/independence of SNP genotypes of DUVB and non-DUVB behavior, respectively: assuming that a queen is informative, she must therefore be heterozygous with regard to a QTL for DUVB behavior and the neighboring marker. If we denote the desired allele for DUVB as Q, the other as q, and both marker alleles as M1 and M2, then the haplotypes can be either QM1/qM2 or qM1/QM2. The drones were likely to have a genetic disposition for non-DUVB behavior, represented by the haplotype qM1 with probability d, where d is the allele frequency of M1 in the drones. The SNP genotypes then differ between DUVB and non-DUVB family members. The following represents an ideal case:
| SNP genotype | DUVB | non-DUVB |
|---|---|---|
| Haplotype of the queen: QM1/qM2 | ||
| M1M1 | d | 0 |
| M1M2 | (1-d) | d |
| M2M2 | 0 | (1-d) |
| Haplotype of the queen: qM1/QM2 | ||
| M1M1 | 0 | d |
| M1M2 | d | (1-d) |
| M2M2 | (1-d) | 0 |
For clarity’s sake, we demonstrate our approach for SNP AMB-00573174 as an example, because the linkage phase between this marker and the putative QTL was obviously the same in all informative families. The allele frequency of M1 in the drones (d) is, in this case, obviously close to 1. The haplotypes of the queens seem to be QM1/qM2 because here, the DUVB bees tend to have the genotype M1M1, while the non-DUVB bees have the genotype M1M2 and only a few bees (or none) have M2M2. For this SNP, there were 2 uninformative families and 182 progenies from 10 informative families, 78 of which demonstrated DUVB and 104 of which did not (Table 1). 77 of the DUVB workers had genotype M1M1, while M1M2 occurred only once in this group. Among the non-DUVB phenotype, 42 individuals were genotyped as M1M1 and 62 as M1M2. Thus, among the DUVB workers, the genotype M1M1 occurred almost exclusively.
Table 1.
Genotypes for SNP AMB-00573174 as an example of DUVB occurrence in informative and uninformative families. For this SNP, there were 2 uninformative families and 182 progenies from 10 informative families, 78 of which demonstrated DUVB and 104 of which did not. The bees displaying DUVB tend to have the M1M1 genotype, while the non-DUVB bees have the genotype M1M2. Only a few bees if any, have genotype M2M2. Among the DUVB worker bees, genotype M1M1 occurred almost exclusively: 77 had genotype M1M1; M1M2 occurred only once. Of the non-DUVB workers, 42 were genotyped as M1M1 and 62 as M1M2
| #DUVB | #Non-DUVB | |
|---|---|---|
| Uninformative families | ||
| Queen 90175 | ||
| M1M1 | 6 | 6 |
| M1M2 | 0 | 0 |
| M2M2 | 0 | 0 |
| Queen 90182 | ||
| M1M1 | 8 | 8 |
| M1M2 | 0 | 0 |
| M2M2 | 0 | 0 |
| Informative families | ||
| Queen 90173 | ||
| M1M1 | 15 | 10 |
| M1M2 | 1 | 11 |
| M2M2 | 0 | 0 |
| Queen 90174 | ||
| M1M1 | 7 | 2 |
| M1M2 | 0 | 6 |
| M2M2 | 0 | 0 |
| Queen 90176 | ||
| M1M1 | 9 | 7 |
| M1M1 | 0 | 10 |
| M1M1 | 0 | 0 |
| Queen 90179 | ||
| M1M1 | 3 | 2 |
| M1M2 | 0 | 3 |
| M2M2 | 0 | 0 |
| Queen 9180 | ||
| M1M2 | 4 | 0 |
| M1M2 | 0 | 3 |
| M2M2 | 0 | 0 |
| Queen 90181 | ||
| M1M1 | 2 | 0 |
| M1M2 | 0 | 4 |
| M2M2 | 0 | 0 |
| Queen 90185 | ||
| M1M1 | 8 | 5 |
| M1M2 | 0 | 4 |
| M2M2 | 0 | 0 |
| Queen 90187 | ||
| M1M1 | 6 | 2 |
| M1M2 | 0 | 7 |
| M2M2 | 0 | 0 |
| Queen 90189 | ||
| M1M1 | 13 | 7 |
| M1M2 | 0 | 10 |
| M1M2 | 0 | 0 |
| Queen 90191 | ||
| M1M1 | 10 | 7 |
| M1M2 | 0 | 4 |
| M2M2 | 0 | 0 |
| All informative families | ||
| M1M1 | 77 | 42 |
| M1M2 | 1 | 62 |
| M2M2 | 0 | 0 |
As the number of observations is relatively small, we used Fisher’s exact test of association versus no association for each SNP. As the linkage phase can differ from queen to queen, each family was analyzed separately. To combine the results across all families, the error probability values were transformed into pseudo-chi-square values. These pseudo-chi-square values and the respective degrees of freedom were summed up over all families by using the reproductive property of the Chi-square distribution, and the final error probability was then computed:
p i: p value for family i (if informative) based on Fisher’s exact test
X i 2: the corresponding chi-square value, truncating the distribution (with df degrees of freedom) at the (1-pi)-quantil
X 2 total: is the combined test statistic over all F informative families with degrees of freedom also summed up over all F families.
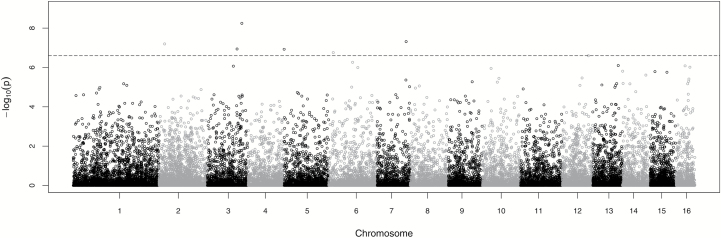
To identify SNPs of importance, we applied the Benjamini and Yekateuli’s (2001) false discovery rate controlling method, which is always valid for P values under any kind of dependency. A Manhattan plot (Figure 2) was constructed using the minus_log_p-values of the 26 007 nonmonomorphic SNPs.
Figure 2.
Manhattan plot using the minus_log_P values of the 26 007 nonmonomorphic SNPs that satisfied all of the filters. The dotted line shows the significance threshold. The 6 SNPs which show significant genome-wide associations with DUVB at the genotype level can be seen above the threshold, at chromosomes 2, 3, 5, 6, and 7.
Identification of Positional and Functional Candidate Genes Related to Varroa Resistance
We carried out BLAST searches to identify SNPs with an effect on Varroa resistance (Altschul et al. 1990), using the SNP flanking sequences as queries to determine their position on the NCBI Map A viewer of the honeybee genome is available here (http://www.ncbi.nlm.nih.gov/projects/mapview/map_search.cgi?taxid=7460). The genomic regions around these SNPs were scanned for putative candidate genes.
Results
Of the 22 000 honey bees that were phenotyped during the behavior bioassays, 768 (3.49%) showed hygienic behavior against Varroa-infested brood cells. These included bees showing both low and high degrees of DUVB behavior. For the analysis, only the 122 top DUVB performing bees and 122 negative controls were used.
A total of 26 007 SNPs were informative in at least 1 family. These SNPs, which are evenly distributed throughout the genome, were included in the association tests. After FDR correction, 6 SNPs showed significant genome-wide associations with DUVB at the genotype level (Table 2). These SNPs were highly significant with P < 0.01, while 14 other SNPs had P < 0.05. Further information about the 6 highly significant SNPs is provided in Table 2. The 14 other promising regions with lower significance will be analyzed in detail in an upcoming project. Information about all of the SNPs used in this assay is deposited in the Dryad repository (DRYAD entry doi:10.5061/dryad.8635cs4h).
Table 2.
SNPs with a significant (P < 0.001) genome-wide association with the trait “detection and uncapping of Varroa-infested brood cells”
| SNP | Nucleotide change | Chr.a | Position (bp) | FDR corrected genotype level association |
|---|---|---|---|---|
| AMB-00457689 | T → C | 3 | 10 425 353 | 0.0016 |
| AMB-00386078 | C → T | 7 | 8 722 970 | 0.0059 |
| AMB-00573174 | G → A | 2 | 1 657 342 | 0.0059 |
| AMB-00913945 | A → G | 3 | 8 984 417 | 0.0066 |
| AMB-01079196 | A → G | 5 | 12 195 | 0.0066 |
| AMB-00745078 | T → C | 6 | 1 398 456 | 0.0081 |
aChromosome.
Based on functional evidence, we selected 4 candidate genes which were spatially associated with 4 of the 6 SNPs with P < 0.01 (Table 2) at the genotype level. These included Adenosine receptor and Cyclin-dependent kinase 5 activator (ador and cdk5alpha, chromosome 3; SNPs: AMB-00457689 ca. 185kb from ador and 790kb from cdk5alpha, AMB-00913945 ca. 65kb from ador and 540kb from cdk5alpha), Octopamine receptor beta-2R (octbeta2R, chromosome 7; SNP: AMB-00386078 ca 70kb from octbeta2R), and Odorant binding protein 1 (obp1, chromosome 2; SNP: AMB-00573174 ca 510kb from obp1). No strong candidates have been identified for SNPs AMB-01079196 on chromosome 5 or AMB-00745078 on chromosome 6.
Discussion
Hygienic behavior is an important factor in the fight against Varroa because it interrupts the mites’ reproductive cycle. The behavior’s initiation depends on the olfactory sensitivity of an individual bee, as well as the odor profile and the stimulus intensity of the abnormal brood (Gramacho and Spivak 2003). Schöning et al. (2012) showed that bees exhibit selective, damage-dependent hygienic behavior against Varroa destructor. Thus, efforts to breed bees for increased hygienic behavior may result in bees with an increased olfactory sensitivity. This would be useful as a mechanism of resistance against Varroa, as well as other brood diseases such as American Foulbrood (Woodrow and Holst 1942) and chalkbrood (Gilliam et al. 1983): these pathogens cause more obvious damage to the bee brood, which should provide a stronger stimulus to bees than Varroa. This strongly supports the molecular genetic elucidation of hygienic behavior as a major goal of honeybee breeding. Our study makes a significant contribution to the achievement of this goal.
The DUVB behavior bioassay used in this study was developed to be suited to the investigation of specific and important component of the hygienic behavior against Varroa parasitized brood. In previous investigations of honeybees’ hygienic behavior as a defense mechanism against brood diseases, freeze-killed brood assays (Spivak 1996; Spivak and Reuter 1998) or pin-killed brood assays (Spivak and Downey 1998; Morais et al. 2009) were used. However, Varroa infestations provide a weaker stimulus for hygienic behavior compared with other brood diseases which cause the brood to literally rot. Thus, the threshold stimulus for the reaction to odors generated by Varroa-infested brood must be lower than that for other brood diseases. The application of our specific assay took this into consideration (Bienefeld et al. 2016).
Previous QTL studies of Varroa resistance behavior have relied on microsatellites (Oxley et al. 2010) or RAPD (Lapidge et al. 2002). Oxley et al. identified 3 significant and 3 suggestive QTLs using a sample of 149 worker bees and a marker set of 437 microsatellites. Lapidge et al. found 7 suggestive QTLs related to hygienic behavior in a sample of 119 sons of an F1 queen using 482 markers. However, it is difficult or impossible to compare the results of these 2 studies because they used different maps. The study of Lapidge et al. is particularly difficult to use in comparison, as their map was based almost exclusively on RAPD markers that were not cross-linked to other maps. A comparison between the QTLs identified in Oxley et al.’s study and our own trait-associated regions in the same chromosomes found no agreement. In chromosome 5, Oxley et al. identified a QTL with the nearest marker, A0058, at around 11Mb, whereas our significantly associated SNP was located at ca. 0.012Mb at the other end of the chromosome. On chromosome 2, we identified an SNP at 1.66Mb. The QTL Oxley et al. found on this chromosome is situated at 14.488Mb, close to the marker K0263, again, at the other end of the chromosome. A major difference between both of these previous studies and our experiment was that they conducted their hygienic behavior bioassays using freeze-killed brood instead of brood that was artificially infested with Varroa. Additionally, they used a honey bee race of Ligustica origin that has a slightly different genetic background. These reasons likely contributed to the differences in the genomic regions identified.
The SNP array described here is not the first applied to the honeybee. Whitfield et al. (2006) also developed a genome-wide SNP assay; however, this assay contained only 1536 SNPs, which were mainly selected based on spacing criteria. Tsuruda et al. (2012) identified 1536 genome-wide SNPs by sequencing honey bee DNA and used them to construct a small-scale SNP-chip. The genotyping platform used was the same one used by Whitfield. Tsuruda et al. applied their chip in a search for a QTL which influences hygienic behavior against Varroa and found a major one on chromosome 9. In our study, we did not discover an associated region on this chromosome.
The candidate genes in the vicinity of significantly trait-associated SNPs were mainly chosen based on functional evidence. Promising functional candidate genes have been identified in 4 out of 6 SNPs where P < 0.01.
Ador, an adenosine receptor, was found 65kb from SNP AMB-00913945 and 185kb from SNP AMB-00457689. Adenosine receptor (AdoR) is an evolutionarily conserved protein that is essential for normal cellular function in Drosophila. Knight et al. (2010) provide evidence that observed defects in associative learning and synaptic function may be attributable to changes in adenosine receptor activation. It is possible that the effects of Apis ador on learning and synaptic function are implicated in the Varroa defense behavior of honeybees.
Cdk5alpha was chosen as a functional candidate near the 2 SNPs AMB-00913945 and AMB-00457689. Cdk5alpha is an activator of cdk5 kinase activity and its expression is restricted to neurons. The complex of ckd5/cdk5alpha is essential for neurite outgrowth during neuronal differentiation in Drosophila and possibly also for neuronal degeneration (Ma and Haddard 1999). Connell-Crowley et al. (2007) show that Drosophila lacking the cdk5 activator, D-p35, display a wide range of defects in embryonic axon patterning. It is possible that cdk5alpha affects the neuronal sensitivity of bees to the external stimuli produced during Varroa infestation. However, the selection of cdk5alpha as a candidate gene should be considered with reservation because it was located 790 and 540kb away from the significant SNPs. This may be too far to justify its selection. It is possible that ador alone or in combination with other still unknown genes the vicinity of SNPs AMB-00913945 and AMB-00457689 affects Varroa resistance. It must be kept in mind that genome analysis in the honey bee has not yet been completed and many genes and their functions remain unknown. This issue should be addressed in future studies.
Insect octopamine receptors carry out many functional roles traditionally associated with vertebrate adrenergic receptors. These include, among others, the modulation of sensory inputs and modulation of memory and learning (Maqueira et al. 2005). Like ador, Apis mellifera octopamine receptor beta-2R near SNP AMB-00386078 may also affect learning and memory, both of which are implicated in the Varroa resistance of honeybees.
The olfactory power of honey bees and other insect species is thought to be generated by the combined action of 2 large protein families, G protein-coupled olfactory receptors (ORs) and odorant-binding proteins (OBPs). In olfactory sensilla, OBPs deliver hydrophobic airborne molecules to ORs (Forêt and Maleszka 2006). It is possible that obp1, which was located near SNP AMB-00573174, plays a comparable role during the identification of Varroa-infested cells, which is assumed to rely on olfactory cues.
Le Conte et al. (2011) identified a set of genes involved in social immunity by analyzing the brain transcriptome of highly Varroa-hygienic bees. There was no direct agreement between the candidate genes for Varroa resistance in their study and our results; however, there were functional similarities between the genes identified in both studies. Le Conte et al. (2011) found that dscam was differentially expressed in the brains of bees with low and high rates of hygienic behavior. The molecular diversity of dscam is essential for mediating axon guidance and the specificity of neuronal wiring (Chen et al. 2006). In the current study, cdk5alpha was identified as a candidate gene for Varroa resistance, with a similar role. This may be interpreted as an indication that these functions are involved in the regulation of Varroa resistance.
In this study, 4 functional candidate genes were identified in the neighborhood of 4 SNPs that were significantly associated with DUVB behavior. The number of families and individuals is surely limited and for some QTL only a few but the majority of queens may actually have been heterozygous—a possibility that our design shares with the so-called daughter-design used for linkage mapping with half-sibs in livestock. Especially those QTL with a low frequency in the selection line, what leads to a low probability of heterozygosity in the queens, or smaller effect may therefore have escaped their detection.
However, the monitored DUVB behavior, which is a measurement of a bee’s sensitivity to highly specific stimuli, is of key importance for Varroa resistance breeding. The genes involved in the detection of “abnormal brood” (due to Varroa or other brood diseases) warrant further research.
Funding
Federal Ministry of Food, Agriculture, and Consumer Protection of Germany (2808HS009); European Union (EFRE – application no. 80137041).
Data Availability
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.7ds27
Acknowledgments
We would like to thank everyone from the beekeeping department at the Institute for Bee Research Hohen Neuendorf involved with the defense behavior bioassays, especially Fred Zautke, Ivonne Kretschmann, Andrea Jäkisch, and Dr. Caspar Schöning. We are grateful to Inga Blunk, FBN, for providing figures.
References
- Allen MF, Ball BV. 1996. The incidence and world distribution of honey bee viruses. Bee World. 77:141–162. [Google Scholar]
- Allen MF, Ball BV, White RF, Antoniw JF. 1986. The detection of acute paralysis virus in Varroa jacobsoni by the use of a simple indirect ELISA. J Apic Res. 25:100–105. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Baker CS. 2013. Journal of heredity adopts joint data archiving policy. J Hered. 104:1. [DOI] [PubMed] [Google Scholar]
- Bakonyi T, Farkas R, Szendroi A, Dobos-Kovács M, Rusvai M. 2002. Detection of acute bee paralysis virus by RT-PCR in honey bee and Varroa destructor field samples: rapid screening of representative Hungarian apiaries. Apidologie. 33:63–74. [Google Scholar]
- Ball BV. 1985. Acute Paralysis Virus isolates from honeybee colonies infested with Varroa jacobsoni . J Apic Res. 24: 115–119. [Google Scholar]
- Ball BV, Allen MF. 1988. The prevalence of pathogens in honey bee colonies infested with the parasitic parasitic mite Varroa jacobsoni . Ann Appl Biol. 113:237–244. [Google Scholar]
- Benjamini Y, Yekateuli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 29:1165–1188. [Google Scholar]
- Bienefeld K, Pirchner F. 1990. Heritabilities for several colony traits in the honeybee (Apis mellifera carnica). Apidologie. 21:175–183. [Google Scholar]
- Bienefeld K, Reinsch N, Thakur RK, 2001. Selection for uncapping of Varroa infested brood cells in the honeybee (Apis mellifera). In: Proc 37th Intern Apimondia Congress Durban, Bucharest: Apimondia Publishing House. p. 12. [Google Scholar]
- Bienefeld K, Zautke F, Gupta P. 2016. A novel method for undisturbed long-term observation of the honey bee (Apis mellifera) behaviour – illustrated by hygienic behaviour towards Varroa infestation. J Apic Res. [Google Scholar]
- Boecking O, Drescher W. 1992. The removal responses of Apis mellifera L. colonies to brood in wax and plastic cells after artificial and natural infestation with Varroa jacobsoni Oud. and to freeze-killed brood. Exp Appl Acarol. 16:321–332. [Google Scholar]
- Boecking O, Bienefeld K, Drescher W. 2000. Heritability of the Varroa-specific hygienic behaviour in honey bees (Hymenoptera: Apidae) J Anim Breed Genet. 117:417–424. [Google Scholar]
- Chen BE, Kondo M, Garnier A, Watson FL, Püettmann-Holgado R, Lamar DR, Schmucker D. 2006. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 125:607–620. [DOI] [PubMed] [Google Scholar]
- Chen Y, Pettis JS, Evans JD, Kramer M, Feldlaufer MF. 2004. Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie. 35:441–448. [Google Scholar]
- Connell-Crowley L, Vo D, Luke L, Giniger E. 2007. Drosophila lacking the Cdk5 activator, p35, display defective axon guidance, age-dependent behavioral deficits and reduced lifespan. Mech Dev. 124:341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forêt S, Maleszka R. 2006. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera). Genome Res. 16:1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries I, Camazine S, Sneyd J. 1994. Population dynamics of Varroa jacobsoni: a model and a review. Bee World. 75:5–28. [Google Scholar]
- Gilliam M, Taber S III, Richardson GV. 1983. Hygienic behavior of honey bees in relation to chalkbrood disease. Apidologie. 14:29–39. [Google Scholar]
- Gramacho KP, Spivak M. 2003. Differences in olfactory sensitivity and behavioral responses among honey bees bred for hygienic behavior. Behav Ecol Sociobiol. 54:472–479. [Google Scholar]
- Harbo JR, Harris JW. 1999. Heritability in honey bees (Hymenoptera: Apidae) of characteristics associated with resistance to Varroa jacobsoni (Mesostigmata: Varroidae). J Econ Entomol. 92:261–265. [Google Scholar]
- Harbo JR, Harris JW, 2005. Suppred mite reproduction explained by the behavior of adult bees. J Apic Res. 44:21–23. [Google Scholar]
- Harbo JR, Harris JW. 2009. Responses to Varroa by honey bees with different levels of Varroa Sensitive Hygiene. J Apic Res. 48:156–161. [Google Scholar]
- Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera . Nature. 443:931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Page RE., Jr 1995. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 139:1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight D, Harvey PJ, Iliadi KG, Klose MK, Iliadi N, Dolezelova E, Charlton MP, Zurovec M, Boulianne GL. 2010. Equilibrative nucleoside transporter 2 regulates associative learning and synaptic function in Drosophila. J Neurosci. 30:5047–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidge KL, Oldroyd BP, Spivak M. 2002. Seven suggestive quantitative trait loci influence hygienic behavior of honey bees. Naturwissenschaften. 89:565–568. [DOI] [PubMed] [Google Scholar]
- Le Conte Y, Alaux C, Martin JF, Harbo JR, Harris JW, Dantec C, Séverac D, Cros-Arteil S, Navajas M. 2011. Social immunity in honeybees (Apis mellifera): transcriptome analysis of varroa-hygienic behaviour. Insect Mol Biol. 20:399–408. [DOI] [PubMed] [Google Scholar]
- Ma E, Haddad G. 1999. A Drosophila CDK5alpha-like molecule and its possible role in response to O(2) deprivation. Biochem Biophys Res Commun. 261:459–463. [DOI] [PubMed] [Google Scholar]
- Maqueira B, Chatwin H, Evans PD. 2005. Identification and characterization of a novel family of Drosophila beta-adrenergic-like octopamine G-protein coupled. J Neurochem. 94:547–560. [DOI] [PubMed] [Google Scholar]
- Martin SJ. 1998. A population dynamic model of the mite Varroa jacobsoni Oud. J Invertebr Pathol. 73:101–106. [DOI] [PubMed] [Google Scholar]
- Martin C, Provost E, Bagneres AG, Roux M, Clement JL, Le Conte Y. 2002. Potential-mechanism for detection by Apis mellifera of the parasitic mite Varroa destructor inside sealed brood cells. Physiol Entomol. 27:175–188. [Google Scholar]
- Milani N. 1999. The resistance of Varroa jacobsoni Oud. to acaricides. Apidologie. 30:229–234. [Google Scholar]
- Momot JP, Rothenbuhler WC. 1971. Behaviour genetics of nest cleaning in honeybees. VI. Interactions of age and genotype of bees, and nectar flow. J Apic Res. 10:11–21. [Google Scholar]
- Morais MM, Francoy TM, Pereira RA, De Jong D, Gonçalves LS. 2009. Africanized honey bees are efficient at detecting, uncapping and removing dead brood. Genet Mol Res. 8:718–724. [DOI] [PubMed] [Google Scholar]
- Moritz RFA. 1988. A reevaluation of the two-locus model hygienic behavior in honey bees, Apis mellifera L. J Hered. 79:257–262. [Google Scholar]
- Navajas M, Migeon A, Alaux C, Martin-Magniette M, Robinson G, Evans J, Cros-Arteil S, Crauser D, Le Conte Y. 2008. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics. 9:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström S, Fries I, Aarhus A, Hansen H, Korpela S, 1999. Virus infections in Nordic honey bee colonies with no, low or severe Varroa jacobsoni infections. Apidologie. 30:475–484. [Google Scholar]
- Oxley PR, Spivak M, Oldroyd BP. 2010. Six quantitative trait loci influence task thresholds for hygienic behaviour in honeybees (Apis mellifera). Mol Ecol. 19:1452–1461. [DOI] [PubMed] [Google Scholar]
- Rath W, Drescher W, 1990. Response of Apis cerana Fabr. towards brood infested with Varroa jacobsoni Oud. and infestation rate of colonies in Thailand. Apidologie. 21:311–321. [Google Scholar]
- Rinderer JE, Harris JW, Hunt GJ, de Guzman LI, 2010. Breeding for resistance to Varroa destructor in North America. Apidologie. 41:409–424. [Google Scholar]
- Rothenbuhler WC, 1964. Behavior genetics of nest cleaning in honey bees. IV. Responses of F1 and backcross generations to disease-killed brood. Am Zool. 12:578–583. [DOI] [PubMed] [Google Scholar]
- Schneider P, Drescher W, 1987. The influence of varroa-jacobsoni oud on weight, development of weight and hypopharyngeal glands, and longevity of Apis mellifera . Apidologie. 18:101–109. [Google Scholar]
- Schöning C, Gisder S, Geiselhardt S, Kretschmann I, Bienefeld K, Hilker M, Genersch E. 2012. Evidence for damage-dependent hygienic behaviour towards Varroa destructor-parasitised brood in the western honey bee, Apis mellifera . J Exp Biol. 215:264–271. [DOI] [PubMed] [Google Scholar]
- Solignac M, Vautrin D, Baudry E, Mougel F, Loiseau A, Cornuet JM. 2004. A microsatellite-based linkage map of the honeybee, Apis mellifera L. Genetics. 167:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak M, 1996. Hygienic behavior and defense against Varroa jacobsoni . Apidologie 27:245–260. [Google Scholar]
- Spivak M, Downey DL. 1998. Field assays for hygienic behavior in honey bees (Hymenoptera: Apidae). J Econ Entomol. 91:64–70. [Google Scholar]
- Spivak M, Reuter GS. 1998. Performance of hygienic honey bee colonies in a commercial apiary. Apidologie. 29:291–302. [Google Scholar]
- Spivak M, Rebecca Masterman R, Rocco Ross R, Karen A, Mesce KA. 2003. Hygienic behavior in the honey bee (Apis mellifera L.) and the modulatory role of octopamine. J Neurobiol. 55:341–354. [DOI] [PubMed] [Google Scholar]
- Spötter A, Gupta P, Nürnberg G, Reinsch N, Bienefeld K. 2012. Development of a 44K SNP assay focussing on the analysis of a varroa-specific defence behaviour in honey bees (Apis mellifera carnica). Mol Ecol Resour. 12:323–332. [DOI] [PubMed] [Google Scholar]
- Thakur RK, Bienefeld K, Keller R. 1997. Varroa defense behavior in A. mellifera carnica. Am Bee J. 137:143–148. [Google Scholar]
- Thakur RK, Bienefeld K, Keller R, Zautke F. 1998. Studies on age dependence of defence behaviour against Varroa jacobsoni in the honey bee (Apis mellifera carnica). Apidologie. 29:431–433. [Google Scholar]
- Thompson VC. 1964. Behaviour genetics of nest cleaning in honey bees. III. Effect of age of bees of a resistant line on their response to disease-killed brood. J Apic Res. 3:25–30. [Google Scholar]
- Tsuruda JM, Harris JW, Bourgeois L, Danka RG, Hunt GJ. 2012. High-resolution linkage analyses to identify genes that influence varroa sensitive hygiene behavior in honey bees. PLoS One. 7:e48276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner K. 1999. Varroacides and their residues in bee products. Apidologie. 30:235–248. [Google Scholar]
- Whitfield CW, Behura SK, Berlocher SH, Clark AG, Johnston JS, Sheppard WS, Smith DR, Suarez AV, Weaver D, Tsutsui ND. 2006. Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera . Science. 314:642–645. [DOI] [PubMed] [Google Scholar]
- Woodrow AW, Holst EC, 1942. The mechanismof colony resistance to American foulbrood. J Econ Entomol. 35:327–330. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data deposited at Dryad: http://dx.doi.org/doi:10.5061/dryad.7ds27